Single-Stranded DNA Viruses in Antarctic Cryoconite Holes
Abstract
1. Introduction
2. Materials and Methods
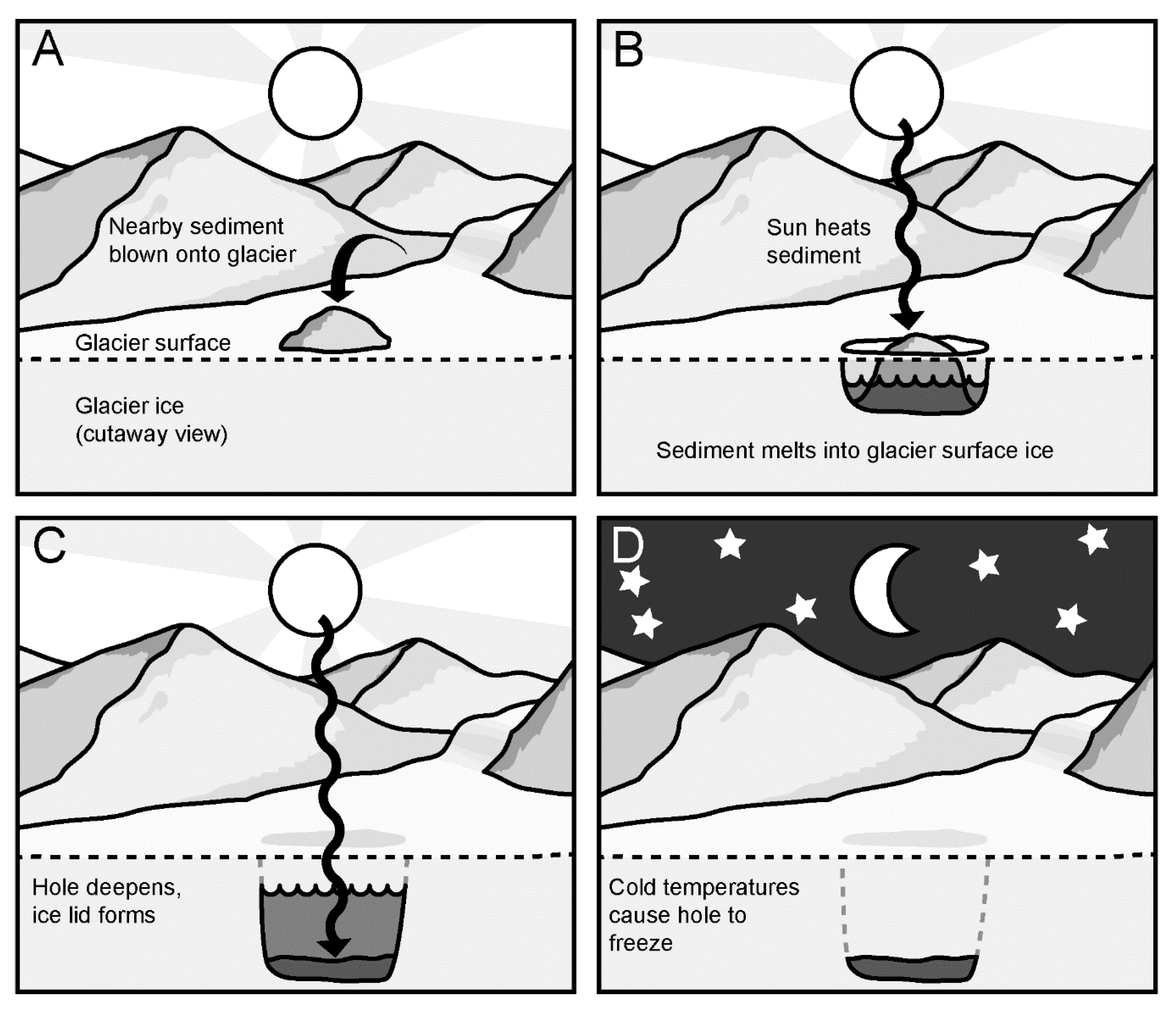
2.1. Sample Collection
2.2. Sample Processing and Nucleic Acid Extraction
2.3. High-Throughput DNA Sequencing and Data Analysis
2.4. Sequence Similarity Network Analysis
2.5. Genome Characterization and Phylogenetic Analysis
3. Results and Discussion
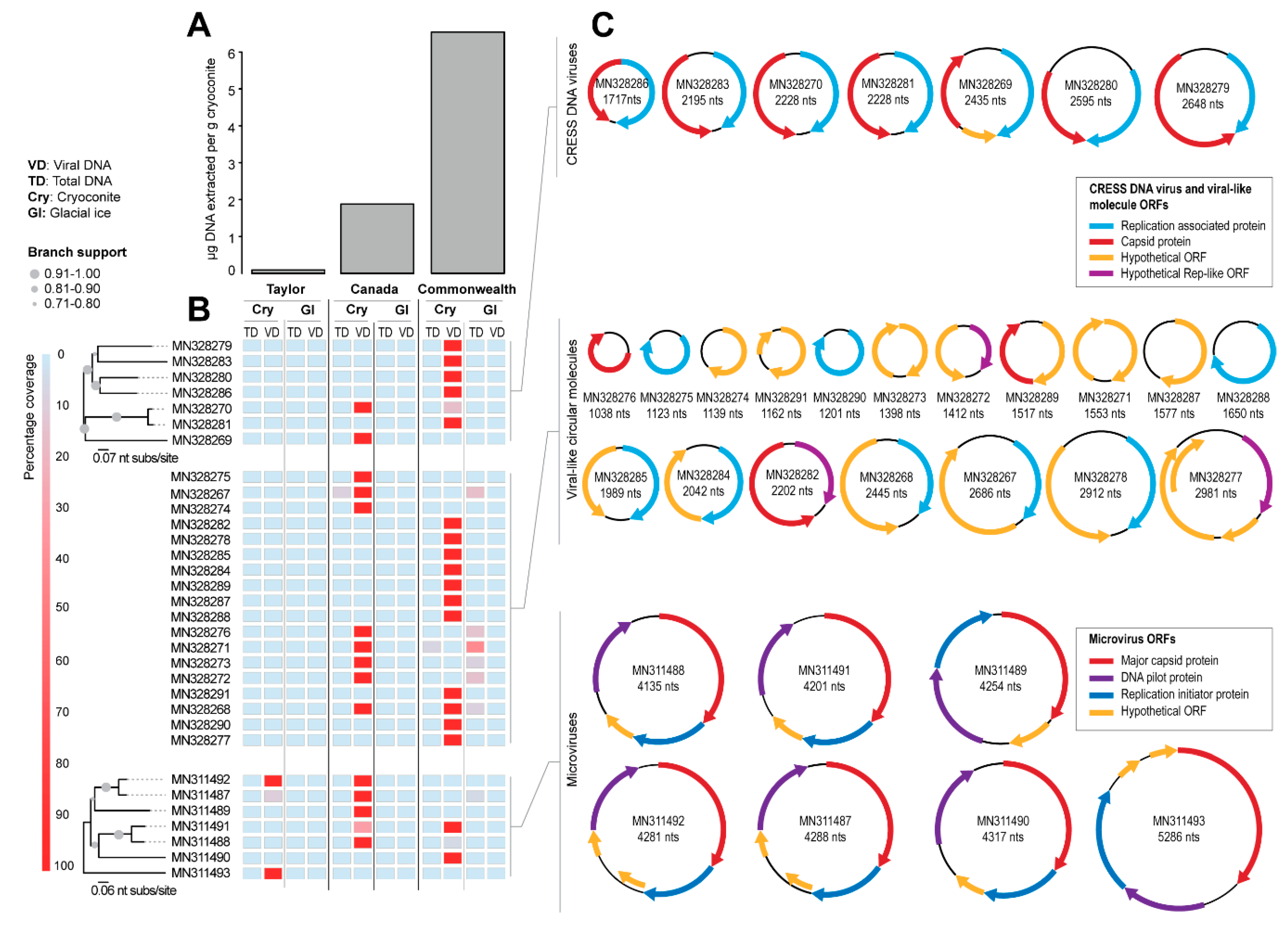
3.1. Identification of Viral Genomes
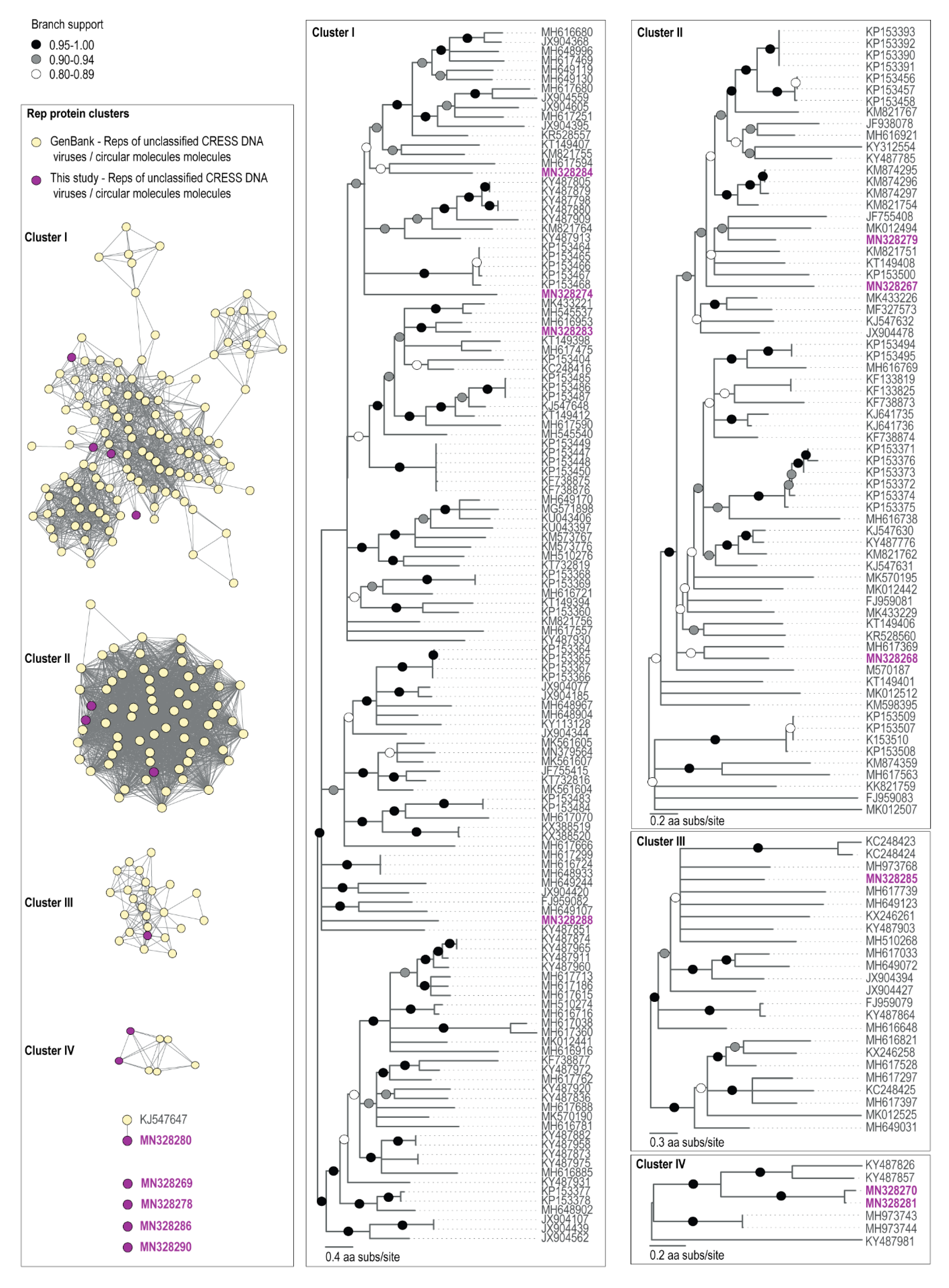
3.2. Diversity and Relationships of CRESS Viruses and Viral-Like Circular Molecules
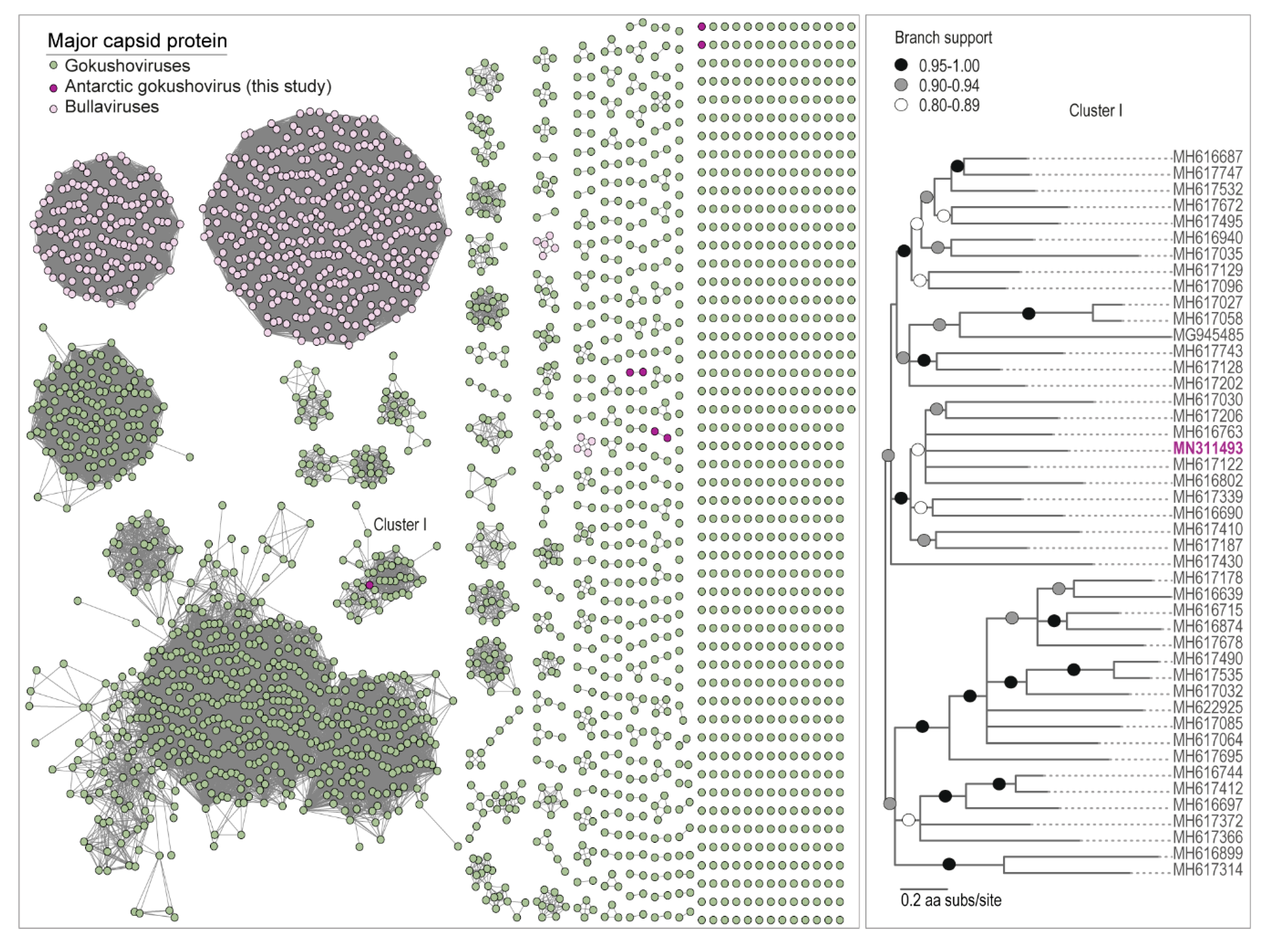
3.3. Microvirus Diversity
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brussaard, C.P.; Wilhelm, S.W.; Thingstad, F.; Weinbauer, M.G.; Bratbak, G.; Heldal, M.; Kimmance, S.A.; Middelboe, M.; Nagasaki, K.; Paul, J.H.; et al. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008, 2, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Middelboe, M.; Brussaard, C.P.D. Marine Viruses: Key Players in Marine Ecosystems. Viruses 2017, 9, 302. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Prangishvili, D.; Lindell, D. Roles of viruses in the environment. Environ. Microbiol. 2009, 11, 2771–2774. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Trubl, G.; Jang, H.B.; Roux, S.; Emerson, J.B.; Solonenko, N.; Vik, D.R.; Solden, L.; Ellenbogen, J.; Runyon, A.T.; Bolduc, B.; et al. Soil Viruses Are Underexplored Players in Ecosystem Carbon Processing. MSystems 2018, 3. [Google Scholar] [CrossRef]
- Anesio, A.M.; Bellas, C.M. Are low temperature habitats hot spots of microbial evolution driven by viruses? Trends Microbiol. 2011, 19, 52–57. [Google Scholar] [CrossRef]
- Cavicchioli, R. Microbial ecology of Antarctic aquatic systems. Nat. Rev. Microbiol. 2015, 13, 691–706. [Google Scholar] [CrossRef]
- Laybourn-Parry, J.; Pearce, D.A. The biodiversity and ecology of Antarctic lakes: Models for evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 2273–2289. [Google Scholar] [CrossRef]
- Rastrojo, A.; Alcami, A. Viruses in Polar Lake and Soil Ecosystems. Adv. Virus Res. 2018, 101, 39–54. [Google Scholar] [CrossRef]
- Wharton, R.A., Jr.; McKay, C.P.; Simmons, G.M., Jr.; Parker, B.C. Cryoconite holes on glaciers. Bioscience 1985, 35, 499–503. [Google Scholar] [CrossRef]
- Fountain, A.G.; Nylen, T.H.; Tranter, M.; Bagshaw, E. Temporal variations in physical and chemical features of cryoconite holes on Canada Glacier, McMurdo Dry Valleys, Antarctica. J. Geophys. Res.-Biogeosci. 2008, 113. [Google Scholar] [CrossRef]
- Fountain, A.G.; Tranter, M.; Nylen, T.H.; Lewis, K.J.; Mueller, D.R. Evolution of cryoconite holes and their contribution to meltwater runoff from glaciers in the McMurdo Dry Valleys, Antarctica. J. Glaciol. 2004, 50, 35–45. [Google Scholar] [CrossRef]
- Gribbon, P.W.F. Cryoconite Holes on Sermikavsak, West Greenland. J. Glaciol. 1979, 22, 177–181. [Google Scholar] [CrossRef]
- Steinbock, O. Cryoconite holes and their biological significance. Z. Gletscherkunde 1939, 24, 1–21. [Google Scholar]
- Cameron, K.A.; Hodson, A.J.; Osborn, A.M. Structure and diversity of bacterial, eukaryotic and archaeal communities in glacial cryoconite holes from the Arctic and the Antarctic. FEMS Microbiol. Ecol. 2012, 82, 254–267. [Google Scholar] [CrossRef]
- Telling, J.; Anesio, A.M.; Tranter, M.; Fountain, A.G.; Nylen, T.; Hawkings, J.; Singh, V.B.; Kaur, P.; Musilova, M.; Wadham, J.L. Spring thaw ionic pulses boost nutrient availability and microbial growth in entombed Antarctic Dry Valley cryoconite holes. Front. Microbiol. 2014, 5, 694. [Google Scholar] [CrossRef]
- Bagshaw, E.A.; Wadham, J.L.; Tranter, M.; Perkins, R.; Morgan, A.; Williamson, C.J.; Fountain, A.G.; Fitzsimons, S.; Dubnick, A. Response of Antarctic cryoconite microbial communities to light. FEMS Microbiol. Ecol. 2016, 92, fiw076. [Google Scholar] [CrossRef]
- Anesio, A.M.; Hodson, A.J.; Fritz, A.; Psenner, R.; Sattler, B. High microbial activity on glaciers: Importance to the global carbon cycle. Glob. Chang. Biol. 2009, 15, 955–960. [Google Scholar] [CrossRef]
- Cook, J.M.; Hodson, A.J.; Anesio, A.M.; Hanna, E.; Yallop, M.; Stibal, M.; Telling, J.; Huybrechts, P. An improved estimate of microbially mediated carbon fluxes from the Greenland ice sheet. J. Glaciol. 2012, 58, 1098–1108. [Google Scholar] [CrossRef]
- Bagshaw, E.A.; Tranter, M.; Fountain, A.G.; Welch, K.; Basagic, H.J.; Lyons, W.B. Do Cryoconite Holes have the Potential to be Significant Sources of C, N, and P to Downstream Depauperate Ecosystems of Taylor Valley, Antarctica? Arct. Antarct. Alp. Res. 2013, 45, 440–454. [Google Scholar] [CrossRef]
- Darcy, J.L.; Gendron, E.M.S.; Sommers, P.; Porazinska, D.L.; Schmidt, S.K. Island Biogeography of Cryoconite Hole Bacteria in Antarctica’s Taylor Valley and Around the World. Front. Ecol. Evol. 2018, 6. [Google Scholar] [CrossRef]
- Sommers, P.; Darcy, J.L.; Porazinska, D.L.; Gendron, E.M.S.; Fountain, A.G.; Zamora, F.; Vincent, K.; Cawley, K.M.; Solon, A.J.; Vimercati, L.; et al. Comparison of Microbial Communities in the Sediments and Water Columns of Frozen Cryoconite Holes in the McMurdo Dry Valleys, Antarctica. Front. Microbiol. 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Stanish, L.F.; O’Neill, S.P.; Gonzalez, A.; Legg, T.M.; Knelman, J.; McKnight, D.M.; Spaulding, S.; Nemergut, D.R. Bacteria and diatom co-occurrence patterns in microbial mats from polar desert streams. Environ. Microbiol. 2013, 15, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Anesio, A.M.; Mindl, B.; Laybourn-Parry, J.; Hodson, A.J.; Sattler, B. Viral dynamics in cryoconite holes on a high Arctic glacier (Svalbard). J. Geophys. Res.-Biogeosci. 2007, 112. [Google Scholar] [CrossRef]
- Bellas, C.M.; Anesio, A.M.; Telling, J.; Stibal, M.; Tranter, M.; Davis, S. Viral impacts on bacterial communities in Arctic cryoconite. Environ. Res. Lett. 2013, 8, 045021. [Google Scholar] [CrossRef]
- Säwström, C.; Laybourn-Parry, J.; Granéli, W.; Anesio, A.M. Heterotrophic bacterial and viral dynamics in Arctic freshwaters: Results from a field study and nutrient-temperature manipulation experiments. Polar Biol. 2007, 30, 1407–1415. [Google Scholar] [CrossRef]
- Bellas, C.M.; Anesio, A.M.; Barker, G. Analysis of virus genomes from glacial environments reveals novel virus groups with unusual host interactions. Front. Microbiol. 2015, 6, 656. [Google Scholar] [CrossRef]
- Roux, S.; Adriaenssens, E.M.; Dutilh, B.E.; Koonin, E.V.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Lavigne, R.; Brister, J.R.; Varsani, A.; et al. Minimum Information about an Uncultivated Virus Genome (MIUViG). Nat. Biotechnol. 2019, 37, 29–37. [Google Scholar] [CrossRef]
- Paez-Espino, D.; Eloe-Fadrosh, E.A.; Pavlopoulos, G.A.; Thomas, A.D.; Huntemann, M.; Mikhailova, N.; Rubin, E.; Ivanova, N.N.; Kyrpides, N.C. Uncovering Earth’s virome. Nature 2016, 536, 425–430. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zayed, A.A.; Conceicao-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1109–1123. [Google Scholar] [CrossRef]
- Yau, S.; Seth-Pasricha, M. Viruses of Polar Aquatic Environments. Viruses 2019, 11, 189. [Google Scholar] [CrossRef] [PubMed]
- Smeele, Z.E.; Ainley, D.G.; Varsani, A. Viruses associated with Antarctic wildlife: From serology based detection to identification of genomes using high throughput sequencing. Virus Res. 2018, 243, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Kramer, R.; Van Goethem, M.W.; Makhalanyane, T.P.; Hogg, I.; Cowan, D.A. Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome 2017, 5, 83. [Google Scholar] [CrossRef] [PubMed]
- Zablocki, O.; Van Zyl, L.; Adriaenssens, E.M.; Rubagotti, E.; Tuffin, M.; Cary, S.C.; Cowan, D. High-level diversity of tailed phages, eukaryote-associated viruses, and virophage-like elements in the metaviromes of antarctic soils. Appl. Environ. Microbiol. 2014, 80, 6888–6897. [Google Scholar] [CrossRef] [PubMed]
- Swanson, M.M.; Reavy, B.; Makarova, K.S.; Cock, P.J.; Hopkins, D.W.; Torrance, L.; Koonin, E.V.; Taliansky, M. Novel bacteriophages containing a genome of another bacteriophage within their genomes. PLoS ONE 2012, 7, e40683. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, D.; Lopez-Bueno, A.; Aguirre de Carcer, D.; De los Rios, A.; Alcami, A.; Quesada, A. Ecosystem function decays by fungal outbreaks in Antarctic microbial mats. Sci. Rep. 2016, 6, 22954. [Google Scholar] [CrossRef]
- Zawar-Reza, P.; Arguello-Astorga, G.R.; Kraberger, S.; Julian, L.; Stainton, D.; Broady, P.A.; Varsani, A. Diverse small circular single-stranded DNA viruses identified in a freshwater pond on the McMurdo Ice Shelf (Antarctica). Infect. Genet. Evol. 2014, 26, 132–138. [Google Scholar] [CrossRef]
- Lopez-Bueno, A.; Tamames, J.; Velazquez, D.; Moya, A.; Quesada, A.; Alcami, A. High diversity of the viral community from an Antarctic lake. Science 2009, 326, 858–861. [Google Scholar] [CrossRef]
- Aguirre de Carcer, D.; Lopez-Bueno, A.; Alonso-Lobo, J.M.; Quesada, A.; Alcami, A. Metagenomic analysis of lacustrine viral diversity along a latitudinal transect of the Antarctic Peninsula. FEMS Microbiol. Ecol. 2016, 92, fiw074. [Google Scholar] [CrossRef]
- Aguirre de Carcer, D.; Lopez-Bueno, A.; Pearce, D.A.; Alcami, A. Biodiversity and distribution of polar freshwater DNA viruses. Sci. Adv. 2015, 1, e1400127. [Google Scholar] [CrossRef]
- Lauro, F.M.; DeMaere, M.Z.; Yau, S.; Brown, M.V.; Ng, C.; Wilkins, D.; Raftery, M.J.; Gibson, J.A.; Andrews-Pfannkoch, C.; Lewis, M.; et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 2011, 5, 879–895. [Google Scholar] [CrossRef] [PubMed]
- Tschitschko, B.; Williams, T.J.; Allen, M.A.; Paez-Espino, D.; Kyrpides, N.; Zhong, L.; Raftery, M.J.; Cavicchioli, R. Antarctic archaea-virus interactions: Metaproteome-led analysis of invasion, evasion and adaptation. ISME J. 2015, 9, 2094–2107. [Google Scholar] [CrossRef] [PubMed]
- Smeele, Z.E.; Burns, J.M.; Van Doorsaler, K.; Fontenele, R.S.; Waits, K.; Stainton, D.; Shero, M.R.; Beltran, R.S.; Kirkham, A.L.; Berngartt, R.; et al. Diverse papillomaviruses identified in Weddell seals. J. Gen. Virol. 2018, 99, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Olivares, F.; Tapia, R.; Galvez, C.; Meza, F.; Barriga, G.P.; Borras-Chavez, R.; Mena-Vasquez, J.; Medina, R.A.; Neira, V. Novel penguin Avian avulaviruses 17, 18 and 19 are widely distributed in the Antarctic Peninsula. Transbound. Emerg. Dis. 2019. [Google Scholar] [CrossRef]
- Wille, M.; Aban, M.; Wang, J.; Moore, N.; Shan, S.; Marshall, J.; Gonzalez-Acuna, D.; Vijaykrishna, D.; Butler, J.; Wang, J.; et al. Antarctic Penguins as Reservoirs of Diversity for Avian Avulaviruses. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Buck, C.B.; Van Doorslaer, K.; Peretti, A.; Geoghegan, E.M.; Tisza, M.J.; An, P.; Katz, J.P.; Pipas, J.M.; McBride, A.A.; Camus, A.C.; et al. The Ancient Evolutionary History of Polyomaviruses. PLoS Pathog. 2016, 12, e1005574. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Kraberger, S.; Austin, C.; Farkas, K.; Bergeman, M.; Paunil, E.; Davison, W.; Varsani, A. Fish polyomaviruses belong to two distinct evolutionary lineages. J. Gen. Virol. 2018, 99, 567–573. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Varsani, A.; Koonin, E.V.; Krupovic, M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat. Commun. 2019, 10, 3425. [Google Scholar] [CrossRef]
- Rosario, K.; Duffy, S.; Breitbart, M. A field guide to eukaryotic circular single-stranded DNA viruses: Insights gained from metagenomics. Arch. Virol. 2012, 157, 1851–1871. [Google Scholar] [CrossRef]
- Labonte, J.M.; Suttle, C.A. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 2013, 7, 2169–2177. [Google Scholar] [CrossRef]
- Labonte, J.M.; Hallam, S.J.; Suttle, C.A. Previously unknown evolutionary groups dominate the ssDNA gokushoviruses in oxic and anoxic waters of a coastal marine environment. Front. Microbiol. 2015, 6, 315. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Mettel, K.A.; Benner, B.E.; Johnson, R.; Scott, C.; Yusseff-Vanegas, S.Z.; Baker, C.C.M.; Cassill, D.L.; Storer, C.; Varsani, A.; et al. Virus discovery in all three major lineages of terrestrial arthropods highlights the diversity of single-stranded DNA viruses associated with invertebrates. PeerJ 2018, 6, e5761. [Google Scholar] [CrossRef] [PubMed]
- Steel, O.; Kraberger, S.; Sikorski, A.; Young, L.M.; Catchpole, R.J.; Stevens, A.J.; Ladley, J.J.; Coray, D.S.; Stainton, D.; Dayaram, A.; et al. Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand. Infect. Genet. Evol. 2016, 43, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Schmidlin, K.; Fontenele, R.S.; Walters, M.; Varsani, A. Unravelling the Single-Stranded DNA Virome of the New Zealand Blackfly. Viruses 2019, 11, 532. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Galatowitsch, M.L.; Arguello-Astorga, G.R.; van Bysterveldt, K.; Kraberger, S.; Stainton, D.; Harding, J.S.; Roumagnac, P.; Martin, D.P.; Lefeuvre, P.; et al. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect. Genet. Evol. 2016, 39, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Krupovic, M.; Daly, R.A.; Borges, A.L.; Nayfach, S.; Schulz, F.; Sharrar, A.; Matheus Carnevali, P.B.; Cheng, J.F.; Ivanova, N.N.; et al. Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth’s biomes. Nat. Microbiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Creasy, A.; Rosario, K.; Leigh, B.A.; Dishaw, L.J.; Breitbart, M. Unprecedented Diversity of ssDNA Phages from the Family Microviridae Detected within the Gut of a Protochordate Model Organism (Ciona robusta). Viruses 2018, 10, 404. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Lacorte, C.; Lamas, N.S.; Schmidlin, K.; Varsani, A.; Ribeiro, S.G. Single Stranded DNA Viruses Associated with Capybara Faeces Sampled in Brazil. Viruses 2019, 11, 710. [Google Scholar] [CrossRef]
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.-Y.S.; Varsani, A.; Krishnamurthy, S.R.; Pesavento, P.A.; et al. Discovery of several thousand highly diverse circular DNA viruses. BioRxiv 2019. [Google Scholar] [CrossRef]
- Cherwa, J.E.; Fane, B.A. Microviridae. In Virus Taxonomy; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: Waltham, MA, USA, 2012; pp. 385–393. [Google Scholar] [CrossRef]
- Varsani, A.; Frankfurter, G.; Stainton, D.; Male, M.F.; Kraberger, S.; Burns, J.M. Identification of a polyomavirus in Weddell seal (Leptonychotes weddellii) from the Ross Sea (Antarctica). Arch. Virol. 2017, 162, 1403–1407. [Google Scholar] [CrossRef]
- Fahsbender, E.; Burns, J.M.; Kim, S.; Kraberger, S.; Frankfurter, G.; Eilers, A.A.; Shero, M.R.; Beltran, R.; Kirkham, A.; McCorkell, R.; et al. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol. 2017, 3, vex017. [Google Scholar] [CrossRef] [PubMed]
- Levy, J. How big are the McMurdo Dry Valleys? Estimating ice-free area using Landsat image data. Antarct. Sci. 2013, 25, 119–120. [Google Scholar] [CrossRef]
- Matsuoka, K.; Skoglund, A.; Deschwanden, A.V. Quantarctica: A free GIS Package for Research, Education, and Operation in Antarctica; Quantarctica (Norwegian Polar Institute): Norway; 2013. Available online: https://quantarctica.npolar.no/downloads/ (accessed on 15 September 2019).
- Fountain, A.G.; Lyons, W.B.; Burkins, M.B.; Dana, G.L.; Doran, P.T.; Lewis, K.J.; McKnight, D.M.; Moorhead, D.L.; Parsons, A.N.; Priscu, J.C.; et al. Physical Controls on the Taylor Valley Ecosystem, Antarctica. BioScience 1999, 49, 961–971. [Google Scholar] [CrossRef]
- Porazinska, D.L.; Fountain, A.G.; Nylen, T.H.; Tranter, M.; Virginia, R.A.; Wall, D.H. The Biodiversity and biogeochemistry of cryoconite holes from McMurdo Dry Valley glaciers, Antarctica. Arct. Antarct. Alp. Res. 2004, 36, 84–91. [Google Scholar] [CrossRef]
- Sommers, P.; Darcy, J.L.; Gendron, E.M.S.; Stanish, L.F.; Bagshaw, E.A.; Porazinska, D.L.; Schmidt, S.K. Diversity patterns of microbial eukaryotes mirror those of bacteria in Antarctic cryoconite holes. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.E.; Virginia, R.A.; Wall, D.H.; Parsons, A.N.; Powers, L.E.; Burkins, M.B. Variation in biogeochemistry and soil biodiversity across spatial scales in a polar desert ecosystem. Ecology 2004, 85, 3105–3118. [Google Scholar] [CrossRef]
- Nylen, T.H.; Fountain, A.G.; Doran, P.T. Climatology of katabatic winds in the McMurdo dry valleys, southern Victoria Land, Antarctica. J. Geophys. Res.-Atmos. 2004, 109. [Google Scholar] [CrossRef]
- Šabacká, M.; Priscu, J.C.; Basagic, H.J.; Fountain, A.G.; Wall, D.H.; Virginia, R.A.; Greenwood, M.C. Aeolian flux of biotic and abiotic material in Taylor Valley, Antarctica. Geomorphology 2012, 155, 102–111. [Google Scholar] [CrossRef]
- Di Pietro, F.; Ortenzi, F.; Tilio, M.; Concetti, F.; Napolioni, V. Genomic DNA extraction from whole blood stored from 15- to 30-years at -20 degrees C by rapid phenol-chloroform protocol: A useful tool for genetic epidemiology studies. Mol. Cell. Probes 2011, 25, 44–48. [Google Scholar] [CrossRef]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab.(LBNL): Berkeley, CA, USA, 2014. [Google Scholar]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef] [PubMed]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 2015, 1854, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Stover, B.C.; Muller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Foreman, C.M.; Sattler, B.; Mikucki, J.A.; Porazinska, D.L.; Priscu, J.C. Metabolic activity and diversity of cryoconites in the Taylor Valley, Antarctica. J. Geophys. Res.-Biogeosci. 2007, 112. [Google Scholar] [CrossRef]
- Stanish, L.F.; Bagshaw, E.A.; McKnight, D.M.; Fountain, A.G.; Tranter, M. Environmental factors influencing diatom communities in Antarctic cryoconite holes. Environ. Res. Lett. 2013, 8, 045006. [Google Scholar] [CrossRef][Green Version]
- Rahlff, J. The Virioneuston: A Review on Viral(-)Bacterial Associations at Air(-)Water Interfaces. Viruses 2019, 11, 191. [Google Scholar] [CrossRef] [PubMed]
- Wigington, C.H.; Sonderegger, D.; Brussaard, C.P.; Buchan, A.; Finke, J.F.; Fuhrman, J.A.; Lennon, J.T.; Middelboe, M.; Suttle, C.A.; Stock, C.; et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 2016, 1, 15024. [Google Scholar] [CrossRef] [PubMed]
- Kepner, R.L., Jr.; Wharton, R.A., Jr.; Suttle, C.A. Viruses in Antarctic lakes. Limnol. Oceanogr. 1998, 43, 1754–1761. [Google Scholar] [CrossRef]
- Sawstrom, C.; Anesio, M.A.; Graneli, W.; Laybourn-Parry, J. Seasonal viral loop dynamics in two large ultraoligotrophic Antarctic freshwater lakes. Microb. Ecol. 2007, 53, 1–11. [Google Scholar] [CrossRef]
- Zhao, L.; Rosario, K.; Breitbart, M.; Duffy, S. Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses With Small Genomes and a Diverse Host Range. Adv. Virus Res. 2019, 103, 71–133. [Google Scholar] [CrossRef]
- Wang, H.; Ling, Y.; Shan, T.; Yang, S.; Xu, H.; Deng, X.; Delwart, E.; Zhang, W. Gut virome of mammals and birds reveals high genetic diversity of the family Microviridae. Virus Evol. 2019, 5, vez013. [Google Scholar] [CrossRef]
- Brentlinger, K.L.; Hafenstein, S.; Novak, C.R.; Fane, B.A.; Borgon, R.; McKenna, R.; Agbandje-McKenna, M. Microviridae, a family divided: Isolation, characterization, and genome sequence of phiMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J. Bacteriol. 2002, 184, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Krupovic, M.; Poulet, A.; Debroas, D.; Enault, F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS ONE 2012, 7, e40418. [Google Scholar] [CrossRef]
- Kraberger, S.; Cook, C.N.; Schmidlin, K.; Fontenele, R.S.; Bautista, J.; Smith, B.; Varsani, A. Diverse single-stranded DNA viruses associated with honey bees (Apis mellifera). Infect. Genet. Evol. 2019, 71, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Male, M.F.; Kraberger, S.; Stainton, D.; Kami, V.; Varsani, A. Cycloviruses, gemycircularviruses and other novel replication-associated protein encoding circular viruses in Pacific flying fox (Pteropus tonganus) faeces. Infect. Genet. Evol. 2016, 39, 279–292. [Google Scholar] [CrossRef] [PubMed]
| Virus Group | Name | Accession |
|---|---|---|
| CRESS DNA viruses | Antarctic virus CAA 003 44 | MN328269 |
| Antarctic virus CAA 003 54 | MN328270 | |
| Antarctic virus COCH21 47 | MN328279 | |
| Antarctic virus COCH21 51 | MN328280 | |
| Antarctic virus COCH21 74 | MN328281 | |
| Antarctic virus COCH21 78 | MN328283 | |
| Antarctic virus COCH21 111 | MN328286 | |
| Microviruses | Antarctic microvirus CAA 003 V 1 | MN311487 |
| Antarctic microvirus CAA 003 V 4 | MN311488 | |
| Antarctic microvirus CAA 003 V 9 | MN311489 | |
| Antarctic microvirus COCH21 V SP 16 | MN311491 | |
| Antarctic microvirus COCH21 V SP 13 | MN311490 | |
| Antarctic microvirus TYR 006 V 25 | MN311492 | |
| Antarctic microvirus TYR 006 V SP 13 | MN311493 | |
| Circular molecules | Antarctic circular molecule CAA 003 32 | MN328267 |
| Antarctic circular molecule CAA 003 40 | MN328268 | |
| Antarctic circular molecule CAA 003 97 | MN328271 | |
| Antarctic circular molecule CAA 003 107 | MN328272 | |
| Antarctic circular molecule CAA 003 115 | MN328273 | |
| Antarctic circular molecule CAA 003 147 | MN328274 | |
| Antarctic circular molecule CAA 003 151 | MN328275 | |
| Antarctic circular molecule CAA 003 179 | MN328276 | |
| Antarctic circular molecule COCH21 37 | MN328277 | |
| Antarctic circular molecule COCH21 39 | MN328278 | |
| Antarctic circular molecule COCH21 77 | MN328282 | |
| Antarctic circular molecule COCH21 94 | MN328284 | |
| Antarctic circular molecule COCH21 102 | MN328285 | |
| Antarctic circular molecule COCH21 141 | MN328287 | |
| Antarctic circular molecule COCH21 149 | MN328288 | |
| Antarctic circular molecule COCH21 162 | MN328289 | |
| Antarctic circular molecule COCH21 215 | MN328290 | |
| Antarctic circular molecule COCH21 226 | MN328291 |
| Accession Number | Name | Endonuclease Domain | SF3 Helicase Domain | |||||
|---|---|---|---|---|---|---|---|---|
| Motif I | Motif II | Motif III | Walker A | Walker B | Motif C | Arg Finger | ||
| MN328269 | Antarctic virus CAA 003 44 | FLTWPK | IHYHVC | GAVGYTGK | GVGFHGKSKFGE | VFDYE | VFAN | MSEDRW |
| MN328270 | Antarctic virus CAA 003 54 | LLTFAQ | AHFHAV | RAVEYVAK | GPSRYGKTVLAR | VLDDL | VLTN | |
| MN328279 | Antarctic virus COCH21 47 | ILTIPA | VHWQLL | AAEEYCGK | GVTGTGKSRTAW | VIDEF | ITSN | ALMRRL |
| MN328283 | Antarctic virus COCH21 78 | CFTWNN | PHLQGY | QNDRYCRK | GDSGCGKTRSVN | LVDDV | VTSQ | ALLRRF |
| MN328281 | Antarctic virus COCH21 74 | LLTFAQ | AHFHAV | RAVEYVAK | GPSRYGKTVLAR | VLDDL | VLTN | |
| MN328280 | Antarctic virus COCH21 51 | CVTIHI | IHWQMY | LAIEYCKK | GRSGLGKTQFAI | IFDDM | FTSN | AIRRRC |
| MN328286 | Antarctic virus COCH21 111 | AWTIYG | LHYQGQ | GSELYCQK | PDGNAGKTCFAK | IVDVK | VFSN | LSKDRW |
| MN328267 | Antarctic circular molecule CAA 003 32 | IATMPH | LHWQFV | AAIAYVWK | GRTGTGKSRRAW | VIDEF | ITSN | AFLRRL |
| MN328268 | Antarctic circular molecule CAA 003 40 | LLTIRQ | VHWQVL | AADEYVWK | GATGTGKSRLAW | VLDEF | ITSN | ALLRRM |
| MN328274 | Antarctic circular molecule CAA 003 147 | CYTLNN | PHLQGY | QNVTYCSK | GPPGTGKSRKAR | IIDDI | VTSN | AIQRRY |
| MN328278 | Antarctic circular molecule COCH21 39 | VFTKHF | IHWQGY | EAREYCMK | TIGGKGKTRLAT | IFDIS | FFSN | LSLDRV |
| MN328284 | Antarctic circular molecule COCH21 94 | CFTLNN | PHLQGF | QNRDYCIK | GQTGCGKTRSAT | IIDDF | VTSQ | AIMRRV |
| MN328285 | Antarctic circular molecule COCH21 102 | ALTFWD | IHYQSY | ENIAYCSK | GPTGVGKSHQAF | FNDFR | VTSS | QLLRRF |
| MN328288 | Antarctic circular molecule COCH21 149 | MWTLNN | PHLQGA | EALDYCVK | GPTGTGKSRSVL | FIDDF | ITSN | PLHRRF |
| MN328290 | Antarctic circular molecule COCH21 215 | CFTWNN | PHYQGY | EAIAYCTK | SQGNAGKTTFTK | VFDIN | VFSN | LSVDRL |
| Name | Accession Number | Rep | CP | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Nearest from Dataset | Nearest from GenBank | Nearest from Dataset | Nearest from GenBank | ||||||
| Sequence ID | Pairwise ID | Sequence ID | Pairwise ID | Sequence ID | Pairwise ID | Sequence ID | Pairwise ID | ||
| Antarctic virus CAA_003 44 | MN328269 | MN328290 | 26% | KF738885 | 31% | MN328276 | 47% | MH552476 | 31% |
| Antarctic virus CAA 003 54 | MN328270 | MN328281 | 94% | KY487857 | 50% | MN328281 | 87% | KY487857 | 52% |
| Antarctic virus COCH21 47 | MN328279 | MN328268 | 55% | KM821754 | 61% | MN328286 | 29% | MH552476 | 30% |
| Antarctic virus COCH21 5 | MN328280 | MN328268 | 30% | KJ547647 | 94% | MN328281 | 21% | KJ547647 | 78% |
| Antarctic virus COCH21 74 | MN328281 | MN328268 | 26% | KY487857 | 49% | MN328270 | 87% | KY487857 | 50% |
| Antarctic virus COCH21 78 | MN328283 | MN328284 | 40% | MH616953 | 66% | MN328276 | 23% | KY487851 | 25% |
| Antarctic virus COCH21 111 | MN328286 | MN328278 | 31% | MH617452 | 41% | MN328279 | 29% | KY487835 | 35% |
| Antarctic circular molecule CAA 003 32 | MN328267 | MN328268 | 55% | KM821754 | 61% | ||||
| Antarctic circular molecule CAA 003 40 | MN328268 | MN328267 | 55% | KM874297 | 63% | ||||
| Antarctic circular molecule CAA 003 147 | MN328274 | MN328284 | 41% | JX904420 | 46% | ||||
| Antarctic circular molecule COCH21 39 | MN328278 | MN328286 | 31% | MH616877 | 41% | ||||
| Antarctic circular molecule COCH21 94 | MN328284 | MN328288 | 42% | JX904420 | 45% | ||||
| Antarctic circular molecule COCH21 102 | MN328285 | MN328274 | 33% | KY487903 | 48% | ||||
| Antarctic circular molecule COCH21 149 | MN328288 | MN328284 | 42% | MH648933 | 45% | ||||
| Antarctic circular molecule COCH21 215 | MN328290 | MN328286 | 31% | KX534391 | 38% | ||||
| Antarctic circular molecule CAA 003 179 | MN328276 | MN328269 | 47% | MH552476 | 28% | ||||
| Antarctic circular molecule COCH21 77 | MN328282 | MN328276 | 27% | MK012465 | 27% | ||||
| Antarctic circular molecule COCH21 162 | MN328289 | MN328269 | 27% | MH616939 | 31% | ||||
| Name | Accession Number | Nearest Neighbor within Dataset | Nearest Neighbor within Database | ||
|---|---|---|---|---|---|
| Sequence | Pairwise % | Sequence | Pairwise % | ||
| Antarctic microvirus CAA 003 V 1 | MN311487 | MN311491 | 56% | MK765582 | 59% |
| Antarctic microvirus CAA 003 V 4 | MN311488 | MN311491 | 90% | MH617700 | 61% |
| Antarctic microvirus CAA 003 V 9 | MN311489 | MN311492 | 53% | MK765646 | 54% |
| Antarctic microvirus COCH21 V SP 13 | MN311490 | MN311489 | 51% | MH617350 | 60% |
| Antarctic microvirus COCH21 V SP 16 | MN311491 | MN311488 | 90% | MH617700 | 61% |
| Antarctic microvirus TYR 006 V 25 | MN311492 | MN311487 | 92% | MK765582 | 59% |
| Antarctic microvirus TYR 006 V SP 13 | MN311493 | MN311492 | 29% | MH617122 | 64% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sommers, P.; Fontenele, R.S.; Kringen, T.; Kraberger, S.; Porazinska, D.L.; Darcy, J.L.; Schmidt, S.K.; Varsani, A. Single-Stranded DNA Viruses in Antarctic Cryoconite Holes. Viruses 2019, 11, 1022. https://doi.org/10.3390/v11111022
Sommers P, Fontenele RS, Kringen T, Kraberger S, Porazinska DL, Darcy JL, Schmidt SK, Varsani A. Single-Stranded DNA Viruses in Antarctic Cryoconite Holes. Viruses. 2019; 11(11):1022. https://doi.org/10.3390/v11111022
Chicago/Turabian StyleSommers, Pacifica, Rafaela S. Fontenele, Tayele Kringen, Simona Kraberger, Dorota L. Porazinska, John L. Darcy, Steven K. Schmidt, and Arvind Varsani. 2019. "Single-Stranded DNA Viruses in Antarctic Cryoconite Holes" Viruses 11, no. 11: 1022. https://doi.org/10.3390/v11111022
APA StyleSommers, P., Fontenele, R. S., Kringen, T., Kraberger, S., Porazinska, D. L., Darcy, J. L., Schmidt, S. K., & Varsani, A. (2019). Single-Stranded DNA Viruses in Antarctic Cryoconite Holes. Viruses, 11(11), 1022. https://doi.org/10.3390/v11111022






