Reconstructing the Dissemination Dynamics of the Major HIV-1 Subtype B Non-Pandemic Lineage Circulating in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. HIV-1 BCAR pol Sequence Dataset
2.2. Evolutionary, Phylogeographic, and Demographic Analyses
3. Results
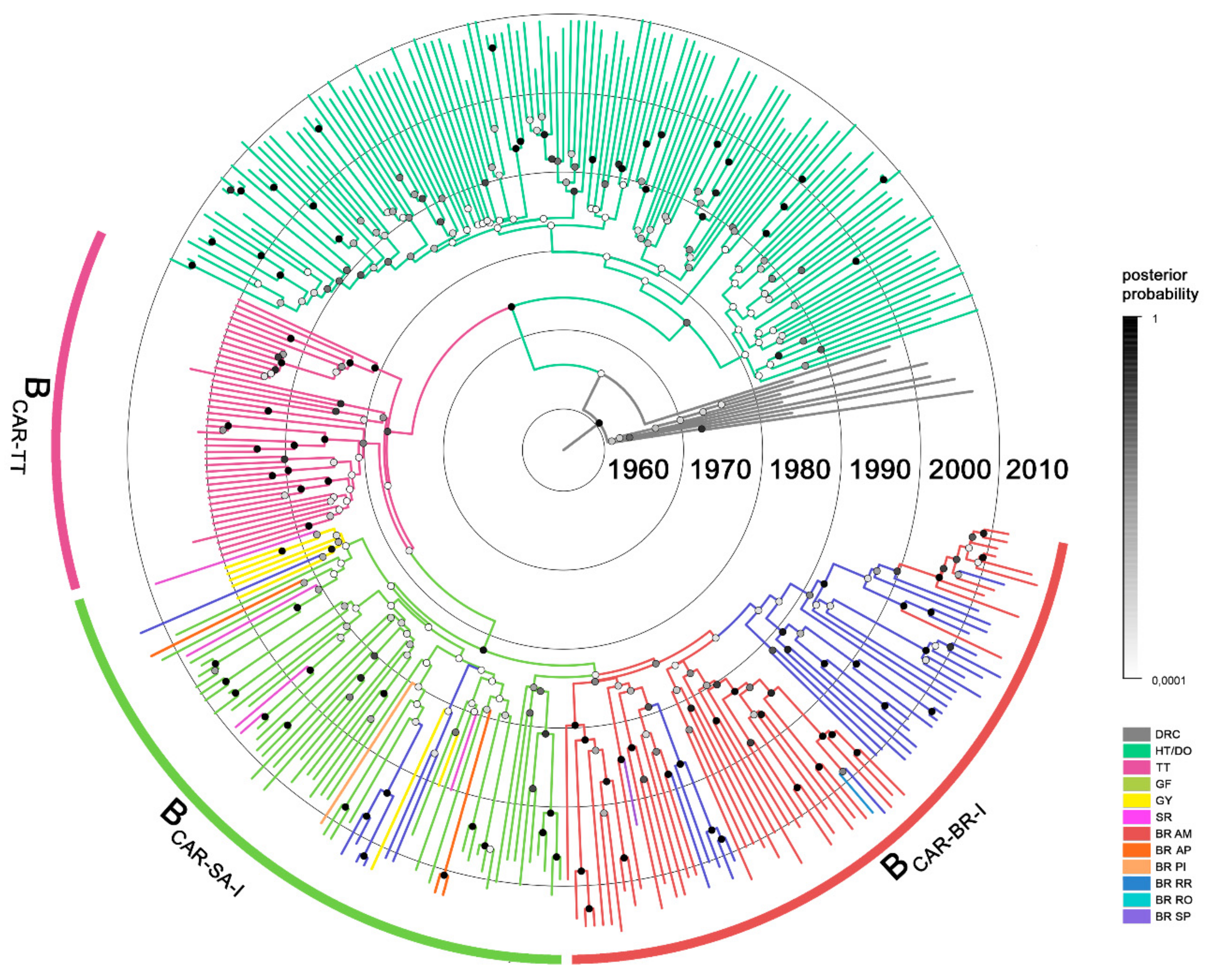
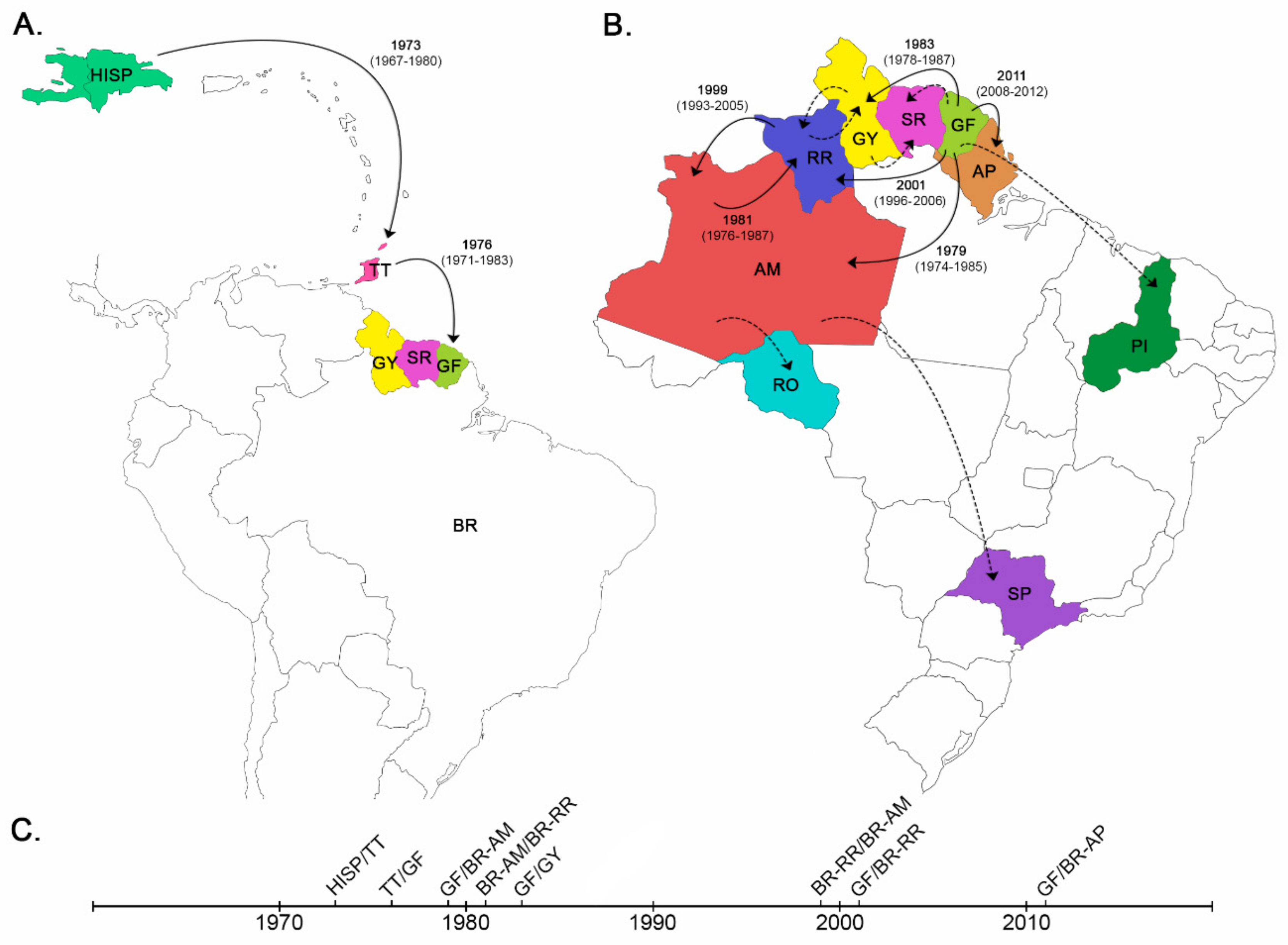
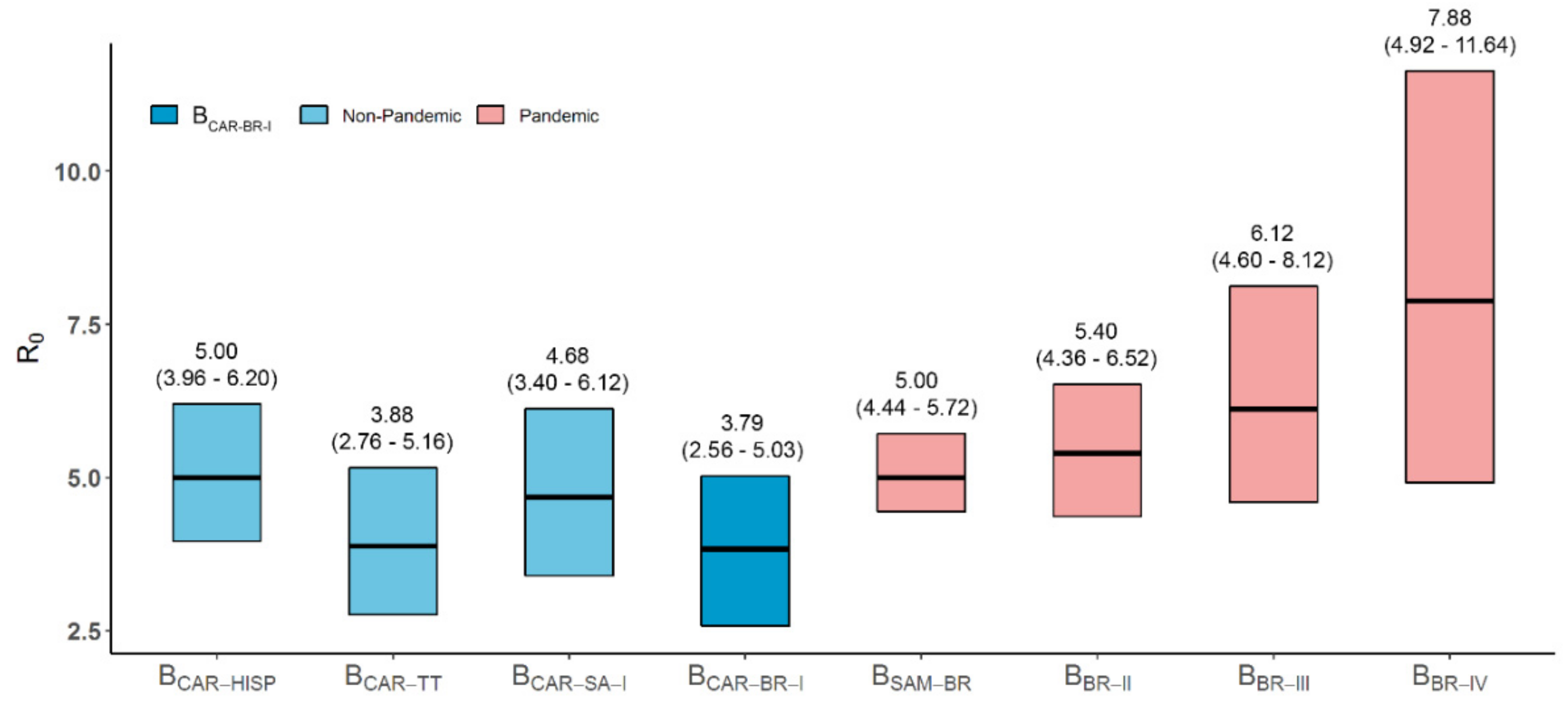
3.1. Dispersal Pattern of the HIV-1 BCAR Strains from the Caribbean to Brazil
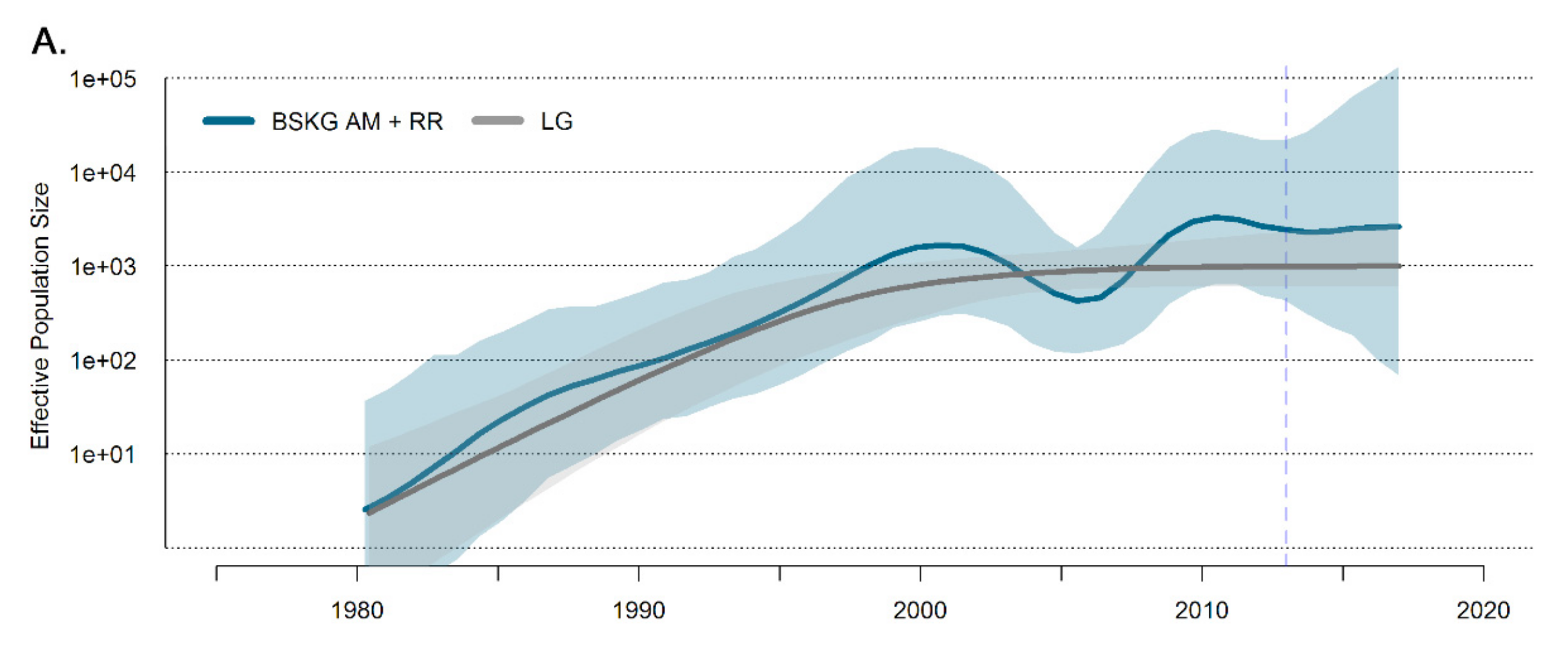
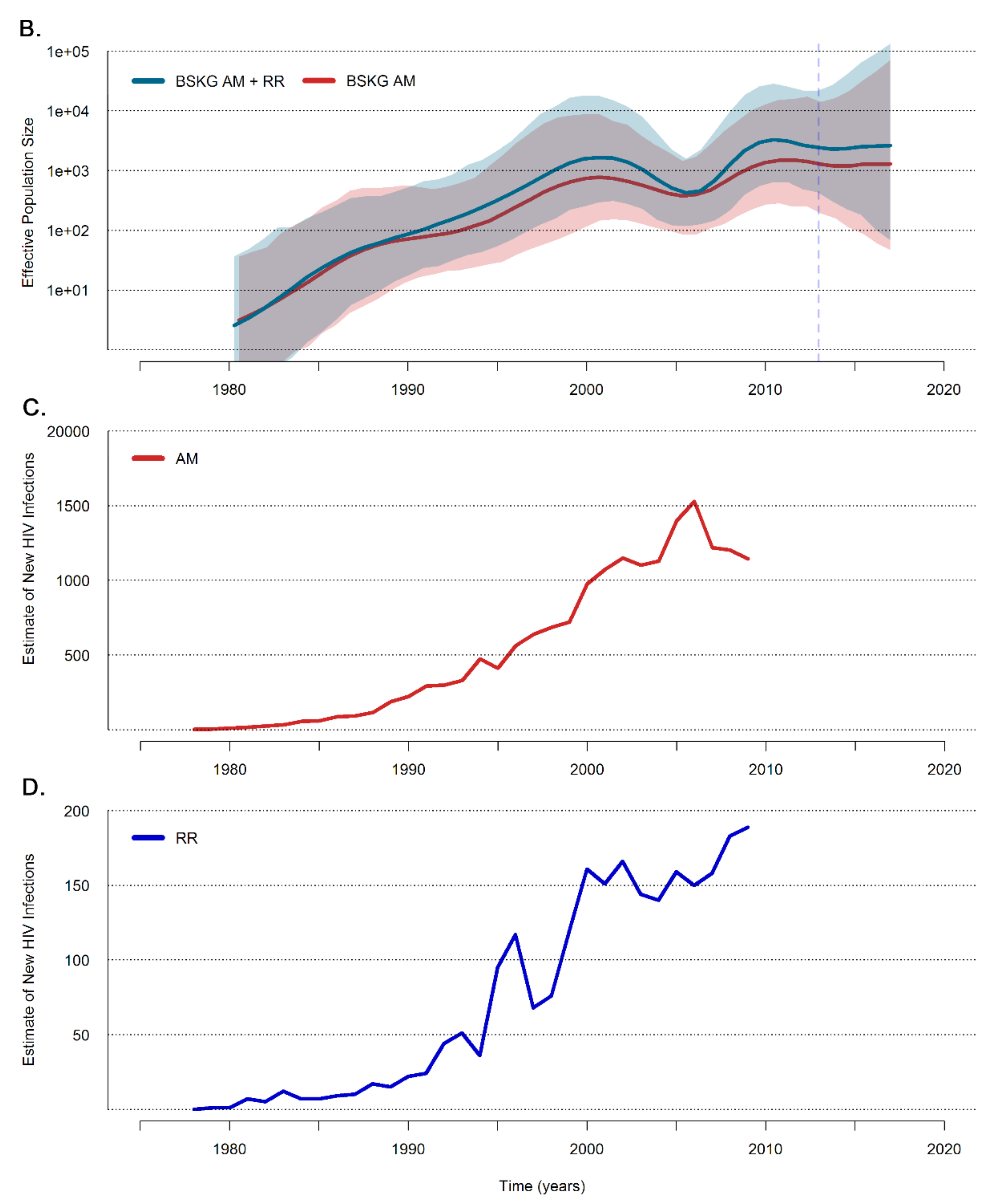
3.2. Dissemination of the HIV-1 BCAR-BR-I Lineage in Northern Brazil
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Brazilian, Ministry of Health. AIDS Epidemiological Bulletin. Available online: http://www.aids.gov.br/sites/default/files/anexos/publicacao/2015/58534/boletim_aids_11_2015_web_pdf_19105.pdf (accessed on 28 August 2019). (in Portuguese)
- Gilbert, M.T.; Rambaut, A.; Wlasiuk, G.; Spira, T.J.; Pitchenik, A.E.; Worobey, M. The emergence of HIV/AIDS in the Americas and beyond. Proc. Natl. Acad. Sci. USA 2007, 104, 18566–18570. [Google Scholar] [CrossRef] [PubMed]
- Cabello, M.; Mendoza, Y.; Bello, G. Spatiotemporal dynamics of dissemination of non-pandemic HIV-1 subtype B clades in the Caribbean region. PLoS ONE 2014, 9, e106045. [Google Scholar] [CrossRef] [PubMed]
- Junqueira, D.M.; de Medeiros, R.M.; Matte, M.C.; Araujo, L.A.; Chies, J.A.; Ashton-Prolla, P.; Almeida, S.E. Reviewing the history of HIV-1: Spread of subtype B in the Americas. PLoS ONE 2011, 6, e27489. [Google Scholar] [CrossRef][Green Version]
- Bello, G.; Nacher, M.; Divino, F.; Darcissac, E.; Mir, D.; Lacoste, V. The HIV-1 Subtype B Epidemic in French Guiana and Suriname Is Driven by Ongoing Transmissions of Pandemic and Non-pandemic Lineages. Front. Microbiol. 2018, 9, 1738. [Google Scholar] [CrossRef] [PubMed]
- Divino, F.; de Lima Guerra Corado, A.; Gomes Naveca, F.; Stefani, M.M.; Bello, G. High Prevalence and Onward Transmission of Non-Pandemic HIV-1 Subtype B Clades in Northern and Northeastern Brazilian Regions. PLoS ONE 2016, 11, e0162112. [Google Scholar] [CrossRef] [PubMed]
- Esashika Crispim, M.A.; da Guarda Reis, M.N.; Fraiji, N.; Bello, G.; Stefani, M.M.A. Detection of human immunodeficiency virus Type 1 phylogenetic clusters with multidrug resistance mutations among 2011 to 2017 blood donors from the highly endemic Northern Brazilian Amazon. Transfusion 2019, 59, 2593–2601. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Nicholls, G.K.; Rodrigo, A.G.; Solomon, W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 2002, 161, 1307–1320. [Google Scholar] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Suchard, M.A.; Rambaut, A. Many-core algorithms for statistical phylogenetics. Bioinformatics 2009, 25, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Ho, S.Y.; Phillips, M.J.; Rambaut, A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Lam, T.T.; Carvalho, L.M.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef] [PubMed]
- Hue, S.; Pillay, D.; Clewley, J.P.; Pybus, O.G. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proc. Natl. Acad. Sci. USA 2005, 102, 4425–4429. [Google Scholar] [CrossRef] [PubMed]
- Zehender, G.; Ebranati, E.; Lai, A.; Santoro, M.M.; Alteri, C.; Giuliani, M.; Palamara, G.; Perno, C.F.; Galli, M.; Lo Presti, A.; et al. Population dynamics of HIV-1 subtype B in a cohort of men-having-sex-with-men in Rome, Italy. J. Acquir. Immune Defic. Syndr. 2010, 55, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Wong, K.H.; Chan, K.C.; To, S.W.; Chen, Z.; Yam, W.C. Phylodynamics of HIV-1 subtype B among the men-having-sex-with-men (MSM) population in Hong Kong. PLoS ONE 2011, 6, e25286. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, Y.; Martinez, A.A.; Castillo Mewa, J.; Gonzalez, C.; Garcia-Morales, C.; Avila-Rios, S.; Reyes-Teran, G.; Armien, B.; Pascale, J.M.; Bello, G. Human Immunodeficiency Virus Type 1 (HIV-1) Subtype B Epidemic in Panama Is Mainly Driven by Dissemination of Country-Specific Clades. PLoS ONE 2014, 9, e95360. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009, 5, e1000520. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.A.R.; Suchard, M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008, 26, 355–368. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian population dynamics inference: A coalescent-based model for multiple loci. Mol. Biol. Evol. 2013, 30, 713–724. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Suchard, M.A. Genealogical Working Distributions for Bayesian Model Testing with Phylogenetic Uncertainty. Syst. Biol. 2016, 65, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- FigTree v1.4.4. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 28 August 2019).
- d-maps.com. Available online: https://d-maps.com (accessed on 28 August 2019).
- Brazilian, Ministry of Health. Available online: http://www.aids.gov.br/pt-br/taxonomy/term/595 (accessed on 28 August 2019).
- Mir, D.; Cabello, M.; Romero, H.; Bello, G. Phylodynamics of major HIV-1 subtype B pandemic clades circulating in Latin America. AIDS 2015, 29, 1863–1869. [Google Scholar] [CrossRef] [PubMed]
- Corbin, H.P. Brazilian migration to Guyana as a livelihood strategy: A case study approach. Available online: http://www.repositorio.ufpa.br/jspui/bitstream/2011/1966/1/Dissertacao_BrazilianMigrationGuyana.pdf (accessed on 28 August 2019).
- Leonardi, V. Fronteiras Amazonicas do Brasil: Saúde e História Social Brasilia: Paralelo 15; Marco Zero: Sao Paulo, Brazil, 2000. (in Portuguese) [Google Scholar]
- Diniz, A.M.A.; dos Santos, R.O. O vertiginoso crescimento populacional de Roraima e seus impactos socioambientais (in Portuguese). Cad. Geogr. 2005, 15, 23–44. [Google Scholar]
- Vale, A.L.F. Imigração de nordestinos para Roraima. Estud. Avançados 2006, 20, 255–261. (in Portuguese). [Google Scholar] [CrossRef]
- Kadri, M.R.; Schweickardt, J.C. The emergence of Aids in Amazonas. Hist. Cienc. Saude—Manguinhos 2016, 23, 301–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Novitsky, V.; Moyo, S.; Lei, Q.; DeGruttola, V.; Essex, M. Impact of sampling density on the extent of HIV clustering. AIDS Res. Hum. Retrovir. 2014, 30, 1226–1235. [Google Scholar] [CrossRef]
| Clade | Location | Location Code | N | Sampling Time |
| BCAR-BR-I | Brazil/Amazonas | AM | 45 | 2009–2017 |
| Brazil/Roraima | RR | 29 | 2010–2017 | |
| Brazil/Rondônia | RO | 1 | 2015 | |
| Brazil/São Paulo | SP | 1 | 2003 | |
| BCAR-SA-I | Brazil/Amapá | AP | 3 | 2013 |
| Brazil/Roraima | RR | 7 | 2011–2013 | |
| Brazil/Piauí | PI | 1 | 2011 | |
| French Guiana | GF | 46 | 2006–2012 | |
| Guyana | GY | 7 | 2000–2013 | |
| Suriname | SR | 5 | 2000–2009 | |
| BCAR-TT | Trinidad and Tobago | TT | 41 | 2000–2003 |
| BCAR-HISP | Hispaniola | HISP | 130 | 2003–2011 |
| Clade | TMRCA Current Study | TMRCA Ref. [5] | TMRCA Ref. [6] | TMRCA Ref. [2] |
|---|---|---|---|---|
| Subtype B | 1970 (1963–1985) | - | 1969 (1964–1974) | 1966 (1962–1970) |
| BCAR-TT | 1973 (1967–1980) | - | 1973 (1970–1976) | 1973 (1970–1976) |
| BCAR-SA-I | 1976 (1971–1983) | 1977 (1973–1981) | - | - |
| BCAR-BR-I | 1979 (1974–1985) | - | 1978 (1975–1981) | - |
| Location | Complete Dataset (AM = 45 Sequences) | Subset 1 (AM = 22 Sequences) | Subset 2 (AM = 23 Sequences) |
|---|---|---|---|
| AM | 92% | 97% | 93% |
| RR | 1% | 3% | 6% |
| GF | 0 | 0 | 1% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arantes, I.; Esashika Crispim, M.A.; Nogueira da Guarda Reis, M.; Martins Araújo Stefani, M.; Bello, G. Reconstructing the Dissemination Dynamics of the Major HIV-1 Subtype B Non-Pandemic Lineage Circulating in Brazil. Viruses 2019, 11, 909. https://doi.org/10.3390/v11100909
Arantes I, Esashika Crispim MA, Nogueira da Guarda Reis M, Martins Araújo Stefani M, Bello G. Reconstructing the Dissemination Dynamics of the Major HIV-1 Subtype B Non-Pandemic Lineage Circulating in Brazil. Viruses. 2019; 11(10):909. https://doi.org/10.3390/v11100909
Chicago/Turabian StyleArantes, Ighor, Myuki Alfaia Esashika Crispim, Mônica Nogueira da Guarda Reis, Mariane Martins Araújo Stefani, and Gonzalo Bello. 2019. "Reconstructing the Dissemination Dynamics of the Major HIV-1 Subtype B Non-Pandemic Lineage Circulating in Brazil" Viruses 11, no. 10: 909. https://doi.org/10.3390/v11100909
APA StyleArantes, I., Esashika Crispim, M. A., Nogueira da Guarda Reis, M., Martins Araújo Stefani, M., & Bello, G. (2019). Reconstructing the Dissemination Dynamics of the Major HIV-1 Subtype B Non-Pandemic Lineage Circulating in Brazil. Viruses, 11(10), 909. https://doi.org/10.3390/v11100909





