1. Introduction
Zika virus (ZIKV) was first isolated in monkeys in Uganda in 1947, and then detected in
Aedes africanus mosquitoes in 1948. ZIKV belongs to a mosquito-borne flavivirus of the family Flavivirus, which spread from Africa to south-eastern Asia through transmission by
Aedes mosquitoes, such as
A. albopictus, A. Aegypti, and
A. Polynesiensis. ZIKV exhibits genetic variation that is classified into African and Asian lineages [
1]. ZIKV infection causes self-limiting, mild illnesses, such as low-grade fever, headaches, rash, myalgia, arthralgia, and conjunctivitis; fewer than 20 human ZIKV cases were reported from the 1960s to 1980s [
2]. Recently, the ZIKV Asian-lineage strains spread from south-east Asia toward the Pacific, leading to three large outbreaks on Yap island (Micronesia) in 2007, French Polynesia in 2013–2014, in Brazil in 2015–2016, and then across the Americas in 2016 [
3]. The epidemic ZIKV Asian strains evolved into highly virulent and neurotropic variants, resulting in a dramatic increase in cases of congenital, ZIKV-associated microcephaly in newborns and Guillain–Barre syndrome (GBS) in adults during these three outbreaks [
3]. The epidemic ZIKV Asian strains have also been transmitted via vertical, sexual, and blood transfusion routes [
4]. Therefore, the WHO Emergency Committee announced that ZIKV is considered a “Public Health Emergency of International Concern” (PHEIC) in November 2016 [
5]. Nowadays, the epidemic ZIKV Asian strains have broadened worldwide; laboratory-confirmed symptomatic ZIKV cases are still reported [
6]. ZIKV’s emergence has become an extraordinary and enduring challenge to public health; thus, there is a great need to understand the features of the epidemic ZIKV Asian strains.
ZIKV, like other flaviviruses, such as Dengue virus, Japanese encephalitis virus (JEV), and West Nile virus, possesses a single-strand, positive-sense RNA genome in a small, enveloped virion [
7]. The flavivirus genome, nearly 11 kb in length, encodes a large open reading frame (ORF) flanked by two noncoding regions at the 5’ and 3’-ends, which translates into a large polyprotein that is divided into three structural proteins (capsid (C), membrane (prM/M), and envelope (E)) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) through the proteolytic process of the viral proteases (NS2B–NS3) and host cell proteases. E protein is the surface protein for receptor binding and cell membrane fusion. NS3 is a multidomain protein with an N-terminal protease (NS3pro), C-terminal RNA triphosphatase (NS3 RTPase), and helicase (NS3hel). NS5 consists of an N-terminal methyltransferase (NS5 MTase) and C-terminal RNA polymerase (NS5RdRp). Phylogenetic analysis of ZIKV African and Asian-lineage strains isolated from 1947 to 2017 indicates that some mutations accumulated in the epidemic ZIKV Asian strains in 2015–2016 [
8,
9]. Epidemic ZIKV Asian strains contain 75 amino acid substitutions compared with their African lineage strains, five of which are located in C, nine in prM, 10 in E, four in NS1, five in NS2A, two in NS2B, nine in NS3, one in NS4A, nine in NS4B, and 21 in NS5 [
8,
9]. The serine-to-asparagine substitution (Ser
139→Asn
139 (S139N0) in prM increases ZIKV infectivity in neural progenitor cells and the severity of fetal microcephaly in a mouse model [
10]. The alanine-to-valine substitution (A
984→V
984 [A984V]) in NS1 induces the inhibition of interferon-β production and leads to viral immune escape [
11]. In addition, comparison between pre-epidemic and epidemic ZIKV Asian strains indicated 24 amino acid substitutions within prM (2), E (3), NS2A (1), NS3 (3), NS4B (7), and NS5 (8), such as T773M in E, Y2082H in NS3, L2451S in NS4B, and T2630V in NS5 [
8,
9]. The mutations in epidemic ZIKV Asian strains in comparison with pre-epidemic Asian and African strains might be determinants for their unique features; considering the increase in virulence and persistent infection, these mutations are worth understandingly thoroughly.
Reverse genetics is an effective tool for preparing replicons and infectious clones of flaviviruses, in order to study their mechanisms of viral replication and pathogenesis and to identify potential antiviral compounds [
12,
13]. The full-length, infectious cDNA clones of flaviviruses under cytomegalovirus’s (CMV’s) promoter-driven transcription have been widely used to generate infectious particles in vitro. However, full-length flavivirus cDNA clones are highly toxic to
Escherichia coli, which causes instability of flavivirus cDNAs and increases the difficulty of constructing flavivirus cDNA clones [
14]. DNA-launched replicons of dengue virus, West Nile virus, and JEV contain the 5’ and 3’-noncoding regions, partial C gene, and the complete gene fragment of all non-structural proteins under the control of the CMV promoter [
12]. CMV promoter-driven viral RNA subgenomes enable self-replication in transfected cells with DNA-launched replicons, but they possess no infectious particle production due to lacking the coding region of structural proteins. The approach using two plasmids with the structural genes (C, prM, and E) and the remaining genome as the replicon has been established to produce single-round infectious particles (SRIPs). The packaging cells carrying the recombinant plasmid encoding the viral structural proteins are transfected with DNA-launched replicons and then self-replicate viral subgenomes and express all viral proteins, allowing viral assembly and release as SRIPs. Several flavivirus, replicon-based SRIPs, including JEV, dengue virus, West Nile virus, and tick-borne encephalitis virus have been generated and demonstrated as safer vaccine candidates [
15,
16,
17]. Subsequently, replicon-based SRIPs are suitable systems for investigating the mechanism of the ZIKV life cycle, ZIKV-host interaction, and virulence.
The ZIKV Natal RGN strain that is associated with microcephaly was recognized as the representative, epidemic, ZIKV Asian strain in 2015–2016 [
18]. The ZIKV Natal RGN strain is detected in brain tissues from fetal autopsy cases with microcephaly in the Natal region of Brazil; its genome has been sequenced using next-generation sequencing (GenBank accession number KU527068) [
15]. The ZIKV Natal RGN strain contains a unique genotype and phenotype; therefore, this study aimed to generate SRIPs of the ZIKV Natal RGN strain using synthetic and reverse genetic technologies. This paper characterizes the infectivity of ZIKV Natal RGN SRIPs in different cell types. Moreover, the paper analyzes the expression pattern of type I interferons and apoptosis-related genes induced by ZIKV Natal RGN SRIPs.
2. Material and Methods
2.1. Cells
TE671 (human, rhabdomyosarcoma/muscle), SF268 (human brain, glioblastoma/astrocytoma), and ARPE-19 (human, retinal pigmented epithelium) cells were cultured in minimum essential medium containing 10% fetal bovine serum, 2 mM glutamine, 1 mM pyruvate, and 1× penicillin-streptomycin at 37 °C with 5% CO2.
2.2. Generation of the ZIKV Natal RGN Replicon
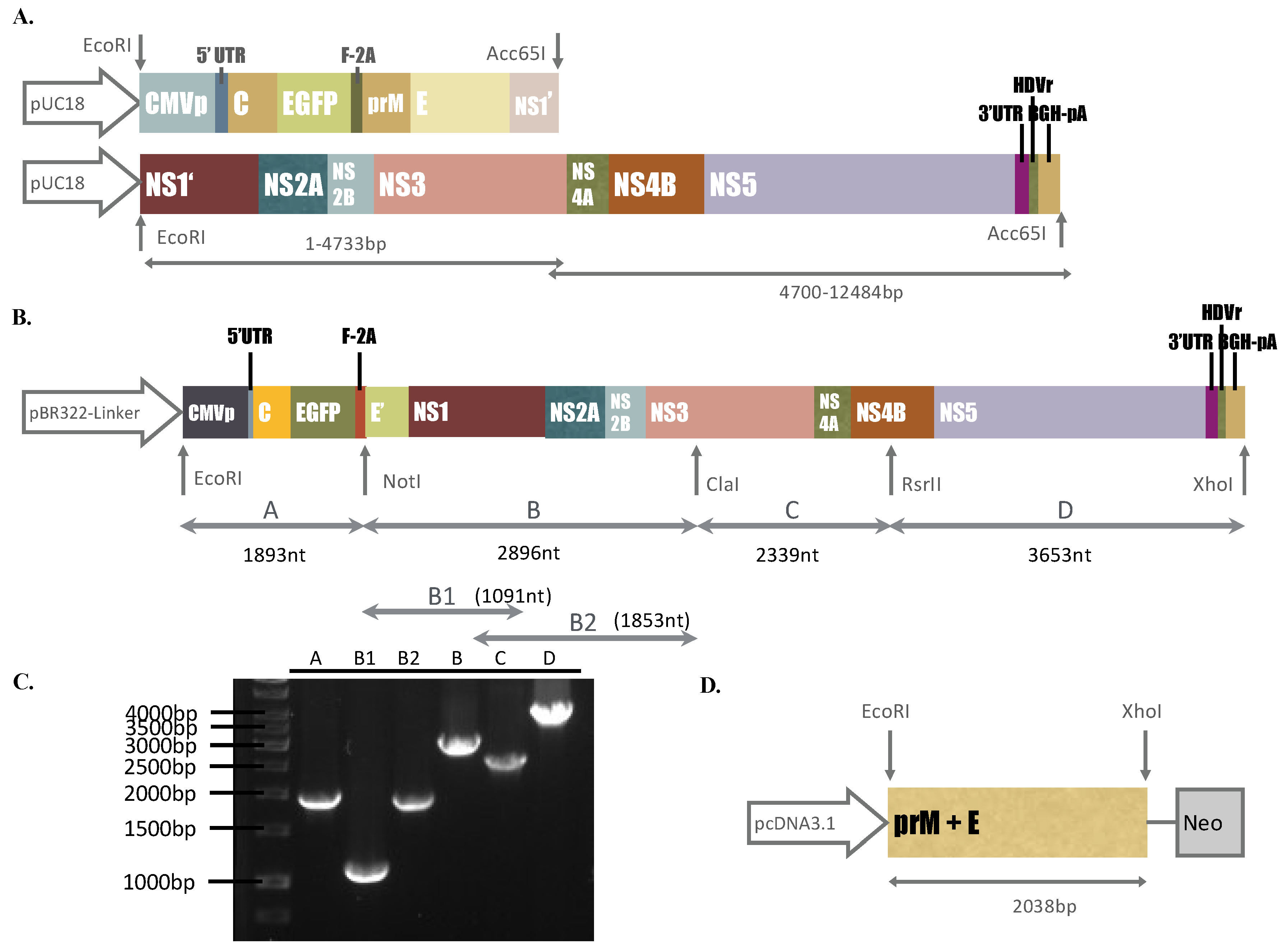
Components of two DNA segments we synthesized, I and II, consisted of the CMV immediate-early promoter (CMVp), cDNA fragments of the ZIKV Natal RGN genome (GenBank accession number KU527068), enhanced green fluorescent protein (EGFP), FMDV-2A (F-2A), hepatitis delta virus ribozyme (HDVr), and bovine growth hormone polyA signal (BGH-pA), which were purchased from Bio Basic Inc.
(Ontario, Canada). The construction of synthesized DNA fragments, plasmid information, and restriction enzyme digestion analysis of the DNA fragments we purchased are shown in
Supplementary Figures S1 and S2 and
Figure 1A. To construct the ZIKV Natal RGN replicon, the plasmid pBR322-Linker, described in our prior report [
15], was chosen as the vector to assemble fragments A, B, C, and D of the CMVp-driven ZIKV Natal RGN replicon with the EGFP reporter via the cloning sites of EcoRI, NotI, ClaI, RsrII, and XhoI (
Figure 1B). Fragments A, C, and D were generated using the Platinum
® PCR Supermix High Fidelity reaction with the DNA segments we synthesized, I and II, as the templates (Life Technology, Carlsbad, CA, USA). Fragment B was produced through two-round PCR with the Gibson Assembly reaction of two first-round PCR products (B1 and B2) as the template (New England Biolabs, Ipswich, MA, USA). The pairs containing the restriction enzyme site(s) indicated, are listed in
Supplementary Table S1. The resultant plasmid carrying the CMVp-driven ZIKV Natal RGN replicon with the EGFP reporter was propagated in
E. coli DH5α and then sequenced by Sanger sequencing assays with sequencing primers (
Supplementary Table S2). The nucleotide and deduced amino acid sequence alignment analysis of the CMVp-driven ZIKV Natal RGN replicon and its parent strain (GenBank accession number KU527068) was performed using the Lasergene DNASTAR Megalign software.
2.3. Functional Analysis of the ZIKV Natal RGN Replicon Using RT- PCR and Immunofluorescent Staining
To detect self-amplifying RNA genomes of the CMVp-driven ZIKV Natal RGN replicon, the synthesis of positive and negative-sense RNA subgenomes in vitro was examined using SYBR Green-based real-time PCR. Total RNAs of TE-671 cells transfected with the ZIKV Natal RGN replicon were extracted using the PureLink Mini Total RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA), reverse transcribed into cDNA with specific-capture primers, and measured using real-time PCR with ZIKV
NS5-specific primer pairs (
Supplementary Table S3), according to our prior report [
19]. Relative levels of self-amplifying ZIKV sense and antisense genomes were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The absolute copy number of the ZIKV genome was determined according to the standard curve of real-time PCR for serial dilutions of the plasmid containing the ZIKV
NS5 gene at a known concentration. To explore the expression of replicon-based EGFP and ZIKV reporter proteins, replicon-transfected cells were initially photographed using immunofluorescence microscopy; then, an immunofluorescence assay (IFA) was performed with primary antibodies against ZIKV NS1 and NS5 (GeneTex, Inc., Taiwan) and secondary AF546 goat anti-rabbit IgG (Thermo Fisher Scientific). The immunofluorescent staining assay was carried out as described in our prior report [
15]. The fluorescence intensity of replicon-based EGFP and ZIKV proteins in ZIKV Natal RGN replicon-transfected cells was counted by Image J.
2.4. Establishment of a Packaging Cell Line Expressing ZIKV prM and E Proteins
To establish the packaging cells expressing ZIKV structural proteins, the
prM/M-E gene was amplified using PCR with specific primers (
Supplementary Table S1) and synthesized DNA segment I as the template. The PCR product of the ZIKV
prM-E gene was digested with EcoRI and XhoI, and then cloned into the EcoRI and XhoI sites of the expression plasmid pcDNA3.1-HisC. The resultant plasmid pcDNA-ZIKV prME was transfected with TE-671 cells at 90% confluence in a 6-well plate with Lipofectamine LTX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s guidelines. The transfected cells were selected in the culture media with 500 µg/mL G418 for 2 weeks; the expression of ZIKV prM and E proteins in a stably transfected cell line (packaging cell line) was validated by real-time RT-PCR with ZIKV
E-specific primers (
Supplementary Table S3) and immunofluorescence staining with primary antibodies against ZIKV E protein (GeneTex, Inc.), as described above.
2.5. Production of ZIKV Natal RGN SRIPs
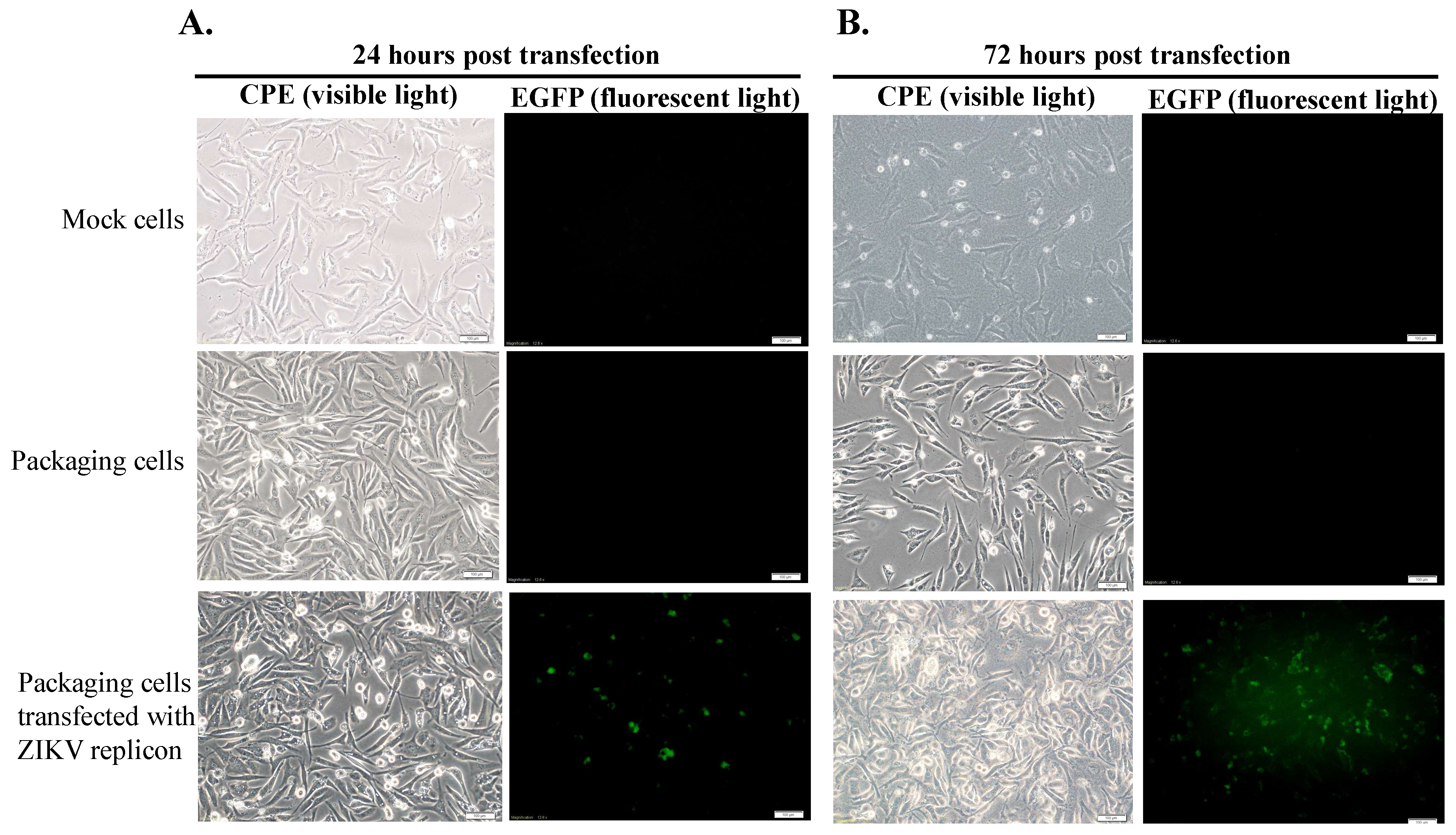
The packaging cells grown at 90% confluence in 6-well plates were transfected with or without the ZIKV Natal RGN replicon using Lipofectamine LTX. The cytopathic effect (CPE), replicon-based EGFP and ZIKV proteins, and viral sense and antisense genomes in mock and transfected cells, were measured 24 and 72 h post-transfection using immunofluorescent staining assays, as described above. Moreover, real-time RT-PCR assays with
NS5-specific primer pairs, listed in
Supplementary Table S3, were performed to examine the positive and negative-sense viral genomes in replicon-transfected packaging cells, as well as the viral genome in SRIPs. ZIKV Natal RGN SRIPs in the cultured media of transfected packaging cells were concentrated using the PEG Virus Precipitation Kit (Abcam, Cambridge, MA, USA). The cultured media were centrifuged at 3200×
g for 15 min at 4 °C; then, the supernatant (10 mL) was collected from each and incubated with 2.5 mL PEG solution overnight at 4 °C. The pellets of ZIKV Natal RGN SRIPs were harvested after centrifugation at 3200×
g for 30 min at 4 °C, re-suspended in 100 μL Virus Re-suspension Solution, and then stored at −80 °C.
2.6. Antigenicity, Titer, and Infectivity of ZIKV Natal RGN SRIPs
To analyze the antigenicity of ZIKV Natal RGN SRIPs, 10 μL each of one-fold and 10-fold SRIP stock dilution was spotted onto a nitrocellulose membrane for the dot-blot assay with anti-ZIKV E antibodies. The membrane was subsequently blocked with 5% skim milk in TBST (Tris-Buffered Saline, 0.1% Tween-20) buffer at 4 °C for 2 h, incubated overnight with anti-ZKIV E antibodies (GeneTex, Inc., Taiwan), and then reacted with HRP-conjugated anti-mouse IgG antibodies (Invitrogen, Carlsbad, CA, USA) after washing with TBST. Immunoreactive signals for the E proteins of ZIKV Natal RGN SRIPs were amplified with ECLTM Western Blotting Detection Reagents (GE Healthcare, Chicago, IL, USA), and then imaged by the Multi-function Gel Image System (MultiGel-21) (Gentaur, San Jose, CA, USA). The infectious titer of the ZIKV Natal RGN SRIP stock was determined by a median tissue culture infectious dose (TCID50) assay. Serial dilutions of the ZIKV Natal RGN SRIP stock were added and incubated on the 90% confluent monolayer of packaging cells in 96-well plates. After incubating for 72 h at 37 °C, the CPE in each well was observed and recorded to determine the TCID50 titer of the ZIKV Natal RGN SRIP stock. In the infectivity assay, packaging cells were cultured in 6-well plates overnight and infected with different doses of ZIKV Natal RGN SRIPs at multiplicity of infections (MOIs) of 3, 0.3, 0.03, and 0.003. CPE, replicon-driven EGFP, and ZIKV NS1 expression in the SRIP-infected cells were examined 72 h post-infection, and fluorescence microscopy and immunofluorescent staining were performed as described above.
2.7. Cell Line Susceptibility and Attachment to ZIKV Natal RGN SRIPs
In cell line susceptibility assays, TE-671, SF-268, and ARPE-19 cells were infected with ZIKV Natal RGN SRIPs at a MOI of 0.15 TCID50/cell. After incubating for 72 h, the cells were photographed to examine CPE and explore replicon-driven EGFP reporter expression using immunofluorescence microscopy. Moreover, the cells were fixed with paraformaldehyde at 4 °C for 1 h, and then immunofluorescence staining was performed with anti-ZIKV NS1 antibodies, as mentioned above. In the attachment assay, ZIKV Natal RGN SRIPs at a dose of 105 TCID50/well (MOI = 0.5 TCID50/cell) were added onto the monolayers of ARPE-19, TE671, and SF-268 cells and incubated at 4 °C for 1 h. After washing with cold PBS, ZIKV Natal RGN SRIPs attached on the cell surface were harvested, and the relative levels of extracted viral genomes were quantitated using two-step, SYBR, Green I, real-time RT-PCR with ZIKV NS5-specific primer pairs, as described above.
2.8. Statistical Analysis
All data from 3 independent experiments were analyzed using Student’s t-tests or χ2 tests. Statistical significance for each assay was judged at p < 0.05.
4. Discussion
The ZIKV Natal RGN strain found in brain tissues from fetal autopsy cases with microcephaly in the Natal region of Brazil has not been isolated [
18]. This study, firstly, generated SRIPs of the ZIKV Natal RGN strain using synthetic and reverse genetic technologies (
Figure 1,
Figure 2,
Figure 3 and
Figure 4 and
Supplementary Figures S1–S3). The viral yield of ZIKV Natal RGN SRIPs in prM-E-expressing packaging cells transfected with the ZIKV replicon was 6.25 × 10
6 TCID50/mL (
Figure 4), which indicated the replication capacity of the CMV promoter-launched ZIKV Natal RGN replicon. In addition, a green fluorescence reporter, EGFP, encoded within the ZIKV Natal RGN replicon, represented an observable marker to examine replication of the ZIKV Natal RGN replicon and SRIPs (
Figure 2,
Figure 3, and
Figure 5). In our laboratory, the pBR322 plasmid has been used to construct a JEV replicon that is genetically stable in
E. coli [
15]. The JEV genome sequence encoding for structural proteins (C, prM/M, and E) was toxic in
E. coli, which resulted in genetic instability of the JEV infectious clones and replicons [
15]. Similarly, construction of ZIKV Natal RGN infectious clones was not accomplished due to the genetic instability of ZIKV Natal RGN infectious clones, particularly within the NS1 gene region (
Supplementary Figures S1 and S2). However, the recombinant pcDNA3.1 plasmid containing prM and E genes and the pBR322 plasmid containing 5’ and 3’-UTRs, C, and all NS genes, showed genetic stability. Two ZIKV infectious clones obtained by cloning the entire full-length native cDNA have been reported [
20,
21], and other ZIKV infectious clones were reported using strategies to avoid the toxicity of the ZIKV genome sequence, including in vitro assembly of viral cDNA fragments and insertion of eukaryotic introns [
22,
23,
24,
25,
26,
27]. Particularly, the ZIKV Natal RGN infectious clone was produced using bacterial artificial chromosomes (BACs) [
27], in which a single-copy pBeloBAC11 plasmid containing the full-length genome of the ZIKV Natal RGN strain under the CMV promoter control was used to maintain ZIKV genome stability and reduce toxicity in bacteria. In addition, our prior report demonstrated that JEV SRIPs carrying the EGFP reporter were a more sensitive, effective, and efficient platform for quantitating antiviral efficacy using flow cytometry compared to the traditional plaque assay [
15]. Like JEV SRIPs, the ZIKV Natal RGN SRIPs carrying the EGFP reporter could be applicable as a drug screening platform. Therefore, the reverse genetics system for the production of ZIKV Natal RGN SRIPs provided an alternative, applicable, and rapid approach to studying the biological features of the unique ZIKV strains.
The human rhabdomyosarcoma/muscle TE671 cells were used as packaging cells expressing ZIKV prM and E proteins (
Figure 2), in which transfection with the ZIKV Natal RGN replicon significantly generated recombinant SRIPs with the high yield of 6.25 × 10
6 TCID50/mL (
Figure 4). Interestingly, TE671 cells expressing JEV C, prM, and E proteins were also used as a packaging cell line that was efficient at producing JEV SRIPs post-transfection with the JEV replicon [
15]. The recombinant ZIKV Natal RGN SRIPs showed infectivity and self-replication in the packaging cells (
Figure 5). Two amino acid substitutions within NS5 (L3277R and E3280N) (
Supplementary Figure S3) seemed to have no influence on the activities of NS5 methyltransferase and RNA-dependent RNA polymerase. A recent study indicated that human embryonic kidney 293T cells had been used to establish different flavivirus prM-E-expressing packaging cells, including DENV1–4, ZIKV, JEV, West Nile virus, yellow fever virus, and tick-borne encephalitis virus, and the cells were transfected with the DENV1 reporter replicon to generate a chimeric flavivirus SRIP-based neutralization assay [
28]. Therefore, recombinant ZIKV Natal RGN SRIPs generated in this study could be used to analyze the ZIKV life cycle, elucidate viral pathogenesis, screen antiviral agents, design faster and safer diagnostic tools, and even develop attenuated vaccines against ZIKV infection.
The full-length ZIKV Natal RGN strain genome was directly sequenced using next-generation sequencing with the total RNA extracted from brain tissues from fetal autopsy cases with microcephaly (GenBank accession number KU527068) [
18]. In this study, recombinant ZIKV Natal RGN SRIPs were rescued using reverse genetic and synthetic biology techniques (
Figure 1,
Figure 2,
Figure 3,
Figure 4 and
Figure 5 and
Supplementary Figures S1–S3). Three cell lines, TE671, SF268, and ARPE-19, were further tested for cell susceptibility to ZIKV Natal RGN
SRIPs (
Figure 6). The order of cell susceptibility to ZIKV Natal RGN SRIPs was TE671 > ARPE-19 > SF268 cells. The CPE level, EGFP reporter intensity, and ZIKV NS1 protein expression were most obvious (>50% involvement) in TE671 cells, but were mild in ARPE-19 cells and limited in SF268 cells 72 h post-infection with SRIPs at an MOI of 0.15 TCID50/cell. The results indicated that ZIKV Natal RGN
SRIPs had a broad range of human tissue tropism with differential cell susceptibilities (
Figure 6A). In addition, ZIKV Natal RGN SRIPs had the highest attachment activity to TE671 cells among those three cell lines (
Figure 6B). SRIP attachment to TE671 cells was suggested to be involved in the infectivity of ZIKV Natal RGN in TE671 cells. This finding implied that ZIKV Natal RGN SRIPs had tropism for rhabdomyosarcoma/muscle and retinal pigmented epithelial cells, which was consistent with myalgia, arthralgia, retinitis, chorioretinal atrophy, and pigmentary maculopathy in symptomatic ZIKV infection [
29,
30]. Therefore, the cell line susceptibility results revealed the unique features of ZIKV Natal RGN SRIPs and suggested the possible pathogenesis of ZIKV infection.
5. Conclusions
In conclusion, the approach for generating recombinant ZIKV SRIPs with an EGFP reporter showed the genetic stability of ZIKV prM-E genes in the mammalian expression vector and ZIKV replicon sequence in the low-copy plasmid pBR322, providing a faster and safer platform to study the biological aspects of ZIKV infection. ZIKV Natal RGN SRIPs were rescued by the combination of prM-E-expressing packaging cells and the ZIKV Natal RGN replicon under CMV promoter control, which mimicked self-replicating, positive-sense, single strand RNA viruses. The biological features of ZIKV Natal RGN SRIPs were revealed, including CPE, infectivity, cell susceptibility, and attachment activity. Therefore, recombinant ZIKV Natal RGN SRIPs could be further used to investigate cell tropism, persistent infection, and vaccine candidates, and to establish reliable, effective, and efficient assays for screening antiviral agents and diagnosing viral infections.