Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria and Bacteriophage Growth Conditions
2.2. Morphological Analysis by TEM
2.3. DNA Purification and Whole-Genome Sequencing of Bacteriophage CPS1
2.4. Phylogenetic Analysis
2.5. Molecular Genetic Method
2.6. ATCC 13124 CPF_0486 Expression and Purification
2.7. ATCC 13124 CPF_0486 Protein Characterization
2.8. In Vitro Bacteriophage Adsorption Assays
2.9. Capsule Staining
2.10. Quantification of Capsular Polysaccharides
2.11. Inhibition of Phage Infection by CPS or Monosaccharide
2.12. Statistical Analysis
3. Results
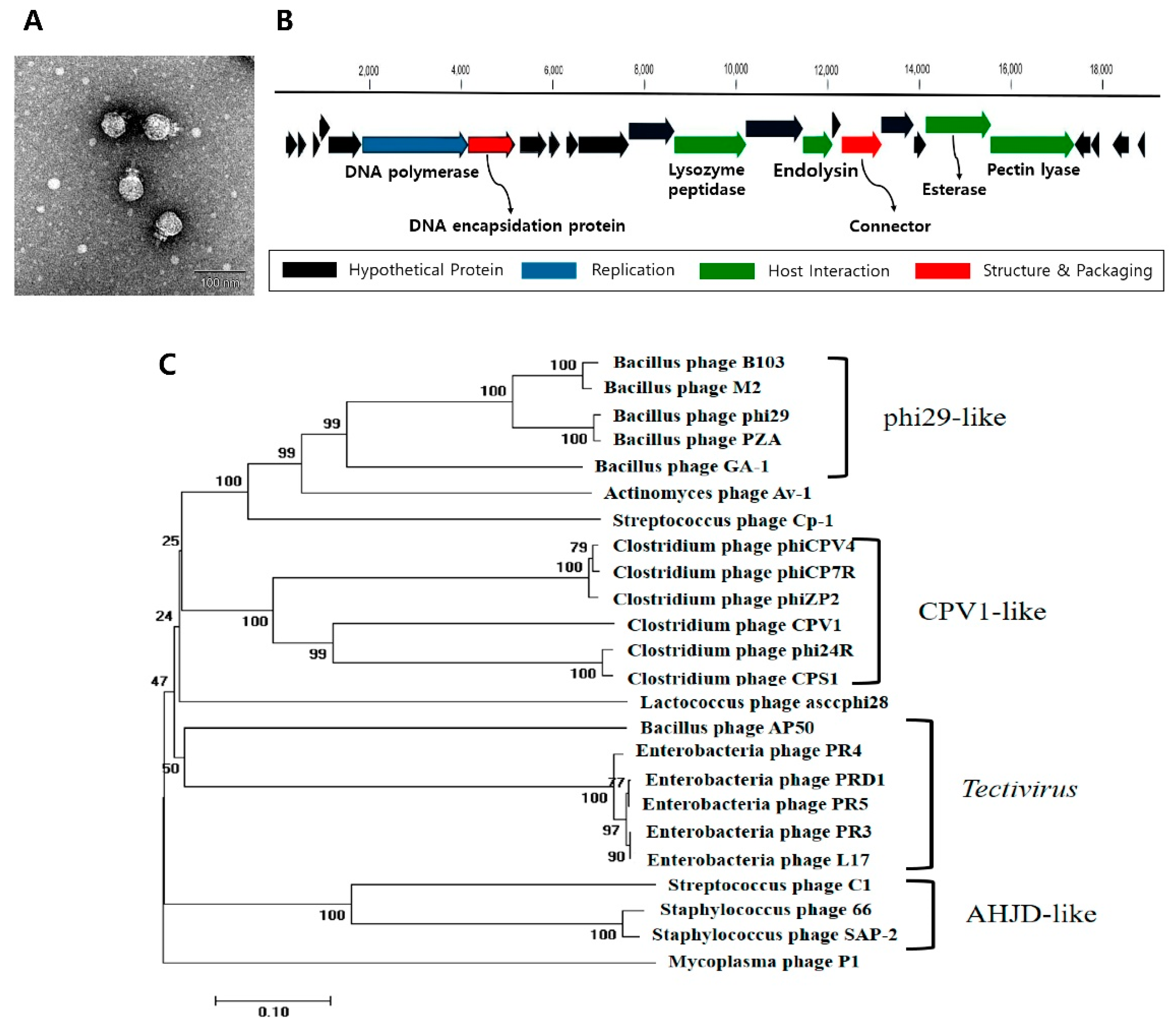
3.1. C. perfringens Bacteriophage CPS1 Is a Member of the Picovirinae
3.2. CPS1 Binds to C. perfringens Cell Surface Polysaccharides
3.3. Screening of a Random Mutant Library of C. perfringens for CPS1 Resistance
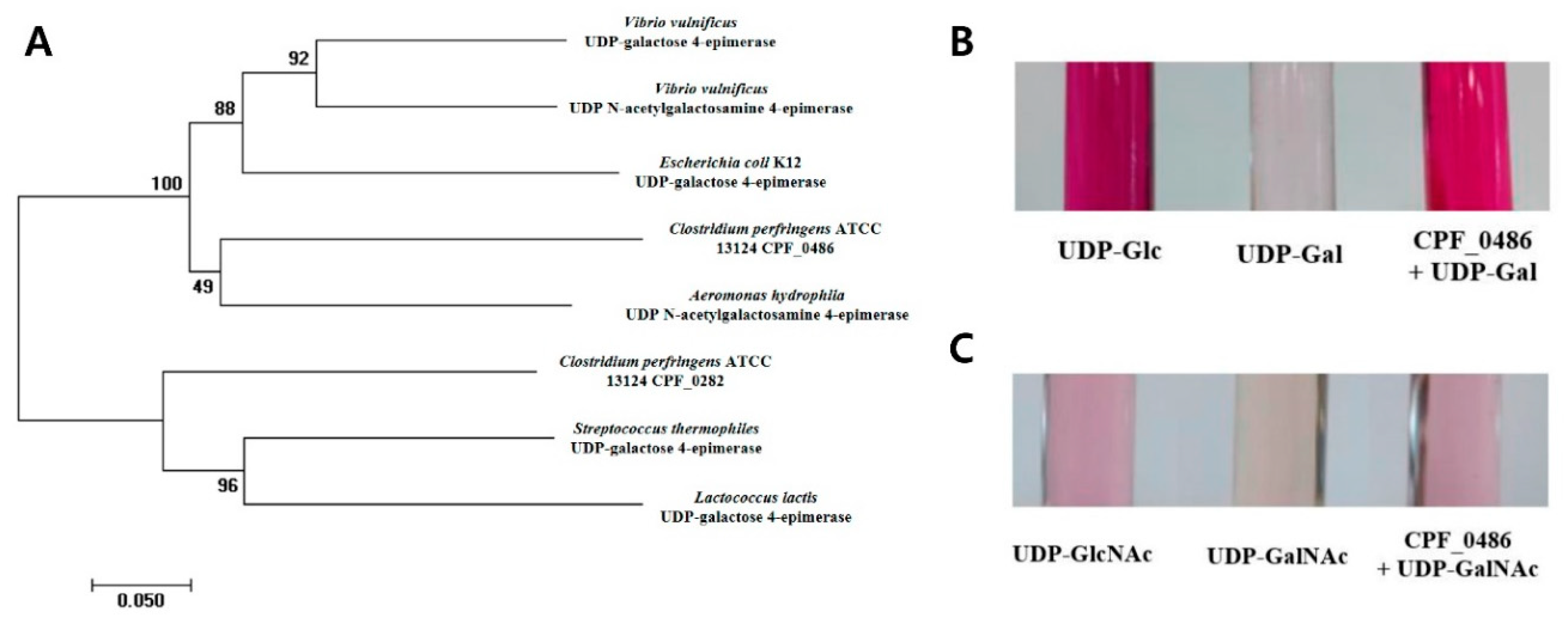
3.4. CPF_0486 Has UDP-glucose 4-epimerase (GalE) and UDP-N-acetylglucosamine 4-epimerase (Gne) Activities
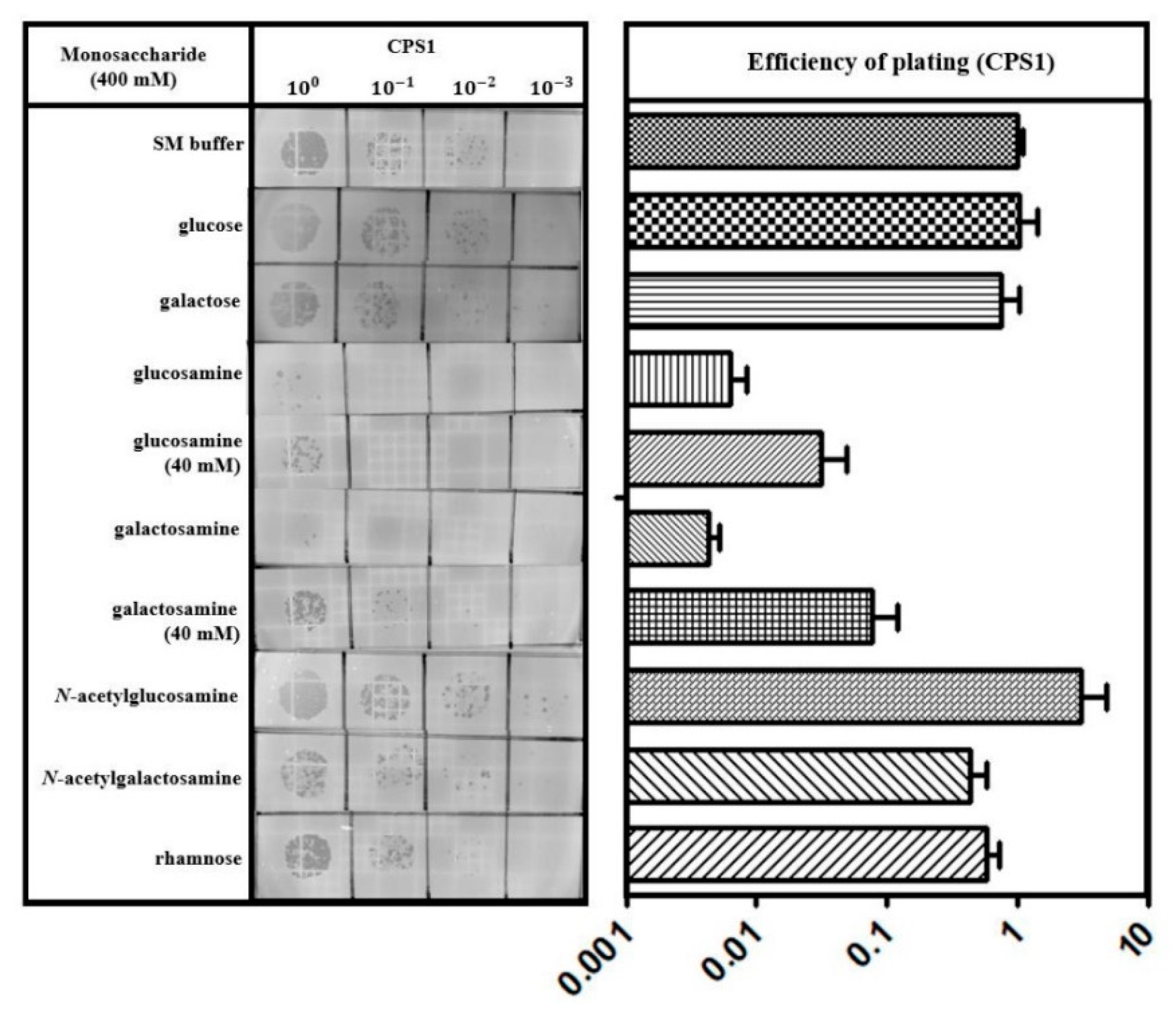
3.5. Capsular Polysaccharides are a Receptor for C. perfringens Phage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rood, J.I.; Cole, S.T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 1991, 55, 621–648. [Google Scholar]
- Sawires, Y.S.; Songer, J.G. Clostridium perfringens: Insight into virulence evolution and population structure. Anaerobe 2006, 12, 23–43. [Google Scholar] [CrossRef] [PubMed]
- Timbermont, L.; Haesebrouck, F.; Ducatelle, R.; Van Immerseel, F. Necrotic enteritis in broilers: An updated review on the pathogenesis. Avian Pathol. 2011, 40, 341–347. [Google Scholar] [CrossRef]
- Olsen, S.J.; MacKinon, L.C.; Goulding, J.S.; Bean, N.H.; Slutsker, L. Surveillance for foodborne-disease outbreaks—United States, 1993–1997. MMWR CDC Surveill. Summ. 2000, 49, 1–62. [Google Scholar] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Ryu, S. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica Serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Kraft, B.L.; Pan, Y.Y.; Wall, S.K.; Saez, A.C.; Ebner, P.D. Development of an anti-Salmonella phage cocktail with increased host range. Foodborne Pathog. Dis. 2010, 7, 1415–1419. [Google Scholar] [CrossRef]
- Shin, H.; Lee, J.H.; Kim, H.; Choi, Y.; Heu, S.; Ryu, S. Receptor diversity and host interaction of bacteriophages infecting Salmonella enterica Serovar Typhimurium. PLoS ONE 2012, 7, e43392. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Baptista, C.; Santos, M.A.; Sao-Jose, C. Phage SPP1 reversible adsorption to Bacillus subtilis cell wall teichoic acids accelerates virus recognition of membrane receptor YueB. J. Bacteriol. 2009, 191, 1726. [Google Scholar] [CrossRef]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage cocktails and the future of phage therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Kim, Y.T.; Ryu, S.; Lee, J.H. Biocontrol and rapid detection of food-borne pathogens using bacteriophages and endolysins. Front. Microbiol. 2016, 7, 474. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, S.; Park, B.; Ryu, S. Core lipopolysaccharide-specific phage SSU5 as an auxiliary component of a phage cocktail for Salmonella biocontrol. Appl. Environ. Microbiol. 2014, 80, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Hyman, P. Phages for phage therapy: Isolation, characterization, and host range breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Chapot-Chartier, M.P. Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages. Front. Microbiol. 2014, 5, 236. [Google Scholar] [CrossRef] [Green Version]
- Silva, J.B.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Szwajcer Dey, E.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar]
- Li, X.H.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kuhner, P.; Peschel, A.; Xia, G.Q.; Winstel, V. An accessory wall teichoic acid glycosyltransferase protects Staphylococcus aureus from the lytic activity of Podoviridae. Sci. Rep. 2015, 5, 17219. [Google Scholar] [CrossRef]
- Seal, B.S. Characterization of bacteriophages virulent for Clostridium perfringens and identification of phage lytic enzymes as alternatives to antibiotics for potential control of the bacterium. Poult. Sci. 2013, 92, 526–533. [Google Scholar] [CrossRef]
- Seal, B.S.; Fouts, D.E.; Simmons, M.; Garrish, J.K.; Kuntz, R.L.; Woolsey, R.; Schegg, K.M.; Kropinski, A.M.; Ackermann, H.W.; Siragusa, G.R. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: Genomic organization and proteomic analysis of the virions. Arch. Virol. 2011, 156, 25–35. [Google Scholar] [CrossRef]
- Morales, C.A.; Oakley, B.B.; Garrish, J.K.; Siragusa, G.R.; Ard, M.B.; Seal, B.S. Complete genome sequence of the podoviral bacteriophage ΦCP24R, which is virulent for Clostridium perfringens. Arch. Virol. 2012, 157, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Kirby, K.S. A new method for the isolation of ribonucleic acids from mammalian tissues. Biochem. J. 1956, 64, 405–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeugin, J.A.; Hartley, J.L. Ethanol precipitation of DNA. Focus 1985, 7, 1–2. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Myers, G.S.; Rasko, D.A.; Cheung, J.K.; Ravel, J.; Seshadri, R.; DeBoy, R.T.; Ren, Q.; Varga, J.; Awad, M.M.; Brinkac, L.M. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006, 16, 1031–1040. [Google Scholar] [CrossRef]
- Jirásková, A.; Vítek, L.; Fevery, J.; Ruml, T.; Branny, P. Rapid protocol for electroporation of Clostridium perfringens. J. Microbiol. Methods 2005, 62, 125–127. [Google Scholar] [CrossRef]
- Konishi, M.; Kawamoto, K.; Izumikawa, M.; Kuriyama, H.; Yamashita, T. Gene transfer into guinea pig cochlea using adeno-associated virus vectors. J. Gene Med. 2008, 10, 610–618. [Google Scholar] [CrossRef]
- Schulz, J.M.; Watson, A.L.; Sanders, R.; Ross, K.L.; Thoden, J.B.; Holden, H.M.; Fridovich-Keil, J.L. Determinants of function and substrate specificity in human UDP-galactose 4′-epimerase. J. Biol. Chem. 2004, 279, 32796–32803. [Google Scholar] [CrossRef] [PubMed]
- Fry, B.N.; Feng, S.; Chen, Y.Y.; Newell, D.G.; Coloe, P.J.; Korolik, V. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 2000, 68, 2594–2601. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huskic, S.; Cisterne, A.; Rothemund, D.; Reeves, P.R. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J. Bacteriol. 2002, 184, 2620–2625. [Google Scholar] [CrossRef] [PubMed]
- Reissig, J.L.; Strominger, J.L.; Leloir, L.F. A modified colorimetric method for the estimation of N-acetylamino sugars. J. Biol. Chem. 1955, 217, 959–966. [Google Scholar]
- Sorensen, M.C.H.; van Alphen, L.B.; Harboe, A.; Li, J.J.; Christensen, B.B.; Szymanski, C.M.; Brondsted, L. Bacteriophage F336 recognizes the capsular phosphoramidate modification of Campylobacter jejuni NCTC11168. J. Bacteriol. 2011, 193, 6742–6749. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Stevens, A.; Jun, D.; Mercer, R.; Lang, A.S.; Beatty, J.T. Quorum-sensing regulation of a capsular polysaccharide receptor for the Rhodobacter capsulatus gene transfer agent (RcGTA). Mol. Microbiol. 2013, 87, 802–817. [Google Scholar] [CrossRef]
- Yang, Y.B.; Chen, J.Q.; Zhao, Y.L.; Bai, J.W.; Ding, W.Y.; Zhou, Y.H.; Chen, X.Y.; Liu, D.; Li, Y.H. Sub-MICs of azithromycin decrease biofilm formation of Streptococcus suis and increase capsular polysaccharide content of S. suis. Front. Microbiol. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Heller, K.; Braun, V. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 1979, 139, 32–38. [Google Scholar]
- Volozhantsev, N.V.; Oakley, B.B.; Morales, C.A.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Popova, A.V.; Zhilenkov, E.L.; Garrish, J.K.; Schegg, K.M. Molecular characterization of podoviral bacteriophages virulent for Clostridium perfringens and their comparison with members of the Picovirinae. PLoS ONE 2012, 7, e38283. [Google Scholar] [CrossRef]
- Kong, M.; Ryu, S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl. Environ. Microbiol. 2015, 81, 2274–2283. [Google Scholar] [CrossRef]
- Vidal, J.E.; Chen, J.; Li, J.; McClane, B.A. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS ONE 2009, 4, e6232. [Google Scholar] [CrossRef] [PubMed]
- Mäkelä, P.H.; Stocker, B.A.D. Genetics of polysaccharide biosynthesis. Annu. Rev. Genet. 1969, 3, 291–322. [Google Scholar] [CrossRef]
- Valiente, E.; Jiménez, N.; Merino, S.; Tomas, J.M.; Amaro, C. Vibrio vulnificus biotype 2 serovar E gne but not galE is essential for lipopolysaccharide biosynthesis and virulence. Infect. Immun. 2008, 76, 1628–1638. [Google Scholar] [CrossRef] [PubMed]
- Soldo, B.; Scotti, C.; Karamata, D.; Lazarevic, V. The Bacillus subtilis Gne (GneA, GalE) protein can catalyse UDP-glucose as well as UDP-N-acetylglucosamine 4-epimerisation. Gene 2003, 319, 65–69. [Google Scholar] [CrossRef]
- Serotype, O.; Vilches, S.; Regue, M.; Merino, S.; Toma, J.M. Role of Gne and GalE in the Virulence of Aeromonas hydrophila. J. Bacteriol. 2007, 189, 540–550. [Google Scholar] [CrossRef]
- Canals, R.; Jimenez, N.; Vilches, S.; Regue, M.; Merino, S.; Tomas, J.M. The UDP N-acetylgalactosamine 4-epimerase gene is essential for mesophilic Aeromonas hydrophila serotype O34 virulence. Infect. Immun. 2006, 74, 537–548. [Google Scholar] [CrossRef]
- Elliott, S.D.; Tai, J.Y. The type-specific polysaccharides of Streptococcus suis. J. Exp. Med. 1978, 148, 1699–1704. [Google Scholar] [CrossRef]
- Anthony, E.E. A note on capsule staining. Science 1931, 73, 319–320. [Google Scholar] [CrossRef]
- Vinogradov, E.; Aubry, A.; Logan, S.M. Structural characterization of wall and lipidated polysaccharides from Clostridium perfringens ATCC 13124. Carbohydr. Res. 2017, 448, 88–94. [Google Scholar] [CrossRef]
- Keogh, B.P.; Pettingill, G. Adsorption of bacteriophage eb7 on Streptococcus cremoris EB7. Appl. Environ. Microbiol. 1983, 45, 1946–1948. [Google Scholar]
- Wendlinger, G.; Loessner, M.J.; Scherer, S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 1996, 142, 985–992. [Google Scholar] [CrossRef]
- Kiu, R.; Caim, S.; Alexander, S.; Pachori, P.; Hall, L.J. Probing genomic aspects of the multi-host pathogen Clostridium perfringens reveals significant pangenome diversity, and a diverse array of virulence factors. Front. Microbiol. 2017, 8, 2485. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, E.; Chun, J.; Kim, M.; Ryu, S. Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1. Viruses 2019, 11, 1002. https://doi.org/10.3390/v11111002
Ha E, Chun J, Kim M, Ryu S. Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1. Viruses. 2019; 11(11):1002. https://doi.org/10.3390/v11111002
Chicago/Turabian StyleHa, Eunsu, Jihwan Chun, Minsik Kim, and Sangryeol Ryu. 2019. "Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1" Viruses 11, no. 11: 1002. https://doi.org/10.3390/v11111002
APA StyleHa, E., Chun, J., Kim, M., & Ryu, S. (2019). Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1. Viruses, 11(11), 1002. https://doi.org/10.3390/v11111002





