Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteriophage Isolation and Propagation
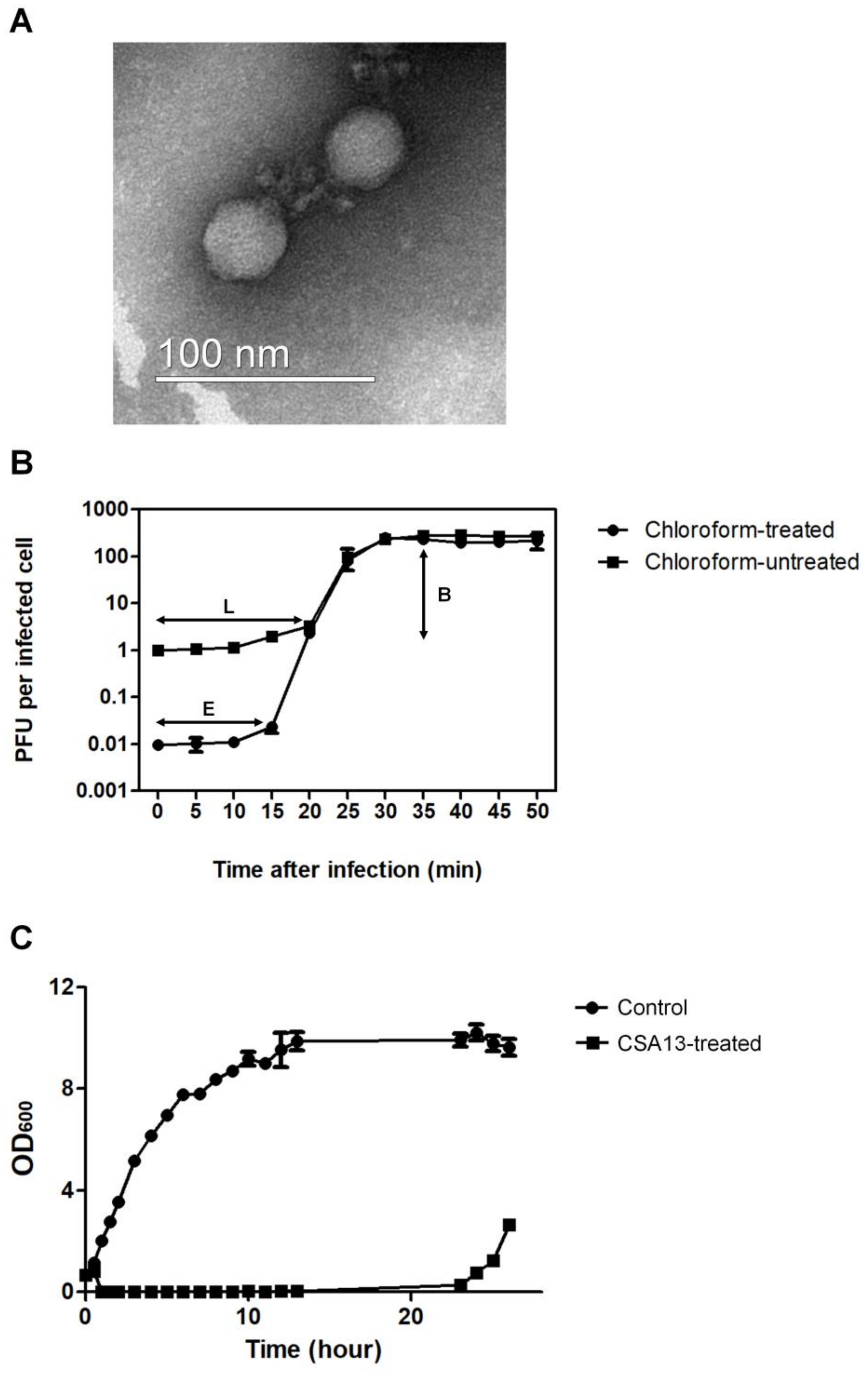
2.2. Transmission Electron Microscopy (TEM) Analysis
2.3. Determination of the Bacteriophage Antimicrobial Spectrum
2.4. Bacterial Inhibition Assay
2.5. One-Step Growth Curve Assay
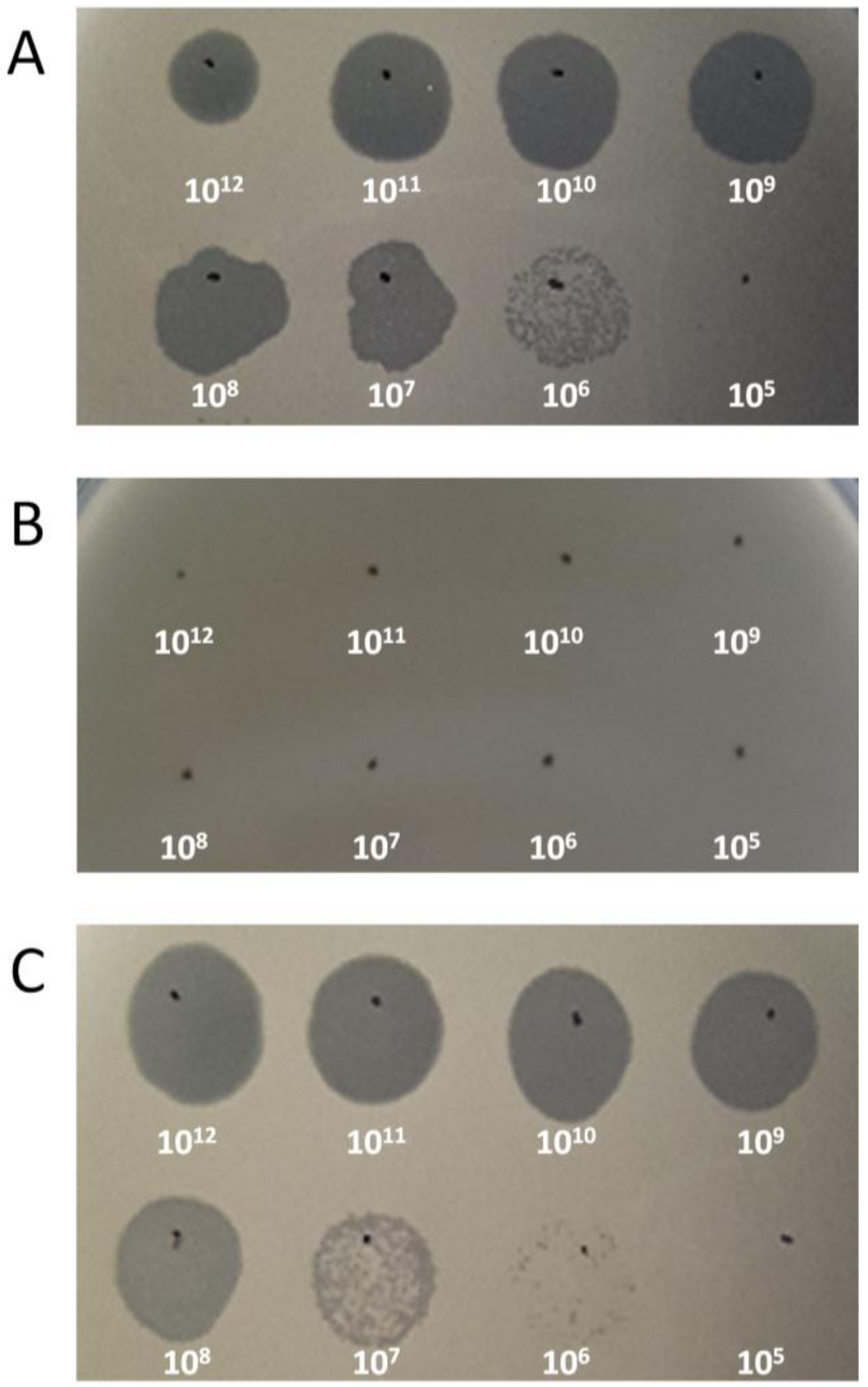
2.6. Receptor Analysis
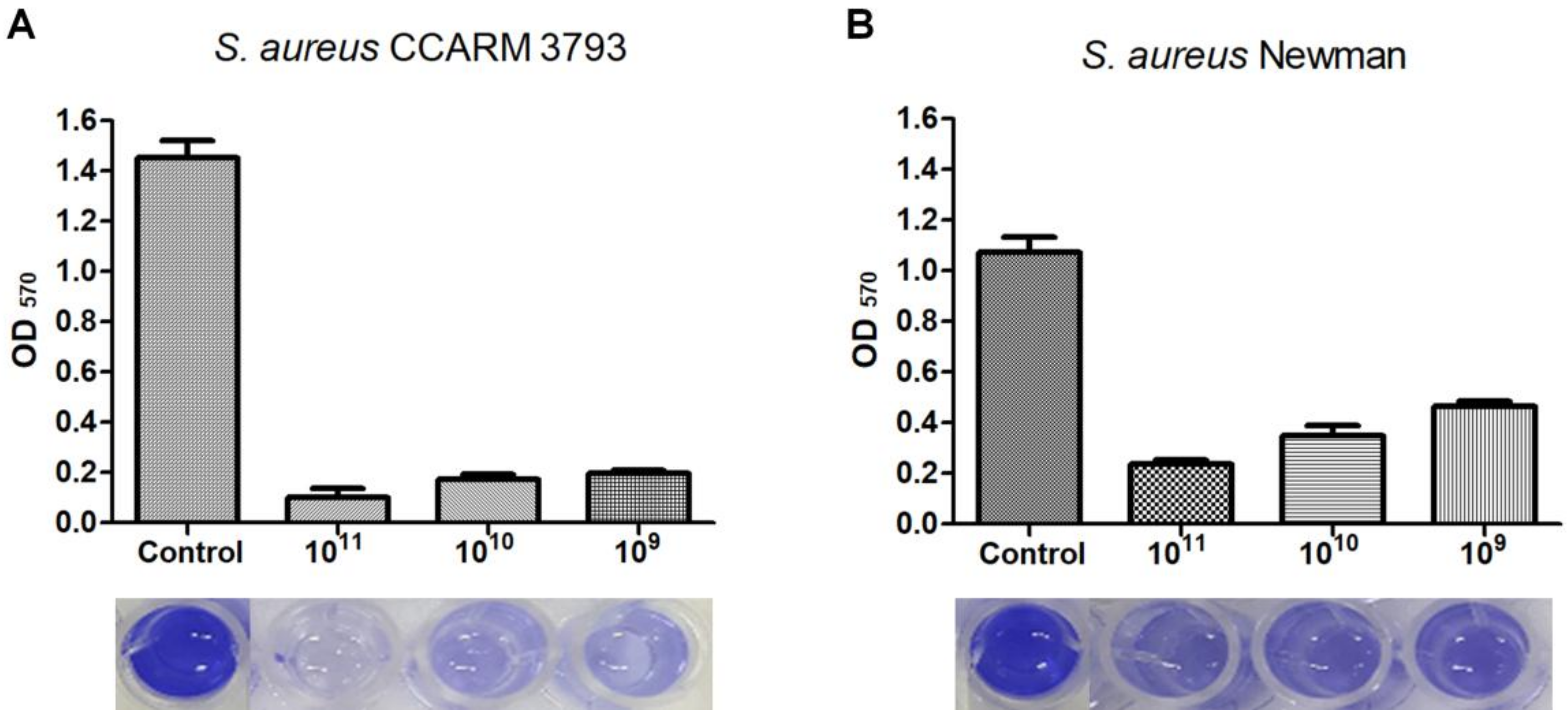
2.7. Biofilm Reduction Assay with Phage CSA13
2.8. Bacteriophage Genomic DNA Purification
2.9. Full-Genome Sequencing of Phage CSA13 and Bioinformatics Analysis
3. Results and Discussion
3.1. Isolation and Physiological Characteristics of S. aureus Phage CSA13
3.2. Receptor Analysis of Phage CSA13
3.3. Host Range of Phage CSA13
3.4. Biofilm Reduction Efficacy of Phage CSA13
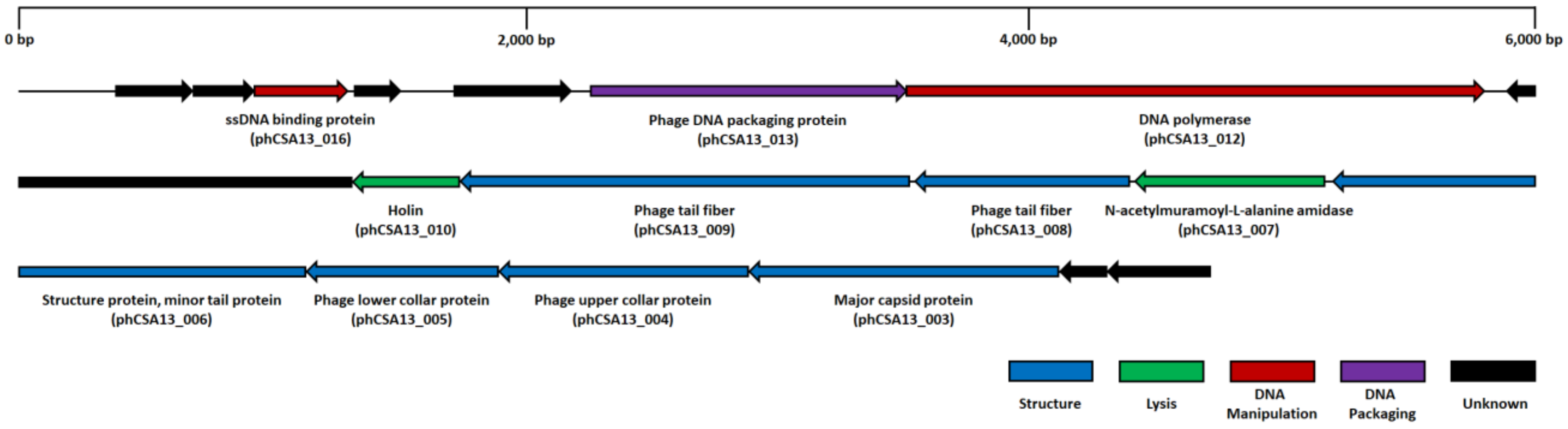
3.5. Genome Analysis of Phage CSA13
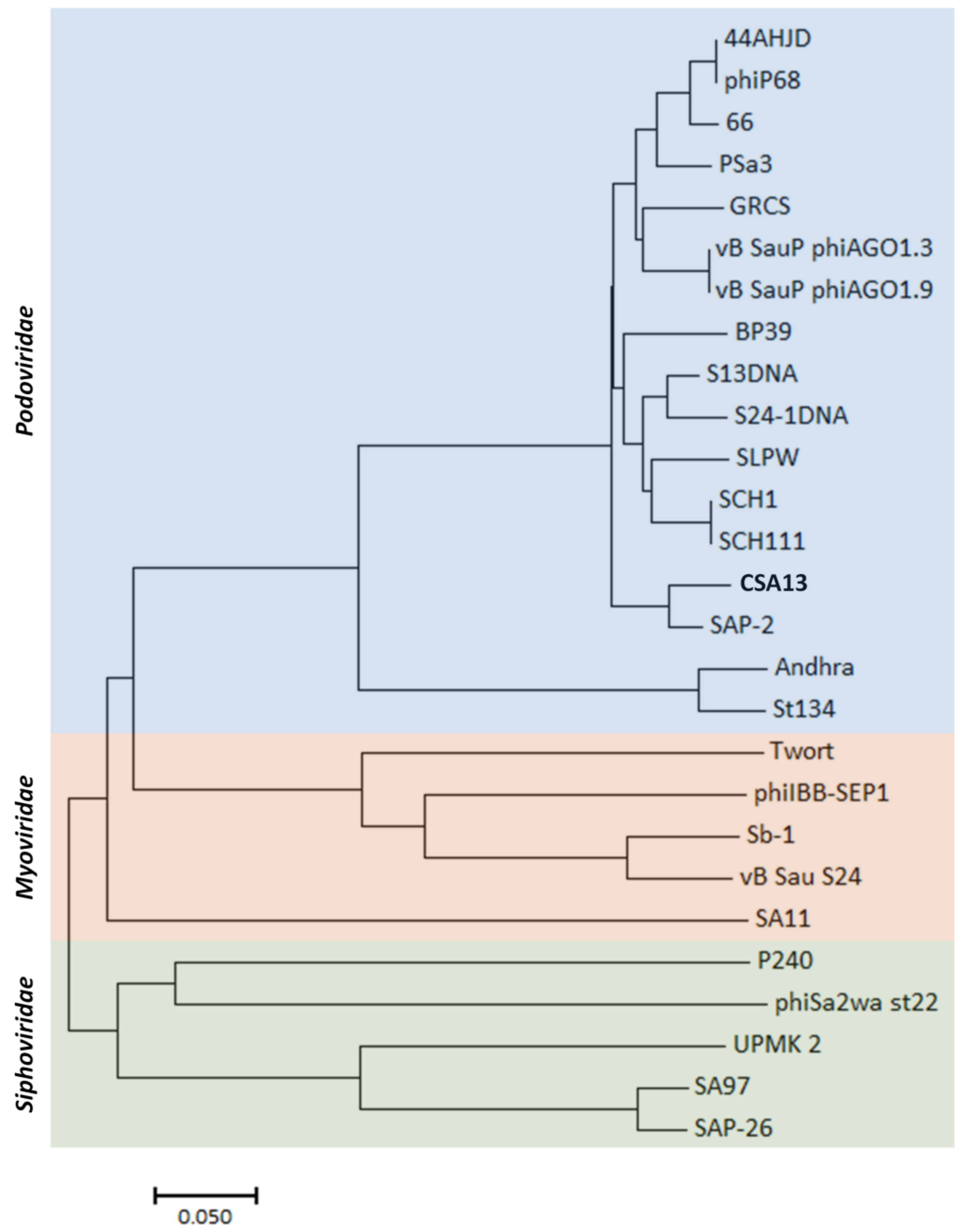
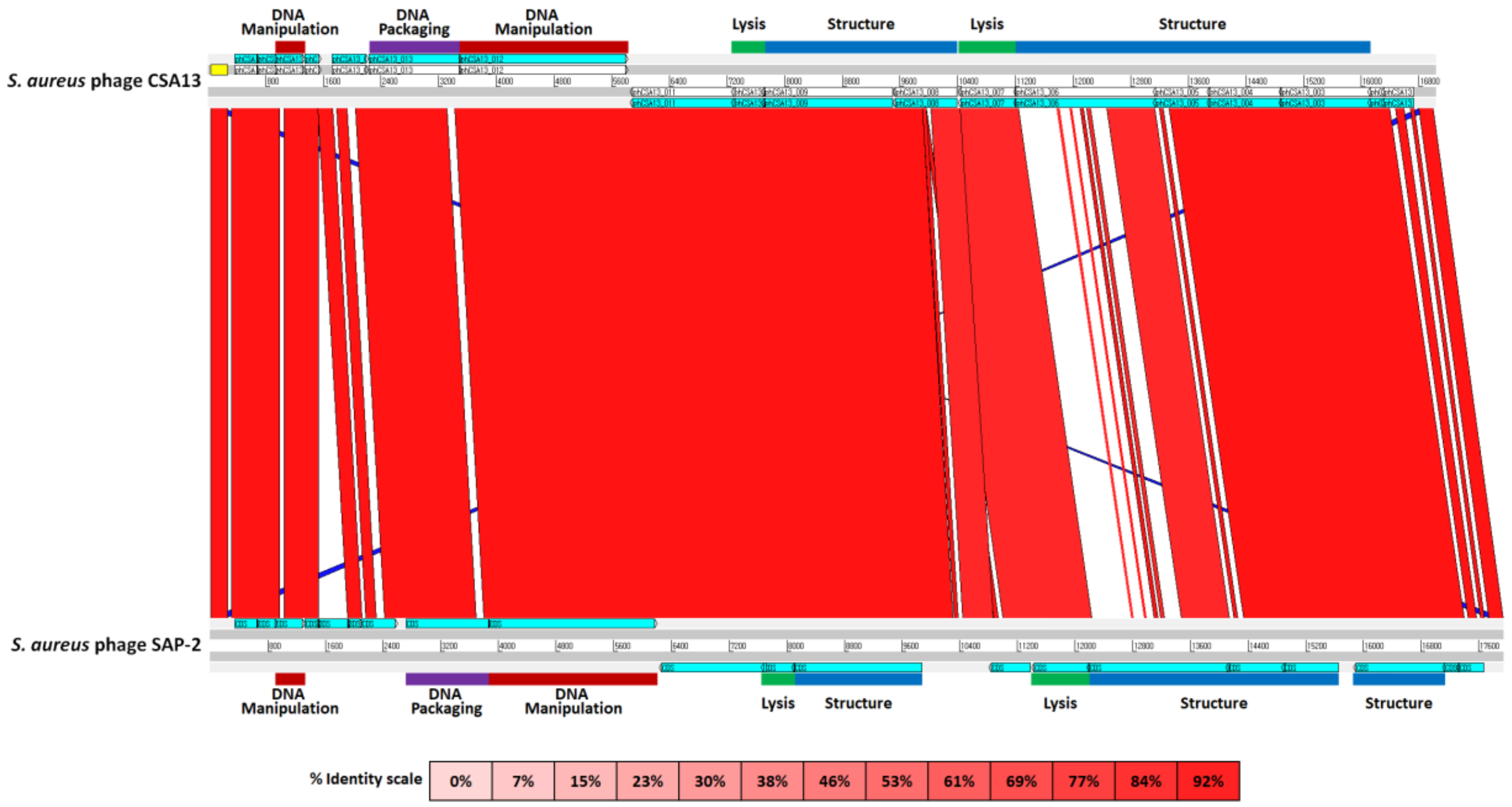
3.6. Comparative Genome Analysis of Phage CSA13
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Liang, Y.; Chen, L.; Wang, W.; Wang, J.; Li, B.; Li, L.; Chen, D.; Xu, Z. Formation and development of staphylococcus biofilm: With focus on food safety. J. Food Saf. 2017, 37, e12358. [Google Scholar] [CrossRef]
- Li, X.-H.; Lee, J.-H. Antibiofilm agents: A new perspective for antimicrobial strategy. J. Microbiol. 2017, 55, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, D.; Rodríguez-Rubio, L.; Martínez, B.; Rodríguez, A.; García, P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 2016, 7, 825. [Google Scholar] [CrossRef] [PubMed]
- Dubey, K.; Chandraker, S.; Sao, S.; Gupta, A.; Dubey, S.K. Bacteriophages as an antibacterial agent: A promising alternative. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 231–234. [Google Scholar] [CrossRef]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage therapy in the food industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Hughes, K.A.; Sutherland, I.W.; Clark, J.; Jones, M.V. Bacteriophage and associated polysaccharide depolymerases—Novel tools for study of bacterial biofilms. J. Appl. Microbiol. 2002, 85, 583–590. [Google Scholar] [CrossRef]
- Donlan, R.M. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009, 17, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Arciola, C.R.; Campoccia, D.; Ravaioli, S.; Montanaro, L. Polysaccharide intercellular adhesin in biofilm: Structural and regulatory aspects. Front. Cell. Infect. Microbiol. 2015, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Bárdy, P.; Pantůček, R.; Benešík, M.; Doškař, J. Genetically modified bacteriophages in applied microbiology. J. Appl. Microbiol. 2016, 121, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Deghorain, M.; Van Melderen, L. The staphylococci phages family: An overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef]
- Rohwer, F.; Edwards, R. The phage proteomic tree: A genome-based taxonomy for phage. J. Bacteriol. 2002, 184, 4529–4535. [Google Scholar] [CrossRef] [PubMed]
- Glazko, G.; Makarenkov, V.; Liu, J.; Mushegian, A. Evolutionary history of bacteriophages with double-stranded DNA genomes. Biol. Direct 2007, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Lee, J.-H.; Shin, H.; Heu, S.; Ryu, S. Characterization and complete genome sequence analysis of staphylococcus aureus bacteriophage sa12. Virus Genes 2013, 47, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.; Parasion, S.; Mizak, L.; Gryko, R.; Bartoszcze, M.; Kocik, J. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against staphylococcus aureus strains. Arch. Virol. 2012, 157, 225–234. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.H.; Shin, H.; Kim, M.; Choi, J.; Kang, D.H.; Heu, S.; Ryu, S. Characterization and comparative genomic analysis of a novel bacteriophage, sfp10, simultaneously inhibiting both salmonella enterica and Escherichia coli o157:H7. Appl. Environ. Microbiol. 2012, 78, 58–69. [Google Scholar] [CrossRef]
- Lu, Z.; Breidt, F.; Fleming, H.P.; Altermann, E.; Klaenhammer, T.R. Isolation and characterization of a lactobacillus plantarum bacteriophage, φjl-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003, 84, 225–235. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Nicholas, K.B.; Nicholas, H.B.J.; Deerfield, D.W. GeneDoc: Analysis and visualization of genetic variation. Embnew. News 1997, 4, 14. [Google Scholar]
- Uchiyama, J.; Takemura-Uchiyama, I.; Kato, S.-I.; Sato, M.; Ujihara, T.; Matsui, H.; Hanaki, H.; Daibata, M.; Matsuzaki, S. In silico analysis of ahjd-like viruses, staphylococcus aureus phages s24-1 and s13′, and study of phage s24-1 adsorption. MicrobiologyOpen 2014, 3, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Taniguchi, M.; Kurokawa, K.; Takemura-Uchiyama, I.; Ujihara, T.; Shimakura, H.; Sakaguchi, Y.; Murakami, H.; Sakaguchi, M.; Matsuzaki, S. Adsorption of staphylococcus viruses s13′ and s24-1 on staphylococcus aureus strains with different glycosidic linkage patterns of wall teichoic acids. J. Gen. Virol. 2017, 98, 2171–2180. [Google Scholar] [CrossRef] [PubMed]
- Wann, E.R.; Dassy, B.; Fournier, J.-M.; Foster, T.J. Genetic analysis of the cap5 locus of staphylococcus aureus. FEMS Microbiol. Lett. 2006, 170, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Swoboda, J.G.; Campbell, J.; Meredith, T.C.; Walker, S. Wall teichoic acid function, biosynthesis, and inhibition. Chembiochem 2010, 11, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Brückner, R. A series of shuttle vectors for bacillus subtilis and Escherichia coli. Gene 1992, 122, 187–192. [Google Scholar] [CrossRef]
- Kelly, D.; McAuliffe, O.; Ross, R.P.; Coffey, A. Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage k and modified derivatives. Lett. Appl. Microbiol. 2012, 54, 286–291. [Google Scholar] [CrossRef]
- Wilcox, S.A.; Toder, R.; Foster, J.W. Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res. 1996, 4, 397–404. [Google Scholar] [CrossRef]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. Genemarks: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef]
- Žiedaitė, G.; Daugelavičius, R.; Bamford, J.K.H.; Bamford, D.H. The holin protein of bacteriophage prd1 forms a pore for small-molecule and endolysin translocation. J. Bacteriol. 2005, 187, 5397–5405. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal w: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000. [Google Scholar]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.-A.; Barrell, B.G.; Parkhill, J. Act: The artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef] [PubMed]
- Son, J.S.; Lee, S.J.; Jun, S.Y.; Yoon, S.J.; Kang, S.H.; Paik, H.R.; Kang, J.O.; Choi, Y.J. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage sap-2 and a derived recombinant cell-wall-degrading enzyme. Appl. Microbiol. Biotechnol. 2010, 86, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.E.; Lo, H.H.; Chen, S.T.; Lee, M.C.; Tseng, Y.H. Wide host range and strong lytic activity of Staphylococcus aureus lytic phage stau2. Appl. Environ. Microbiol. 2011, 77, 756–761. [Google Scholar] [CrossRef]
- O’Flaherty, S.; Ross, R.P.; Flynn, J.; Meaney, W.J.; Fitzgerald, G.F.; Coffey, A. Isolation and characterization of two anti-staphylococcal bacteriophages specific for pathogenic staphylococcus aureus associated with bovine infections. Lett. Appl. Microbiol. 2005, 41, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Shin, H.; Lee, J.-H.; Park, C.; Paik, S.-Y.; Ryu, S. Isolation and genome characterization of the virulent staphylococcus aureus bacteriophage sa97. Viruses 2015, 7, 5225–5242. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Gründling, A.; Peschel, A. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gerlach, D.; Du, X.; Larsen, J.; Stegger, M.; Kühner, P.; Peschel, A.; Xia, G.; Winstel, V. An accessory wall teichoic acid glycosyltransferase protects staphylococcus aureus from the lytic activity of podoviridae. Sci. Rep. 2015, 5, 17219. [Google Scholar] [CrossRef] [PubMed]
- Oku, Y.; Kurokawa, K.; Matsuo, M.; Yamada, S.; Lee, B.-L.; Sekimizu, K. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of staphylococcus aureus cells. J. Bacteriol. 2009, 191, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Kurokawa, K.; Zheng, L.; Jung, D.J.; Tateishi, K.; Jin, J.O.; Ha, N.C.; Kang, H.J.; Matsushita, M.; Kwak, J.Y.; et al. Human serum mannose-binding lectin senses wall teichoic acid glycopolymer of Staphylococcus aureus, which is restricted in infancy. J. Biol. Chem. 2010, 285, 27167–27175. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, C.; Waters, E.M.; Rudkin, J.K.; Schaeffer, C.R.; Lohan, A.J.; Tong, P.; Loftus, B.J.; Pier, G.B.; Fey, P.D.; Massey, R.C.; et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012, 8, e1002626. [Google Scholar] [CrossRef]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin resistance and the biofilm phenotype in staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef]
- Vandersteegen, K.; Kropinski, A.M.; Nash, J.H.E.; Noben, J.-P.; Hermans, K.; Lavigne, R. Romulus and remus, two phage isolates representing a distinct clade within the twortlikevirus genus, display suitable properties for phage therapy applications. J. Virol. 2013, 87, 3237–3247. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Kim, S.; Kim, S.M.; Seol, S.Y.; Kim, J. Characterization of induced Staphylococcus aureus bacteriophage sap-26 and its anti-biofilm activity with rifampicin. Biofouling 2011, 27, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Family—Podoviridae. In Virus Taxonomy; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Elsevier: San Diego, CA, USA, 2012; pp. 63–85. [Google Scholar]
- Borysowski, J.; Weber-Dąbrowska, B.; Górski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16s ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Hambly, E.; Tétart, F.; Desplats, C.; Wilson, W.H.; Krisch, H.M.; Mann, N.H. A conserved genetic module that encodes the major virion components in both the coliphage t4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 2001, 98, 11411–11416. [Google Scholar] [CrossRef] [PubMed]
- Le Marrec, C.; van Sinderen, D.; Walsh, L.; Stanley, E.; Vlegels, E.; Moineau, S.; Heinze, P.; Fitzgerald, G.; Fayard, B. Two groups of bacteriophages infecting streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 1997, 63, 3246–3253. [Google Scholar]
- Tétart, F.; Desplats, C.; Kutateladze, M.; Monod, C.; Ackermann, H.-W.; Krisch, H.M. Phylogeny of the major head and tail genes of the wide-ranging t4-type bacteriophages. J. Bacteriol. 2001, 183, 358–366. [Google Scholar] [CrossRef]
- Catalão, M.J.; Gil, F.; Moniz-Pereira, J.; São-José, C.; Pimentel, M. Diversity in bacterial lysis systems: Bacteriophages show the way. FEMS Microbiol. Rev. 2012, 37, 554–571. [Google Scholar] [CrossRef]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [PubMed]
- Becker, S.C.; Foster-Frey, J.; Stodola, A.J.; Anacker, D.; Donovan, D.M. Differentially conserved staphylococcal sh3b_5 cell wall binding domains confer increased staphylolytic and streptolytic activity to a streptococcal prophage endolysin domain. Gene 2009, 443, 32–41. [Google Scholar] [CrossRef] [PubMed]

| Bacterial Host | Lytic Activity a | Origin |
|---|---|---|
| Staphylococcal strains | ||
| S. aureus RN4220 | T | Laboratory strain |
| S. aureus Newman | I | Human |
| S. aureus ATCC 13301 | C | N/A |
| S. aureus ATCC 23235 | T | Food |
| S. aureus ATCC 33586 | T | Human |
| S. aureus ATCC 33593 | C | Human |
| S. aureus KCTC 1916 | C | Human |
| S. aureus ATCC 6538 | C | Human |
| S. aureus ATCC 29213 | C | Human |
| S. aureus ATCC 12600 | T | Human |
| MRSA CCARM 3793 | C | Human |
| MRSA CCARM 3089 | C | Human |
| MRSA CCARM 3090 | I | Human |
| S. haemolyticus ATCC 29970 | I | Human |
| S. epidermidis ATCC 35983 | C | Human |
| S. hominis ATCC 27844 | C | Human |
| S. warneri ATCC 10209 | T | Antibiosis indicator for snake venoms |
| Other Gram-positive bacteria | ||
| Enterococcus faecalis ATCC 29212 | - | Human |
| Bacillus cereus ATCC 14579 | - | Farmhouse |
| Bacillus subtilis ATCC 23857 | - | N/A |
| Listeria monocytogenes ATCC 19114 | - | Animal |
| Laboratory isolates (S. aureus) | ||
| 129 | C | Animal |
| 130 | T | Animal |
| 131 | T | Animal |
| 134 | C | Animal |
| Clinical isolate 0055 | C | Human |
| Clinical isolate 0136 | C | Human |
| Clinical isolate 0154 | T | Human |
| Clinical isolate 0212 | T | Human |
| Clinical isolate 0600 | C | Human |
| Clinical isolate-FMB_1 | C | Cotton from hospital |
| 77 | T | Human |
| 79 | T | Human |
| 80 | C | Human |
| 81 | T | Human |
| 82 | T | Human |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cha, Y.; Chun, J.; Son, B.; Ryu, S. Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity. Viruses 2019, 11, 54. https://doi.org/10.3390/v11010054
Cha Y, Chun J, Son B, Ryu S. Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity. Viruses. 2019; 11(1):54. https://doi.org/10.3390/v11010054
Chicago/Turabian StyleCha, Yoyeon, Jihwan Chun, Bokyung Son, and Sangryeol Ryu. 2019. "Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity" Viruses 11, no. 1: 54. https://doi.org/10.3390/v11010054
APA StyleCha, Y., Chun, J., Son, B., & Ryu, S. (2019). Characterization and Genome Analysis of Staphylococcus aureus Podovirus CSA13 and Its Anti-Biofilm Capacity. Viruses, 11(1), 54. https://doi.org/10.3390/v11010054






