Molecular Identification of a Novel Hantavirus in Malaysian Bronze Tube-Nosed Bats (Murina aenea)
Abstract
1. Introduction
2. Materials and Methods
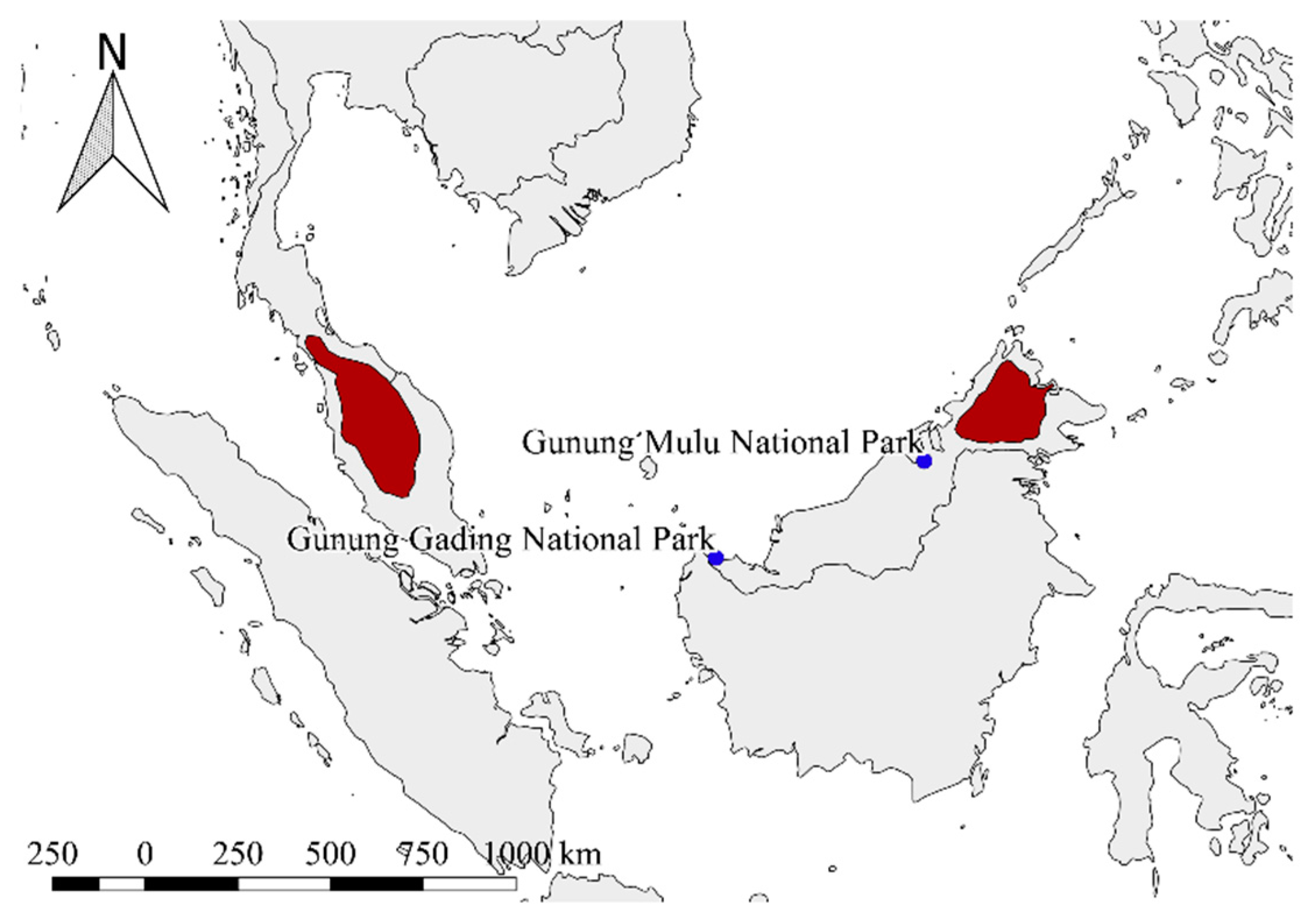
2.1. Sample Collection and Nucleic Acid Extraction
2.2. PCR Screening
2.3. Metagenomic cDNA Preparation and Nanopore Sequencing
2.4. In vitro Virus Propagation
2.5. Phylogenetic Analyses
3. Results and Discussion
3.1. Virus Detection
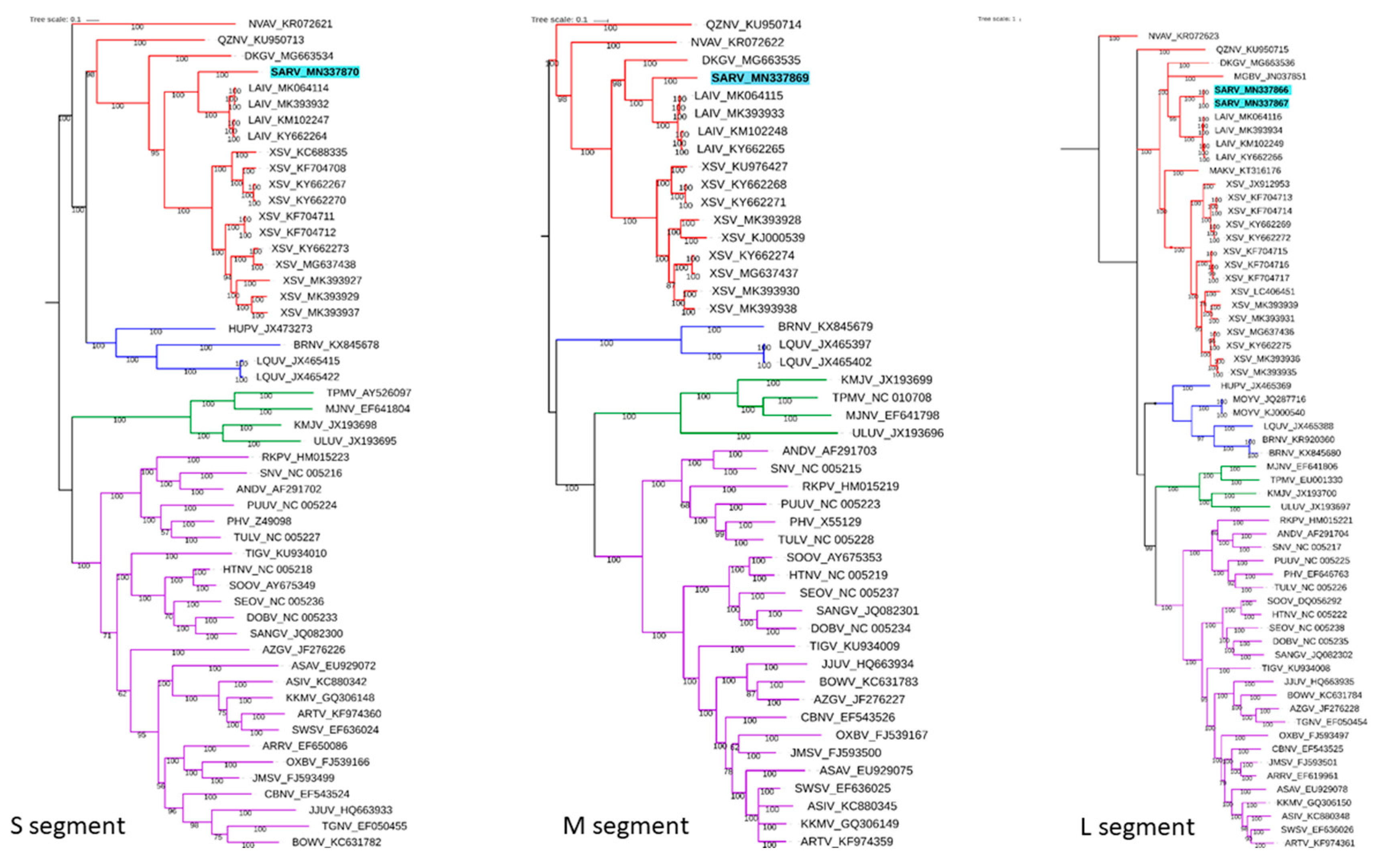
3.2. Sequence and Phylogenetic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Krüger, D.H.; Schönrich, G.; Klempa, B. Human pathogenic hantaviruses and prevention of infection. Hum. Vaccin. 2011, 7, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.C.; Zhang, Y.Z. The evolution and emergence of hantaviruses. Curr. Opin. Virol. 2015, 10, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Witkowski, P.T.; Auste, B.; Nowak, K.; Weber, N.; Fahr, J.; Mombouli, J.V.; Wolfe, N.D.; Drexler, J.F.; Drosten, C.; et al. Hantavirus in bat, Sierra Leone. Emerg. Infect. Dis. 2012, 18, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Sumibcay, L.; Kadjo, B.; Gu, S.H.; Kang, H.J.; Lim, B.K.; Cook, J.A.; Song, J.W.; Yanagihara, R. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Côte d’Ivoire. Virol. J. 2012, 26, 34. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Kikuchi, F.; Bawm, S.; Sơn, N.T.; Lin, K.S.; Tú, V.T.; Aoki, K.; Tsuchiya, K.; Tanaka-Taya, K.; Morikawa, S.; et al. Molecular Phylogeny of Mobatviruses (Hantaviridae) in Myanmar and Vietnam. Viruses 2019, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Aoki, K.; Sơn, N.T.; Tú, V.T.; Kikuchi, F.; Kinoshita, G.; Fukui, D.; Thành, H.T.; Gu, S.H.; Yoshikawa, Y.; et al. Đakrông virus, a novel mobatvirus (Hantaviridae) harbored by the Stoliczka’s Asian trident bat (Aselliscus stoliczkanus) in Vietnam. Sci. Rep. 2019, 15, 10239. [Google Scholar] [CrossRef] [PubMed]
- Arai, S.; Taniguchi, S.; Aoki, K.; Yoshikawa, Y.; Kyuwa, S.; Tanaka-Taya, K.; Masangkay, J.S.; Omatsu, T.; Puentespina, R., Jr.; Watanabe, S.; et al. Molecular phylogeny of a genetically divergent hantavirus harbored by the Geoffroy’s rousette (Rousettus amplexicaudatus), a frugivorous bat species in the Philippines. Infect. Genet. Evol. 2016, 45, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, P.T.; Drexler, J.F.; Kallies, R.; Ličková, M.; Bokorová, S.; Maganga, G.D.; Szemes, T.; Leroy, E.M.; Krüger, D.H.; Drosten, C.; et al. Phylogenetic analysis of a newfound bat-borne hantavirus supports a laurasiatherian host association for ancestral mammalian hantaviruses. Infect. Genet. Evol. 2016, 41, 113–119. [Google Scholar] [CrossRef]
- Francis, C.M. A guide to mammals of Southeast Asia; Princeton University Press: Princeton, NJ, USA, 2008; p. 392. [Google Scholar]
- Hill, J.E. Notes on some tube-nosed bats, genus Murina, from Southeastern Asia, with descriptions of a new species and a new subspecies. Fed. Mus. J. 1964, 8, 48–59. [Google Scholar]
- Sikes, R.S. The Animal Care and Use Committee of the American Society of Mammalogists, 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef]
- Klempa, B.; Fichet-Calvet, E.; Lecompte, E.; Auste, B.; Aniskin, V.; Meisel, H.; Denys, C.; Koivogui, L.; ter Meulen, J.; Krüger, D.H. Hantavirus in African wood mouse, Guinea. Emerg. Infect. Dis. 2006, 12, 838–840. [Google Scholar] [CrossRef] [PubMed]
- Conceição-Neto, N.; Zeller, M.; Lefrère, H.; De Bruyn, P.; Beller, L.; Deboutte, W.; Yinda, C.K.; Lavigne, R.; Maes, P.; van Ranst, M.; et al. Modular approach to customise sample preparation procedures for viral metagenomics: A reproducible protocol for virome analysis. Sci. Rep. 2015, 12, 16532. [Google Scholar] [CrossRef] [PubMed]
- Song, D.H.; Kim, W.K.; Gu, S.H.; Lee, D.; Kim, J.A.; No, J.S.; Lee, S.H.; Wiley, M.R.; Palacios, G.; Song, J.W.; et al. Sequence-Independent, Single-Primer Amplification Next-Generation Sequencing of Hantaan Virus Cell Culture-Based Isolates. Am. J. Trop. Med. Hyg. 2017, 96, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R. Porechop. 2018. Available online: https://github.com/rrwick/Porechop (accessed on 20 January 2019).
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Bennett, S.N.; Sumibcay, L.; Arai, S.; Hope, A.G.; Mocz, G.; Song, J.-W.; Cook, J.A.; Yanagihara, R. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea). PLoS ONE 2009, 4, e6149. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, J.; He, B.; Qin, S.; Xia, L.; Qin, M.; Li, N.; Tu, C. Novel hantavirus identified in black-bearded tomb bats, China. Infect. Genet. Evol. 2015, 31, 158–160. [Google Scholar] [CrossRef] [PubMed]
- Eckerle, I.; Lenk, M.; Ulrich, R.G. More novel hantaviruses and diversifying reservoir hosts—Time for development of reservoir-derived cell culture models? Viruses 2014, 26, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Hamidon, B.B.; Saadiah, S. Seoul hantavirus infection mimicking dengue fever. Med. J. Malays. 2003, 58, 786–787. [Google Scholar]
- Lam, S.K.; Chua, K.B.; Myshrall, T.; Devi, S.; Zainal, D.; Afifi, S.A.; Nerome, K.; Chu, Y.K.; Lee, H.W. Serological evidence of hantavirus infections in Malaysia. Southeast Asian J. Trop. Med. Public Health 2001, 32, 809–813. [Google Scholar]
- Hamdan, N.E.; Ng, Y.L.; Lee, W.B.; Tan, C.S.; Khan, F.A.; Chong, Y.L. Rodent Species Distribution and Hantavirus Seroprevalence in Residential and Forested areas of Sarawak, Malaysia. Trop Life Sci. Res. 2017, 28, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Anderson, D.E.; Zhang, Q.; Tan, C.W.; Lim, B.L.; Luko, K.; Wen, M.; Chia, W.N.; Mani, S.; Wang, L.C.; et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019, 5, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Brook, C.E.; Dobson, A.P. Bats as “special” reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef]
- Hutson, A.M.; Kingston, T.; Francis, C.; Csorba, G.; Bumrungsri, S. Murina aenea. The IUCN Red List of Threatened Species 2008: E.T13936A4366971. 2008. Available online: http://dx.doi.org/10.2305/IUCN.UK.2008.RLTS.T13936A4366971.en (accessed on 20 July 2019).
- Csorba, G.; Görföl, T. (Department of Zoology, Hungarian Natural History Museum, Budapest, Hungary). Personal Communication, 2019.
- Rosli, Q.S.; Khan, F.A.A.; Morni, M.A.; William-Dee, J.; Tingga, R.C.T.; Mohd-Ridwan, A.R. Roosting Behaviour and Site Mapping Of Cave Dwelling Bats In Wind Cave Nature Reserve, Bau, Sarawak, Malaysian Borneo. Malays Appl. Biol. 2018, 47, 231–238. [Google Scholar]
- Morni, M.A.; Khan, F.A.A.; Rosli, Q.S.; William-Dee, J.; Tingga, R.C.T.; Mohd-Ridwan, A.R. Bats Roost Site Preferences in Wind Cave Nature Reserve, Bau, Sarawak. Malays Appl. Biol. 2018, 47, 57–64. [Google Scholar]
| Genomic Segment | Primer Name | Primer Sequence (5′–3′) | Reference |
|---|---|---|---|
| S, M, L | OSM55F | TAGTAGTAGACTCC | [18] |
| S | HVSF1 | TAGTAGTAGACTCCTTRAARAGC | [19] |
| HVSR1906 | TAGTAGTAKRCWCCYTRARAAG | ||
| M | HVMF1 | TAGTAGTAGACWCCGCAAAAG | |
| HVMR3684 | TAGTAGTATRCTCCGCARG | ||
| L | HVLF1 | TAGTAGTAGACTCCRGA | |
| HVLR6561 | TAGTAGTAGTAKRCTCCGRGA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zana, B.; Kemenesi, G.; Buzás, D.; Csorba, G.; Görföl, T.; Khan, F.A.A.; Tahir, N.F.D.A.; Zeghbib, S.; Madai, M.; Papp, H.; et al. Molecular Identification of a Novel Hantavirus in Malaysian Bronze Tube-Nosed Bats (Murina aenea). Viruses 2019, 11, 887. https://doi.org/10.3390/v11100887
Zana B, Kemenesi G, Buzás D, Csorba G, Görföl T, Khan FAA, Tahir NFDA, Zeghbib S, Madai M, Papp H, et al. Molecular Identification of a Novel Hantavirus in Malaysian Bronze Tube-Nosed Bats (Murina aenea). Viruses. 2019; 11(10):887. https://doi.org/10.3390/v11100887
Chicago/Turabian StyleZana, Brigitta, Gábor Kemenesi, Dóra Buzás, Gábor Csorba, Tamás Görföl, Faisal Ali Anwarali Khan, Nurul Farah Diyana Ahmad Tahir, Safia Zeghbib, Mónika Madai, Henrietta Papp, and et al. 2019. "Molecular Identification of a Novel Hantavirus in Malaysian Bronze Tube-Nosed Bats (Murina aenea)" Viruses 11, no. 10: 887. https://doi.org/10.3390/v11100887
APA StyleZana, B., Kemenesi, G., Buzás, D., Csorba, G., Görföl, T., Khan, F. A. A., Tahir, N. F. D. A., Zeghbib, S., Madai, M., Papp, H., Földes, F., Urbán, P., Herczeg, R., Tóth, G. E., & Jakab, F. (2019). Molecular Identification of a Novel Hantavirus in Malaysian Bronze Tube-Nosed Bats (Murina aenea). Viruses, 11(10), 887. https://doi.org/10.3390/v11100887







