Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Serum Panels
2.1.1. Monoinfected Rabbit Sera
2.1.2. Monoinfected Mouse Sera
2.1.3. Human Sera
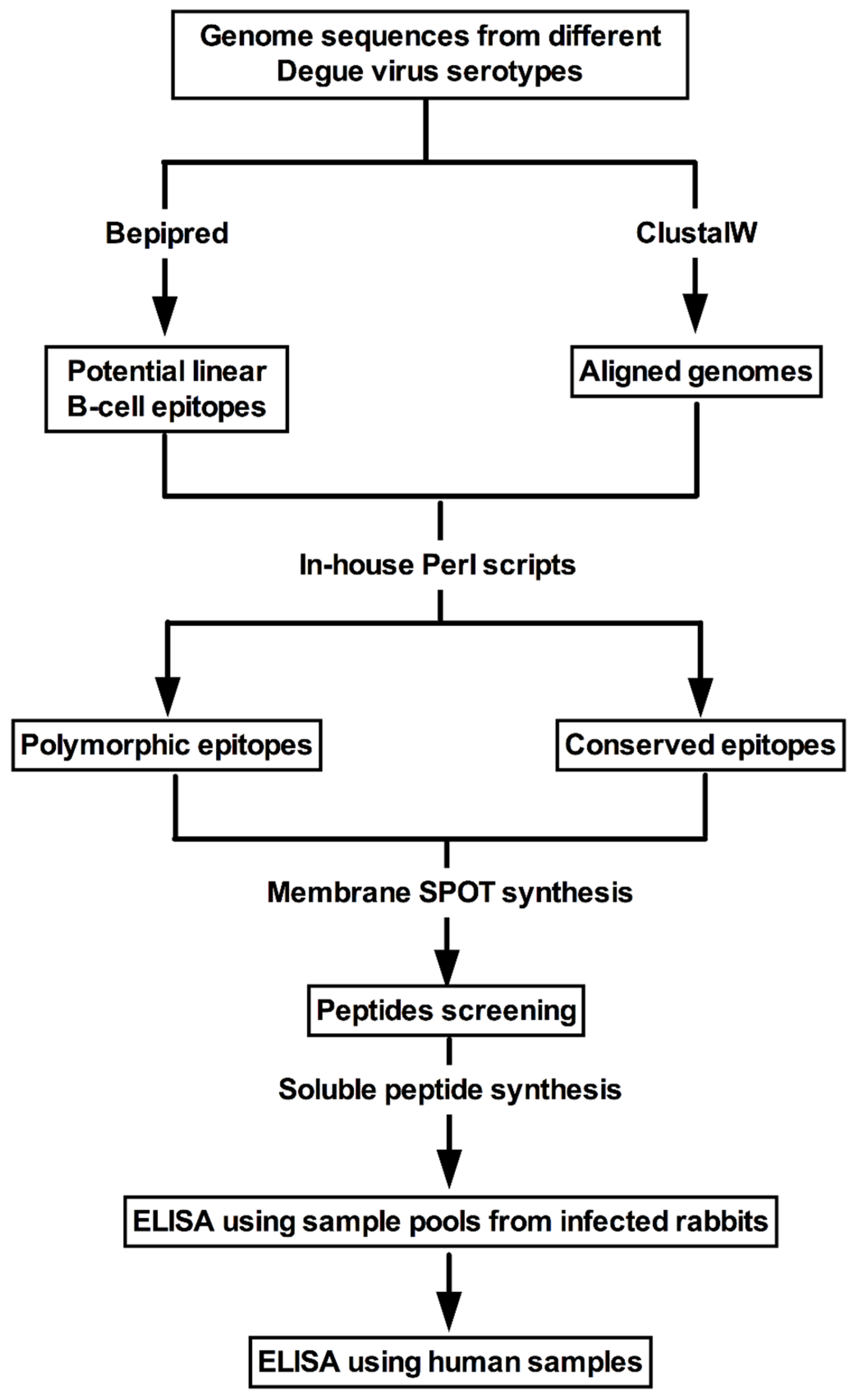
2.2. Proteome Data Acquisition, Linear B-Cell Epitope Mapping and Selection of Conserved and Polymorphic Peptides
2.3. Synthesis, Spot Peptide Array and Immunoblot
2.4. Peptide ELISA Validation with Human Samples
2.5. Statistical Analysis
3. Results
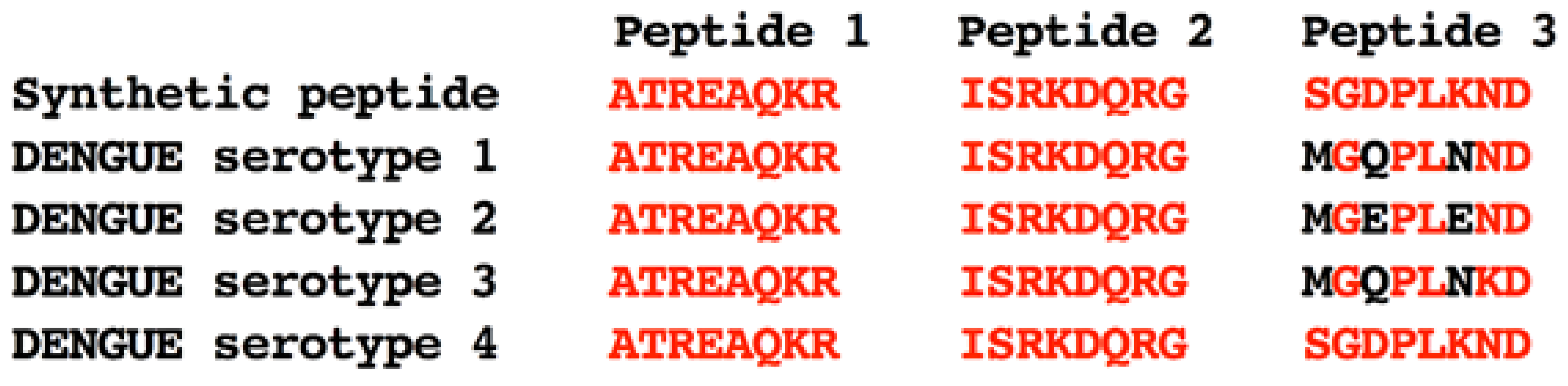
3.1. Prediction and Selection of Peptides
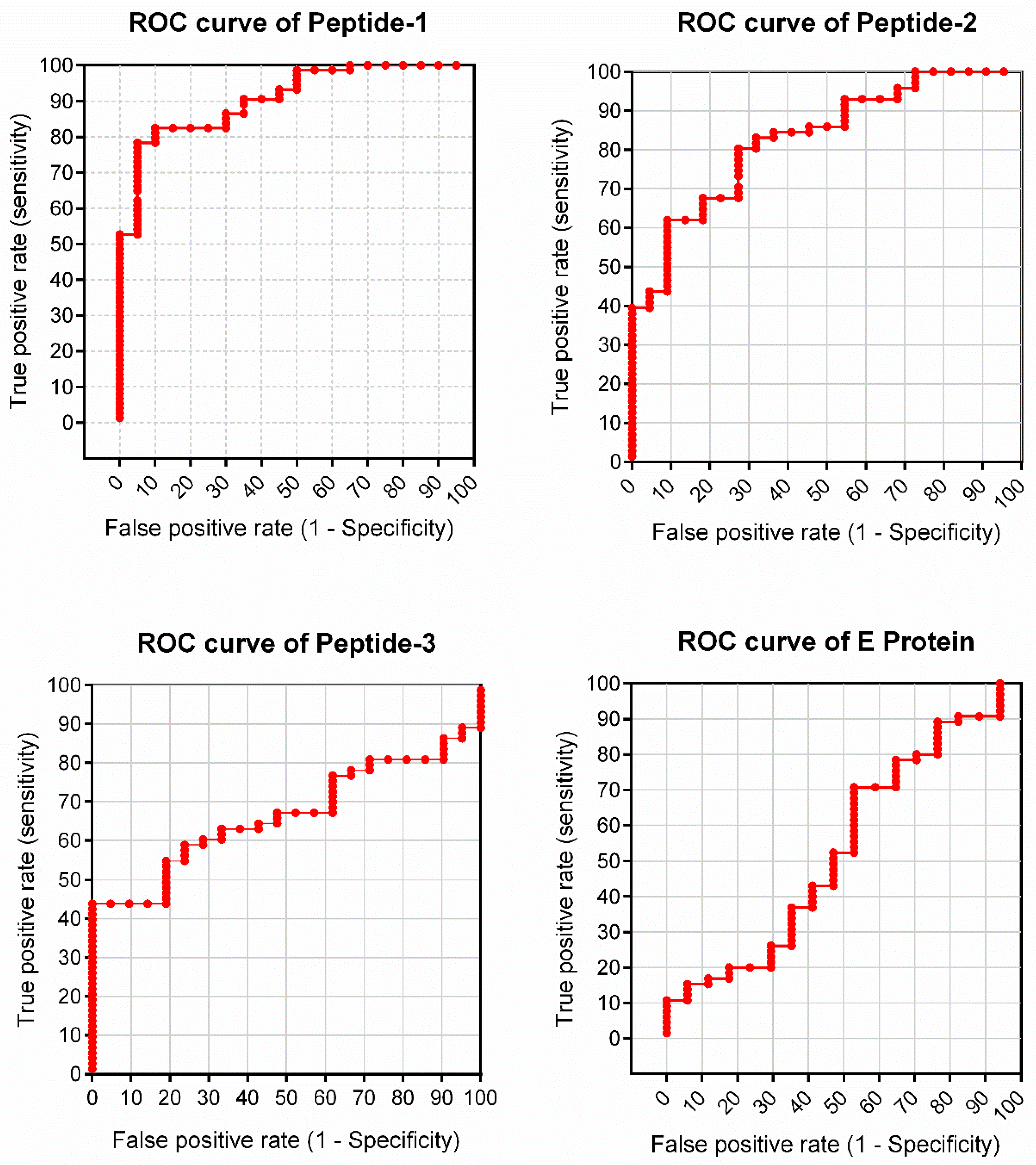
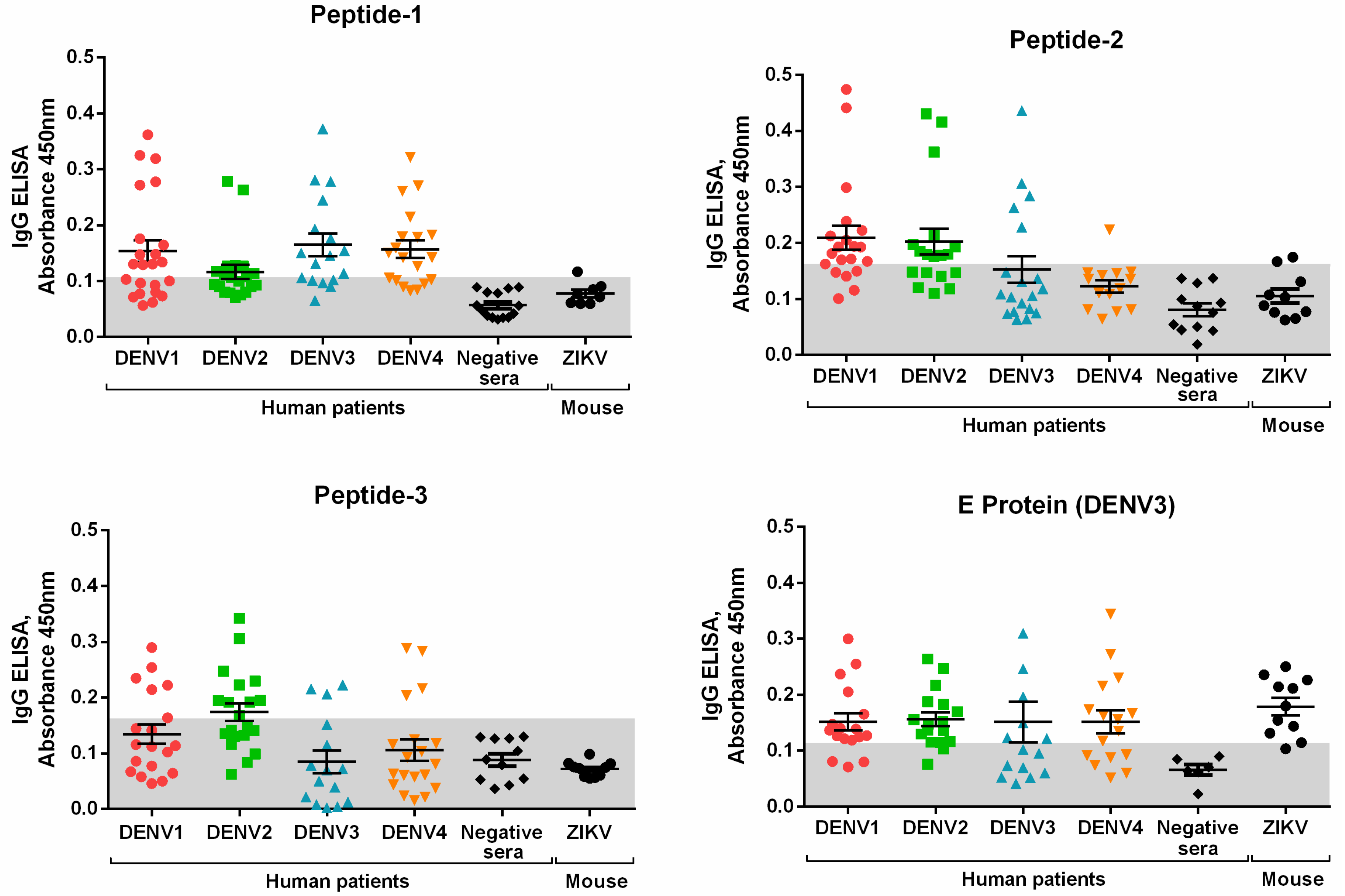
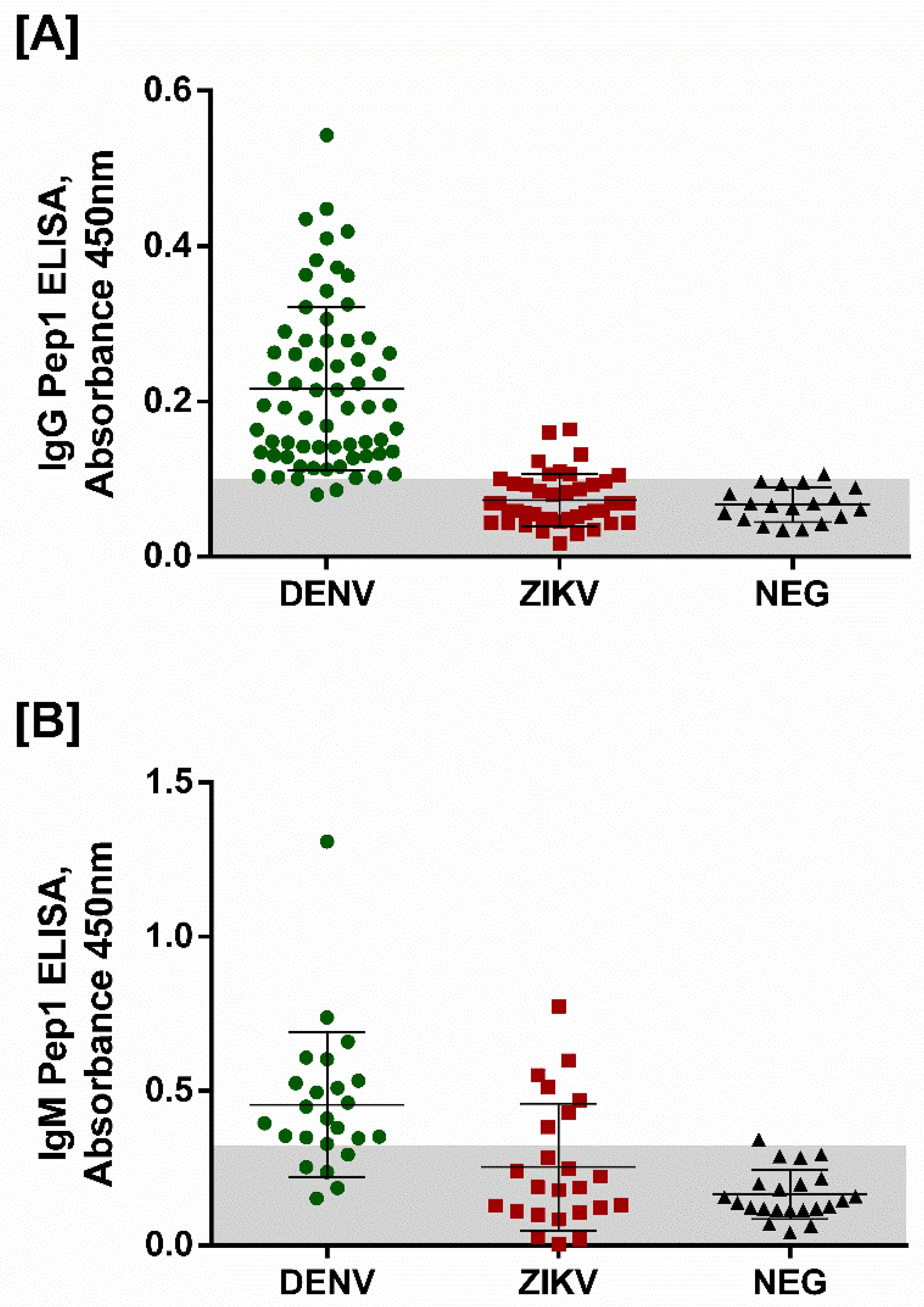
3.2. Peptide Validation with Human Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A


References
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the Global Spatial Limits of Dengue Virus Transmission by Evidence-Based Consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Lee, T.H.; Leo, Y.S. Dengue in the elderly: A review. Expert Rev. Anti. Infect. Ther. 2017, 15, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Brady, O.J.; Johansson, M.A.; Guerra, C.A.; Bhatt, S.; Golding, N.; Pigott, D.M.; Delatte, H.; Grech, M.G.; Leisnham, P.T.; Maciel-de-Freitas, R.; et al. Modelling adult Aedes aegypti and Aedes albopictus survival at different temperatures in laboratory and field settings. Parasit. Vectors 2013, 6, 351. [Google Scholar] [CrossRef]
- Halstead, S.B.; Dans, L.F. Dengue infection and advances in dengue vaccines for children. Lancet Child Adolesc. Heal. 2019, 3, 734–741. [Google Scholar] [CrossRef]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- De Angel, R.M.; Valle, J.R. del Dengue Vaccines: Strongly Sought but Not a Reality Just Yet. PLoS Pathog. 2013, 9, 1–5. [Google Scholar] [CrossRef]
- Screaton, G.; Mongkolsapaya, J.; Yacoub, S.; Roberts, C. New insights into the immunopathology and control of dengue virus infection. Nat. Rev. Immunol. 2015, 15, 745–759. [Google Scholar] [CrossRef]
- Sahili, A.E.; Lescar, J. Dengue virus non-structural protein 5. Viruses 2017, 9, 1–20. [Google Scholar]
- Lindenbach, B.D.; Rice, C.M. Molecular biology of flaviviruses. In Advances in Virus Research; Academic Press: London, UK, 2003; Volume 59, pp. 23–61. ISBN 0065-3527. [Google Scholar]
- Roehrig, J.T. Antigenic Structure of Flavivirus Proteins. In Advances in Virus Research; Academic Press: London, UK, 2003; Volume 59, pp. 141–175. ISBN 0065-3527. [Google Scholar]
- AnandaRao, R.; Swaminathan, S.; Fernando, S.; Jana, A.M.; Khanna, N. A custom-designed recombinant multiepitope protein as a dengue diagnostic reagent. Protein Expr. Purif. 2005, 41, 136–147. [Google Scholar] [CrossRef]
- Chan, H.B.Y.; How, C.H.; Ng, C.W.M. Definitive tests for dengue fever: When and which should I use? Singap. Med. J. 2017, 58, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Artsob, H.; Pelegrino, J.L.; Buchy, P.; Cardosa, M.J.; Devi, S.; Enria, D.A.; Farrar, J.; Gubler, D.J.; Guzman, M.G.; et al. Evaluation of diagnostic tests: Dengue. Nat. Rev. Microbiol. 2010, 8, S30–S37. [Google Scholar] [CrossRef] [PubMed]
- Rathakrishnan, A.; Sekaran, S.D. New development in the diagnosis of dengue infections. Expert Opin. Med. Diagn. 2013, 7, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.W.; Russell, B.J.; Lanciotti, R.S. Serotype-Specific Detection of Dengue Viruses in a Fourplex Real-Time Reverse Transcriptase PCR Assay. J. Clin. Microbiol. 2005, 43, 4977–4983. [Google Scholar] [CrossRef] [PubMed]
- Russell, P.K.; Nisalak, A.; Sukhavac, P.; Vivona, S. A plaque reduction test for dengue virus neutralizing antibodies. J. Immunol. 1967, 99, 285–290. [Google Scholar]
- Roehrig, J.T.; Hombach, J.; Barrett, A.D.T. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 2008, 21, 123–132. [Google Scholar] [CrossRef]
- Bateman, A.; Martin, M.J.; O’Donovan, C.; Magrane, M.; Apweiler, R.; Alpi, E.; Antunes, R.; Arganiska, J.; Bely, B.; Bingley, M.; et al. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Larsen, J.E.P.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006, 2, 2. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Tatusova, T.; Ciufo, S.; Fedorov, B.; O’Neill, K.; Tolstoy, I. RefSeq microbial genomes database: New representation and annotation strategy. Nucleic Acids Res. 2014, 42, D553–D559. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chu, M.C.; O’Rourke, E.J.; Trent, D.W. Genetic relatedness among structural protein genes of dengue 1 virus strains. J. Gen. Virol. 1989, 70, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Tan, B.H.; Yap, E.H.; Chan, Y.C.; Tan, Y.H. Full-length cDNA sequence of dengue type 1 virus (Singapore strain S275/90). Virology 1992, 188, 953–958. [Google Scholar] [CrossRef]
- Puri, B.; Nelson, W.; Porter, K.R.; Henchal, E.A.; Hayes, C.G. Complete nucleotide sequence analysis of a Western Pacific dengue-1 virus strain. Virus Genes 1998, 17, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Blok, J.; McWilliam, S.M.; Butler, H.C.; Gibbs, A.J.; Weiller, G.; Herring, B.L.; Hemsley, A.C.; Aaskov, J.G.; Yoksan, S.; Bhamarapravati, N. Comparison of a dengue-2 virus and its candidate vaccine derivative: Sequence relationships with the flaviviruses and other viruses. Virology 1992, 187, 573–590. [Google Scholar] [CrossRef]
- Deubel, V.; Kinney, R.M.; Trent, D.W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: Comparative analysis of the full-length genome. Virology 1988, 165, 234–244. [Google Scholar] [CrossRef]
- Hahn, Y.S.; Caller, R.; Hunkapiller, T.; Dalrymple, J.M.; Strauss, J.H.; Strauss, E.G. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 1988, 162, 167–180. [Google Scholar] [CrossRef]
- Leitmeyer, K.C.; Vaughn, D.W.; Watts, D.M.; Salas, R.; Villalobos, I.; Ramos, C.; Rico-Hesse, R. Dengue Virus Structural Differences That Correlate with Pathogenesis. J. Virol. 1999, 73, 4738–4747. [Google Scholar]
- Yuan, X.; Geng, L.; Li, X.; Yu, M.; Qin, E. Dengue Virus Type 3 (Strain China/80-2/1980) (DENV-3). Available online: https://www.uniprot.org/proteomes/UP000007198 (accessed on 6 September 2019).
- Osatomi, K.; Sumiyoshi, H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology 1990, 176, 643–647. [Google Scholar] [CrossRef]
- Peyrefitte, C.N.; Couissinier-Paris, P.; Mercier-Perennec, V.; Bessaud, M.; Martial, J.; Kenane, N.; Durand, J.P.A.; Tolou, H.J. Genetic Characterization of Newly Reintroduced Dengue Virus Type 3 in Martinique (French West Indies). J. Clin. Microbiol. 2003, 41, 5195–5198. [Google Scholar] [CrossRef]
- Zhao, B.; Mackow, E.; Buckler-White, A.; Markoff, L.; Chanock, R.M.; Lai, C.J.; Making, Y. Cloning full-length dengue type 4 viral DNA sequences: Analysis of genes coding for structural proteins. Virology 1986, 155, 77–88. [Google Scholar] [CrossRef]
- Klungthong, C.; Zhang, C.; Mammen, M.P.; Ubol, S.; Holmes, E.C. The molecular epidemiology of dengue virus serotype 4 in Bangkok, Thailand. Virology 2004, 329, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Yoong, L.F.; Tan, T.; Anwar, A.; August, T.J.; Too, H.P. Dengue Virus Type 4 (Strain Singapore/8976/1995) (DENV-4). Available online: https://www.uniprot.org/proteomes/UP000007202 (accessed on 6 September 2019).
- Frank, R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—Principles and applications. J Immunol. Methods 2002, 267, 13–26. [Google Scholar] [CrossRef]
- Chang, H.-H.; Huber, R.G.; Bond, P.J.; Grad, Y.H.; Camerini, D.; Maurer-Stroh, S.; Lipsitch, M. Systematic analysis of protein identity between Zika virus and other arthropod-borne viruses. Bull. World Health Organ. 2017, 95, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Mendes, T.A.; Reis Cunha, J.L.; de Almeida Lourdes, R.; Rodrigues Luiz, G.F.; Lemos, L.D.; dos Santos, A.R.; da Câmara, A.C.; Galvão, L.M.; Bern, C.; Gilman, R.H.; et al. Identification of strain-specific B-cell epitopes in Trypanosoma cruzi using genome-scale epitope prediction and high-throughput immunoscreening with peptide arrays. PLoS Negl. Trop. Dis. 2013, 7, e2524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayan, R.; Raja, S.; Kumar, S.; Sambasivam, M.; Jagadeesan, R.; Arunagiri, K.; Krishnasamy, K.; Palani, G. A novel indirect ELISA for diagnosis of dengue fever. Indian J. Med. Res. 2016, 144, 128–133. [Google Scholar]
- Wen, J.; Tang, W.W.; Sheets, N.; Ellison, J.; Sette, A.; Kim, K.; Shresta, S. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat. Microbiol. 2017, 2, 17036. [Google Scholar] [CrossRef] [Green Version]
- Valadon, P.; Scharff, M.D. Enhancement of ELISAs for screening peptides in epitope phage display libraries. J. Immunol. Methods 1996, 197, 171–179. [Google Scholar] [CrossRef]
- Tian, H.; Chen, Y.; Wu, J.; Shang, Y.; Liu, X. Serodiagnosis of sheeppox and goatpox using an indirect ELISA based on synthetic peptide targeting for the major antigen P32. Virol. J. 2010, 7, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Abdelgawad, A.; Hermes, R.; Damiani, A.; Lamglait, B.; Czirják, G.; East, M.; Aschenborn, O.; Wenker, C.; Kasem, S.; Osterrieder, N.; et al. Comprehensive serology based on a peptide ELISA to assess the prevalence of closely related equine herpesviruses in zoo and wild animals. PLoS ONE 2015, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Dubois, M.E.; Hammarlund, E.; Slifka, M.K. Optimization of peptide-based ELISA for serological diagnostics: A retrospective study of human monkeypox infection. Vector-Borne Zoonotic Dis. 2012, 12, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Felix, A.C.; Souza, N.C.S.; Figueiredo, W.M.; Costa, A.A.; Inenami, M.; da Silva, R.M.G.; Levi, J.E.; Pannuti, C.S.; Romano, C.M. Cross reactivity of commercial anti-dengue immunoassays in patients with acute Zika virus infection. J. Med. Virol. 2017, 89, 1477–1479. [Google Scholar] [CrossRef] [PubMed]

| Virus Serotype | Isolate Origin | Strain | Access Number | Reference |
|---|---|---|---|---|
| DENV1 | Brazil | 97-11/1997 | P27909 | [24] |
| DENV1 | Singapore | S275/1990 | P33478 | [25] |
| DENV1 | Nauru | West Pac/1974 | P17763 | [26] |
| DENV2 | Thailand | 16681/1984 | P29990 | [27] |
| DENV2 | Jamaica | 1409/1983 | P07564 | [28] |
| DENV2 | Puerto Rico | PR159-S1/1969 | P12823 | [29] |
| DENV2 | Peru | IQT2913/1996 | Q9WDA6 | [30] |
| DENV3 | China | 80-2/1980 | Q99D35 | [31] |
| DENV3 | Philippines | H87/1956 | P27915 | [32] |
| DENV3 | Martinique | 1243/1999 | Q6YMS3 | [33] |
| DENV4 | Dominica | 814669/1981 | P09866 | [34] |
| DENV4 | Thailand | 0348/1991 | Q2YHF0 | [35] |
| DENV4 | Singapore | 8976/1995 | Q5UCB8 | [36] |
| Peptide | Dengue Virus Specificity | Bepipred Score * |
|---|---|---|
| CPRITETE | Serotype 1 | 1.45 |
| SSQDEKGVT | Serotype 1 | 1.30 |
| TAPGTFKTPEGE | Serotype 1 | 1.53 |
| SSQSTTPET | Serotype 2 | 1.34 |
| IATQQPES | Serotype 2 | 1.38 |
| IPYDPKFE | Serotype 2 | 1.30 |
| QRKKTGKP | Serotype 3 | 1.31 |
| TEDGQGKA | Serotype 3 | 1.51 |
| NAEPDGPT | Serotype 3 | 2.10 |
| MSKEPGVV | Serotype 3 | 1.82 |
| PETPNMDV | Serotype 3 | 1.44 |
| AGATEVDS | Serotype 4 | 1.43 |
| SGDPLKND | Serotype 4 | 1.30 |
| SYDPKFEK | Serotype 4 | 1.78 |
| YHGSYEAP | Serotype 4 | 1.52 |
| AQKRTAAG | Conserved | 1.30 |
| KGGPGHEE | Conserved | 1.64 |
| AGWDTRIT | Conserved | 1.31 |
| QRGSGQVG | Conserved | 1.30 |
| VRKDIPQW | Conserved | 1.42 |
| PEPEKQRT | Conserved | 1.79 |
| PEREKSAA | Conserved | 1.41 |
| ATREAQKR | Conserved | 1.30 |
| ISRKDQRG | Conserved | 1.36 |
| Parameters | Peptides | E Protein | ||
|---|---|---|---|---|
| Peptide-1 | Peptide-2 | Peptide-3 | ||
| Cutoff (2SD) | 0.107 | 0.160 | 0.163 | 0.113 |
| TSe (%) | 59.46 | 45.07 | 31.51 | 70.77 |
| TSp (%) | 95.00 | 90.91 | 100.00 | 41.18 |
| PPV (%) | 97.78 | 94.12 | 100.00 | 82.14 |
| NPV (%) | 38.78 | 33.90 | 29.58 | 26.92 |
| AC (%) | 67.02 | 55.91 | 46.81 | 64.63 |
| AUC | 0.9088 | 0.8278 | 0.6641 | 0.5367 |
| TP | 44 | 32 | 23 | 46 |
| TN | 19 | 20 | 21 | 7 |
| FP | 1 | 2 | 0 | 10 |
| FN | 30 | 39 | 50 | 19 |
| Parameters | Peptide-1 | |
|---|---|---|
| IgG | IgM | |
| Cutoff (2SD) | 0.111 | 0.324 |
| TSe (%) | 87.88 | 79.17 |
| TSp (%) | 93.55 | 82.61 |
| PPV (%) | 93.55 | 70.37 |
| NPV (%) | 87.88 | 88.37 |
| AC (%) | 90.63 | 81.43 |
| AUC | 0.9648 | 0.8542 |
| AUC-CI95% | 0.9379 to 0.9917 | 0.7658 to 0.9425 |
| TP | 58 | 19 |
| TN | 58 | 38 |
| FP | 4 | 8 |
| FN | 8 | 5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Versiani, A.F.; Rocha, R.P.; Mendes, T.A.O.; Pereira, G.C.; Coelho dos Reis, J.G.A.; Bartholomeu, D.C.; da Fonseca, F.G. Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections. Viruses 2019, 11, 1079. https://doi.org/10.3390/v11111079
Versiani AF, Rocha RP, Mendes TAO, Pereira GC, Coelho dos Reis JGA, Bartholomeu DC, da Fonseca FG. Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections. Viruses. 2019; 11(11):1079. https://doi.org/10.3390/v11111079
Chicago/Turabian StyleVersiani, Alice F., Raissa Prado Rocha, Tiago A. O. Mendes, Glauco C. Pereira, Jordana Graziella A. Coelho dos Reis, Daniella C. Bartholomeu, and Flávio G. da Fonseca. 2019. "Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections" Viruses 11, no. 11: 1079. https://doi.org/10.3390/v11111079
APA StyleVersiani, A. F., Rocha, R. P., Mendes, T. A. O., Pereira, G. C., Coelho dos Reis, J. G. A., Bartholomeu, D. C., & da Fonseca, F. G. (2019). Identification of B-Cell Epitopes with Potential to Serologicaly Discrimnate Dengue from Zika Infections. Viruses, 11(11), 1079. https://doi.org/10.3390/v11111079






