Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5′UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Construction of HCV 1b Mutant Virus
2.3. HCV Production and Immunofluorescence
2.4. HCV RNA Quantification
2.5. RNA Binding Assays
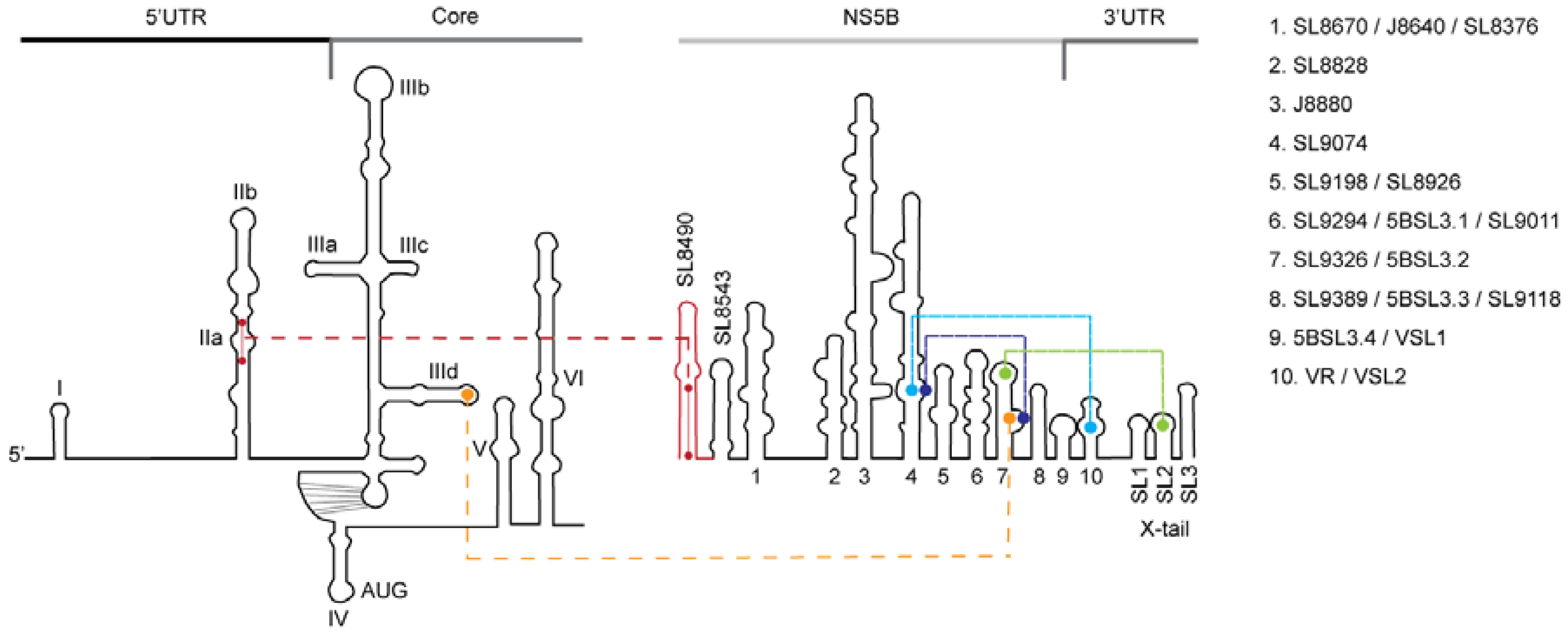
2.6. RNA Structure Models and Data Analysis
3. Results
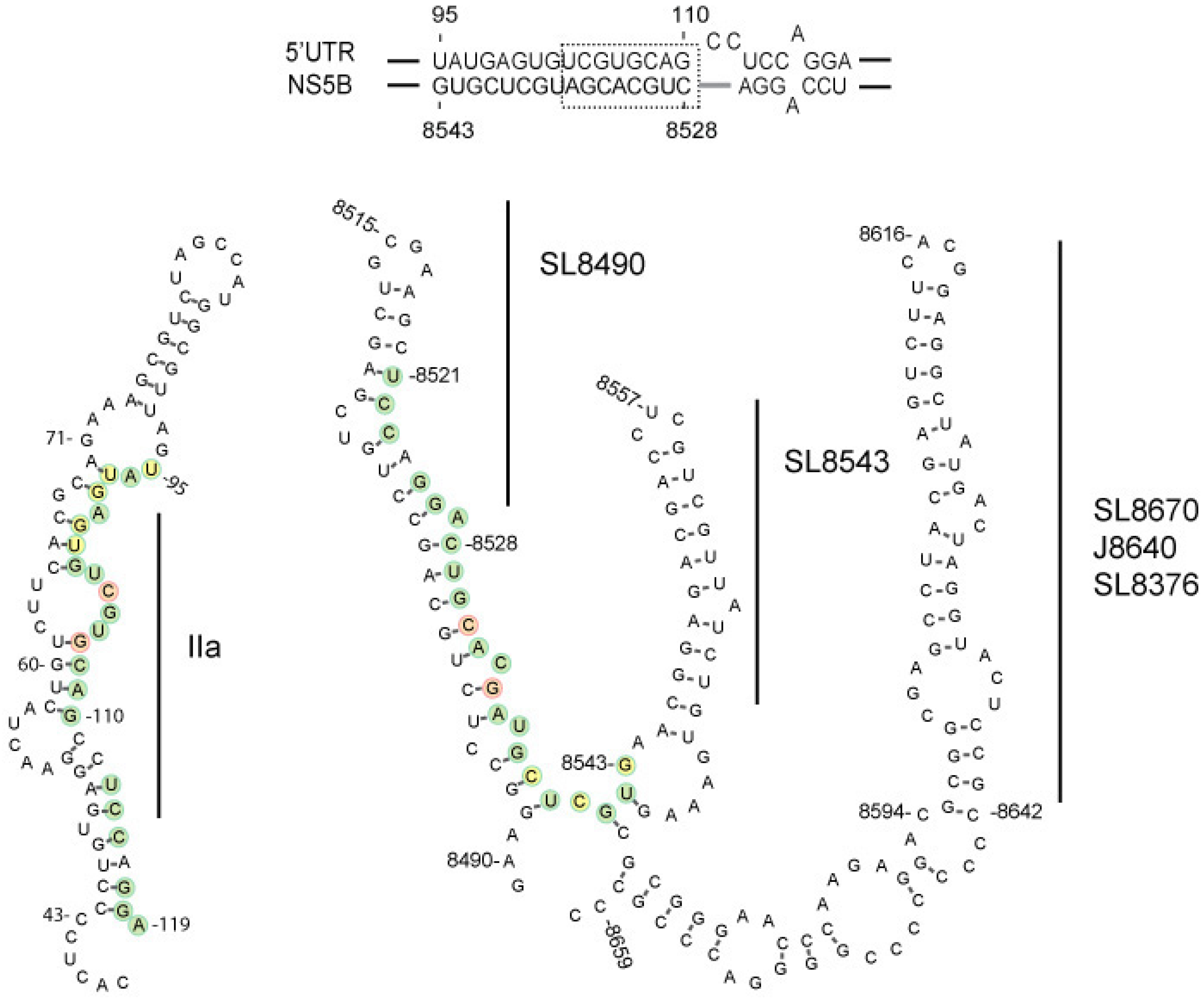
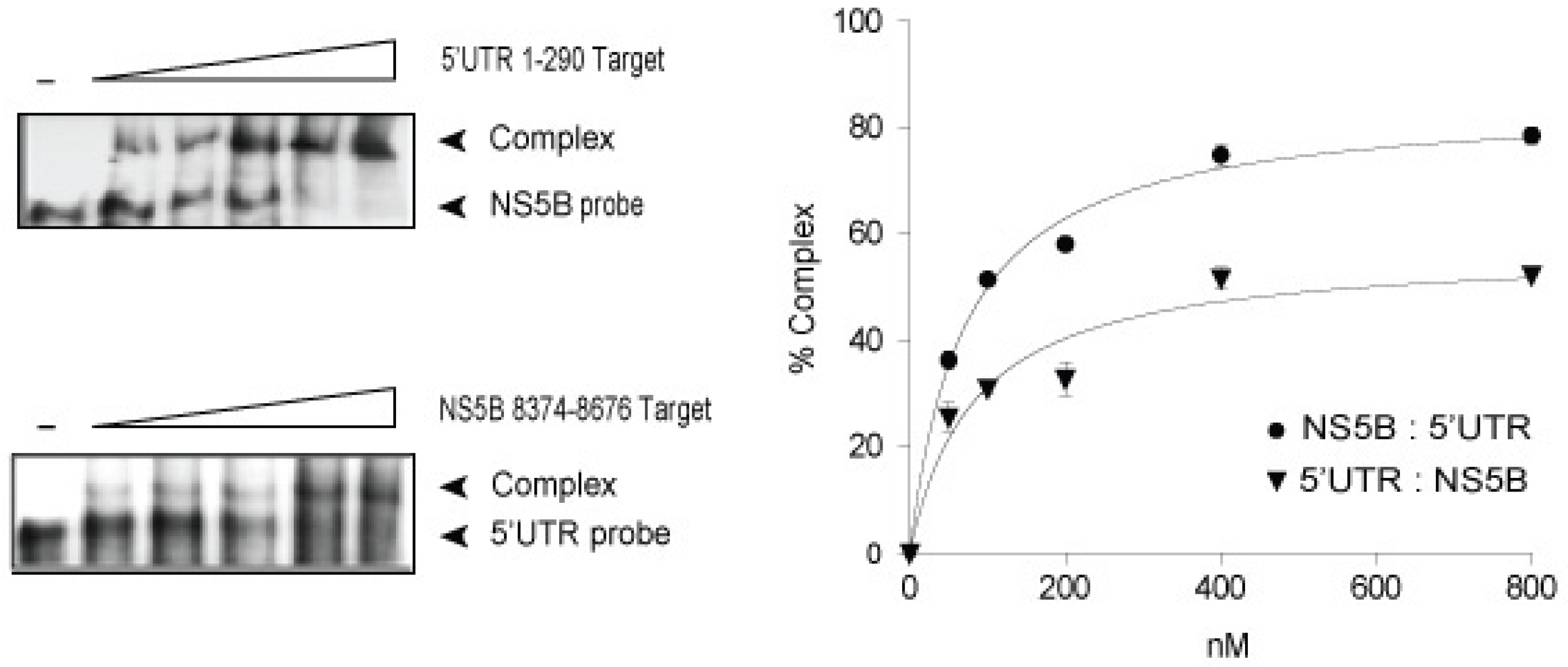
3.1. In vitro Complex Formation between 5′UTR(95–110) and NS5B(8528–8543) RNAs
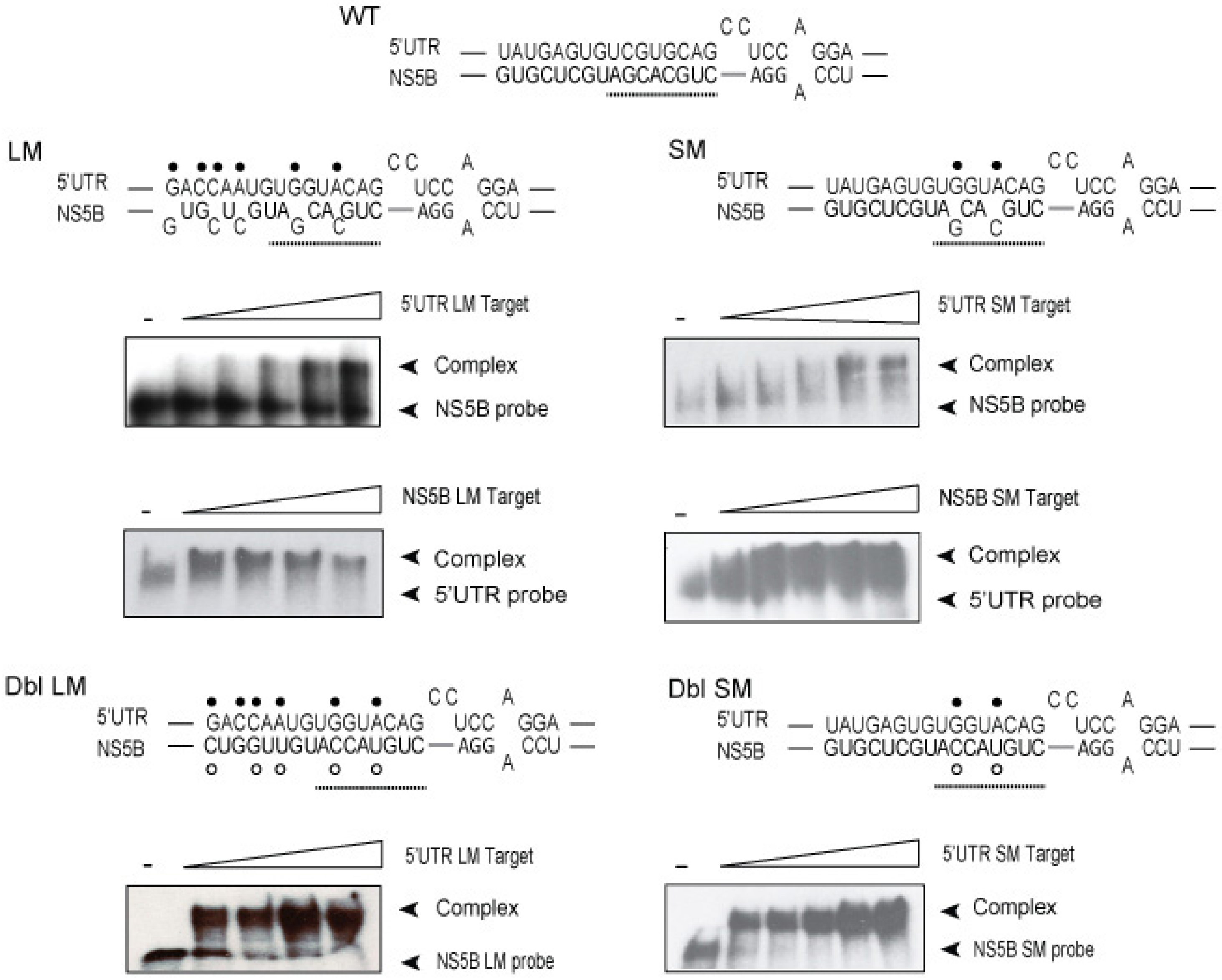
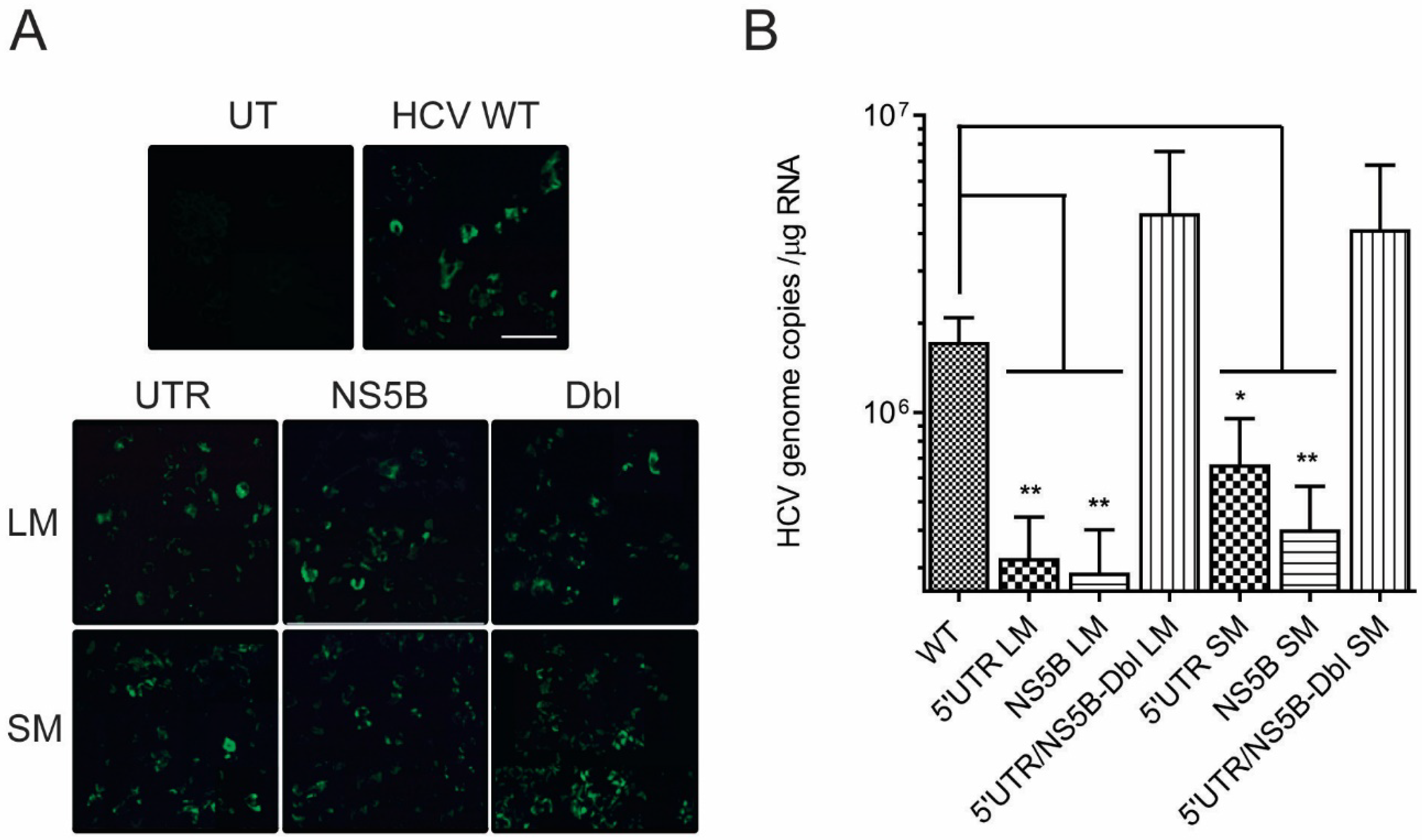
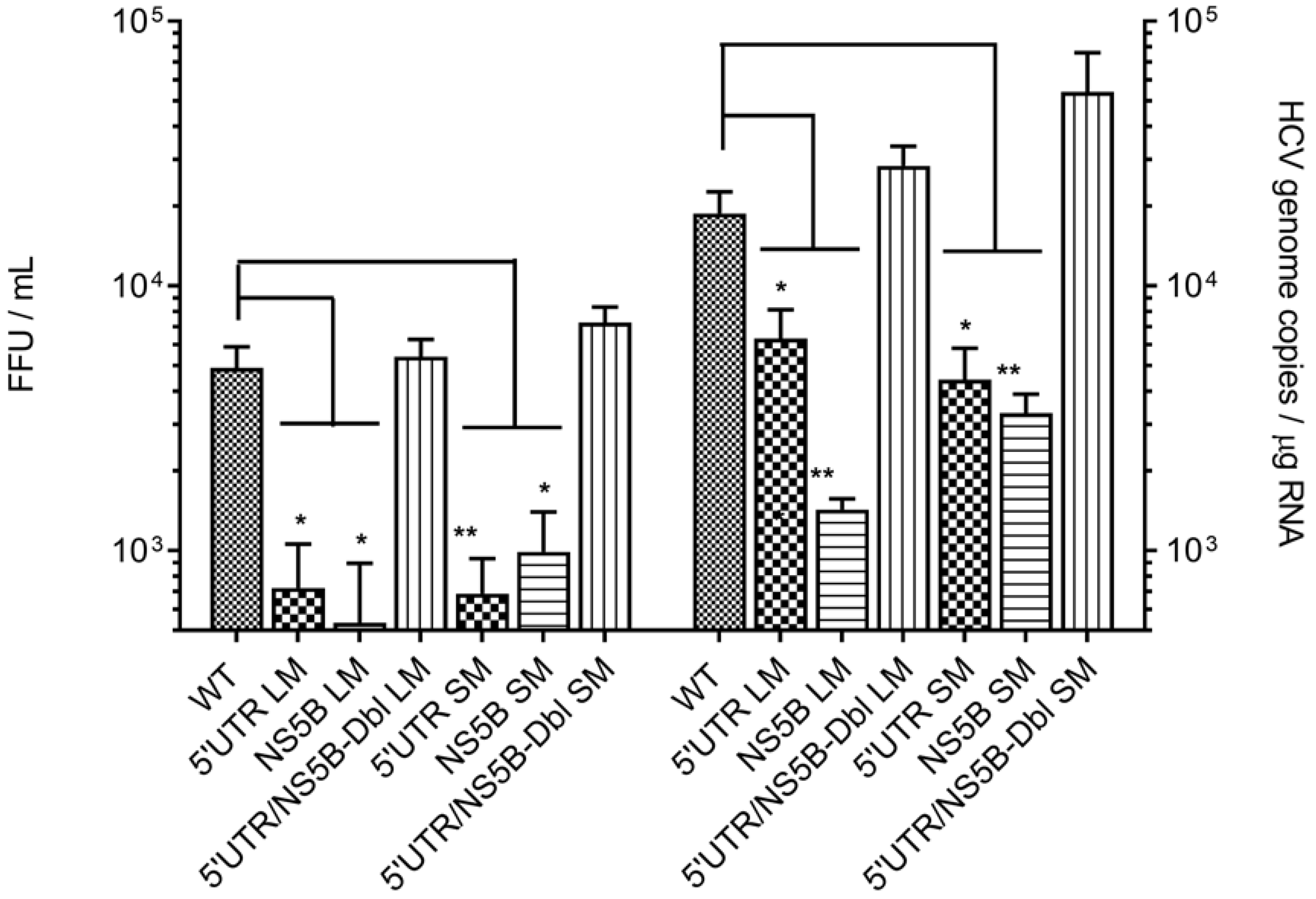
3.2. Disruption of the Distal Interaction between 5′UTR nt 95–110 and NS5B nt 8528–8543 Decreases HCV RNA and Virus Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lindenbach, B.D.; Murry, C.L.; Thiel, J.M.; Rice, C.M.; Knipe, D.M.; Howley, P.M. Flaviviridae. In Virology, Fields; Lippincott, Williams and Wilkins: Philadelphia, PA, USA, 2013; Volume 6, pp. 712–746. [Google Scholar]
- Gu, M.; Rice, C.M. Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function. Curr. Opin. Virol. 2013, 3, 129–136. [Google Scholar] [CrossRef]
- Wang, H.; Tai, A.W. Mechanisms of cellular membrane reorganization to support hepatitis C virus replication. Viruses 2016, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, P.; Di Cara, A. Hairpin RNA: A secondary structure of primary importance. Cell. Mol. Life Sci. 2006, 63, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Pirakitikulr, N.; Kohlway, A.; Lindenbach, B.D.; Pyle, A.M. The coding region of the HCV genome contains a network of regulatory RNA structures. Mol. Cell 2016, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.; Sagan, S.M.; Pezacki, J.P.; Evans, D.J.; Simmonds, P. Bioinformatic and physical characterizations of genome-scale ordered RNA structure in mammalian rna viruses. J. Virol. 2008, 82, 11824–11836. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.B.; Bukh, J.; Kuiken, C.; Muerhoff, A.S.; Rice, C.M.; Stapleton, J.T.; Simmonds, P. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: Updated criteria and genotype assignment web resource. Hepatology 2014, 59, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Fricke, M.; Dunnes, N.; Zayas, M.; Bartenschlager, R.; Niepmann, M.; Marz, M. Conserved RNA secondary structures and long-range interactions in hepatitis C viruses. RNA 2015, 21, 1219–1232. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Quinkert, D.; Bochkarova, O.; Klein, R.; Kezmic, N.; Bartenschlager, R.; Lohmann, V. Identification of determinants involved in initiation of hepatitis C virus RNA synthesis by using intergenotypic replicase chimeras. J. Virol. 2007, 81, 5270–5283. [Google Scholar] [CrossRef]
- Friebe, P.; Lohmann, V.; Krieger, N.; Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001, 75, 12047–12057. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, C.S.; Lee, S.H.; Jang, S.K. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 2002, 290, 105–112. [Google Scholar] [CrossRef]
- Niepmann, M.; Shalamova, L.A.; Gerresheim, G.K.; Rossbach, O. Signals involved in regulation of hepatitis C virus RNA genome translation and replication. Front. Microbiol. 2018, 9, 395. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Blanco, M.A.; Vasudevan, S.G.; Bradrick, S.S.; Nicchitta, C. Flavivirus RNA transactions from viral entry to genome replication. Antivir. Res. 2016, 134, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A.; Evans, D.J.; Simmonds, P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004, 85, 3037–3047. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.L.; Pirakitikulr, N.; Pyle, A.M. Functional RNA structures throughout the hepatitis C virus genome. Curr. Opin. Virol. 2017, 24, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 3692–3697. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.; Ren, S.; Hu, S.; Wang, W.G.; Subramanian, A.; Contreras, D.; Kanagavel, V.; Chung, E.; Ko, J.; Amirtham Jacob Appadorai, R.S.; et al. Systematic analysis of enhancer and critical cis-acting RNA elements in the protein-encoding region of the hepatitis c virus genome. J. Virol. 2013, 87, 5678–5696. [Google Scholar] [CrossRef] [PubMed]
- Thurner, C.; Witwer, C.; Hofacker, I.L.; Stadler, P.F. Conserved RNA secondary structures in flaviviridae genomes. J. Gen. Virol. 2004, 85, 1113–1124. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microrna. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Takamizawa, A.; Mori, C.; Fuke, I.; Manabe, S.; Murakami, S.; Fujita, J.; Onishi, E.; Andoh, T.; Yoshida, I.; Okayama, H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 1991, 65, 1105–1113. [Google Scholar]
- Zuker, M.; Stiegler, P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981, 9, 133–148. [Google Scholar] [CrossRef]
- Mathews, D.H.; Turner, D.H.; Zuker, M. RNA secondary structure prediction. Curr. Protoc. Nucleic Acid Chem. 2007. Chapter 11: Unit 11.2. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Krausslich, H.G.; Mizokami, M.; et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Date, T.; Murayama, A.; Morikawa, K.; Akazawa, D.; Wakita, T. Cell culture and infection system for hepatitis C virus. Nat. Protoc. 2006, 1, 2334–2339. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Katsume, A.; Tanaka, T.; Abe, A.; Inoue, K.; Tsukiyama-Kohara, K.; Kawaguchi, R.; Tanaka, S.; Kohara, M. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 1999, 116, 636–642. [Google Scholar] [CrossRef]
- Fernandez-Miragall, O.; Ramos, R.; Ramajo, J.; Martinez-Salas, E. Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 2006, 12, 223–234. [Google Scholar] [CrossRef] [PubMed]
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers 1990, 29, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Bernhart, S.H.; Tafer, H.; Muckstein, U.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. Partition function and base pairing probabilities of RNA heterodimers. Algorithms Mol. Biol. 2006, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- RNAcofold WebServer. Available online: http://www.rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAcofold.cgi (accessed on 6 November 2018).
- HCV Sequence Database. Available online: https://hcv.lanl.gov/content/sequence/QUICK_ALIGN_HCV/QuickAlign.html (accessed on 3 November 2018).
- Jayaswal, V.; Wong, T.K.; Robinson, J.; Poladian, L.; Jermiin, L.S. Mixture models of nucleotide sequence evolution that account for heterogeneity in the substitution process across sites and across lineages. Syst. Biol. 2014, 63, 726–742. [Google Scholar] [CrossRef]
- Yamamoto, H.; Collier, M.; Loerke, J.; Ismer, J.; Schmidt, A.; Hilal, T.; Sprink, T.; Yamamoto, K.; Mielke, T.; Burger, J.; et al. Molecular architecture of the ribosome-bound hepatitis c virus internal ribosomal entry site rna. EMBO J. 2015, 34, 3042–3058. [Google Scholar] [CrossRef]
- Romero-Lopez, C.; Berzal-Herranz, A. A long-range RNA-RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA 2009, 15, 1740–1752. [Google Scholar] [CrossRef]
- Bartenschlager, R.; Sparacio, S. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 2007, 127, 195–207. [Google Scholar] [CrossRef]
- Saxena, P.; Lomonossoff, G.P. Virus infection cycle events coupled to RNA replication. Annu. Rev. Phytopathol. 2014, 52, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Selisko, B.; Wang, C.; Harris, E.; Canard, B. Regulation of flavivirus RNA synthesis and replication. Curr. Opin. Virol. 2014, 9, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A. Diverse roles and interactions of rna structures during the replication of positive-stranded RNA viruses of humans and animals. J. Gen. Virol. 2015, 96, 1497–1503. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ untranslated regions of the flaviviral genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Brinton, M.A.; Basu, M. Functions of the 3′ and 5′ genome RNA regions of members of the genus flavivirus. Virus Res. 2015, 206, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Boudet, J.; Simorre, J.P.; Bartenschlager, R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 2005, 79, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Berzal-Herranz, A. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012, 69, 103–113. [Google Scholar] [CrossRef] [PubMed]
- You, S.; Rice, C.M. 3′ RNA elements in hepatitis C virus replication: Kissing partners and long poly(u). J. Virol. 2008, 82, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Tuplin, A.; Struthers, M.; Simmonds, P.; Evans, D.J. A twist in the tail: Shape mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012, 40, 6908–6921. [Google Scholar] [CrossRef] [PubMed]
- Diviney, S.; Tuplin, A.; Struthers, M.; Armstrong, V.; Elliott, R.M.; Simmonds, P.; Evans, D.J. A hepatitis C virus cis-acting replication element forms a long-range RNA-RNA interaction with upstream RNA sequences in NS5B. J. Virol. 2008, 82, 9008–9022. [Google Scholar] [CrossRef] [PubMed]
- Rance, E.; Hu, J.; Tanner, J.E.; Alfieri, C. A method for in situ localization of single-strand and double-strand RNA elements contained in the hepatitis C virus genome 3′-untranslated region. Adv. Tech. Biol. Med. 2016, 4, 171–183. [Google Scholar] [CrossRef]
- Varani, G.; McClain, W.H. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000, 1, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Lukavsky, P.J.; Kim, I.; Otto, G.A.; Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003, 10, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.P.; Miyaji, A.; Jefferson, E.A.; Sannes-Lowery, K.A.; Osgood, S.A.; Propp, S.S.; Ranken, R.; Massire, C.; Sampath, R.; Ecker, D.J.; et al. Sar by MS: Discovery of a new class of RNA-binding small molecules for the hepatitis C virus: Internal ribosome entry site IIA subdomain. J. Med. Chem. 2005, 48, 7099–7102. [Google Scholar] [CrossRef] [PubMed]
- Dibrov, S.M.; Ding, K.; Brunn, N.D.; Parker, M.A.; Bergdahl, B.M.; Wyles, D.L.; Hermann, T. Structure of a hepatitis C virus RNA domain in complex with a translation inhibitor reveals a binding mode reminiscent of riboswitches. Proc. Natl. Acad. Sci. USA 2012, 109, 5223–5228. [Google Scholar] [CrossRef] [PubMed]
- Boerneke, M.A.; Dibrov, S.M.; Gu, J.; Wyles, D.L.; Hermann, T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl. Acad. Sci. USA 2014, 111, 15952–15957. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Rios-Marco, P.; Berzal-Herranz, B.; Berzal-Herranz, A. The HCV genome domains 5BSL3.1 and 5BSL3.3 act as managers of translation. Sci. Rep. 2018, 8, 16101. [Google Scholar] [CrossRef] [PubMed]
| RNA Probe | Target RNA | Avg Kd + SD (nM) 1 | Bmax (%) 1 | KdMT/KdWT | % Bmax MT/WT | MFE |
|---|---|---|---|---|---|---|
| NS5B-WT | 5′UTR-LM | 364.7 ± 124.6 | 49.78 ± 7.47 | 5.2 | 58.4 | −6.9 |
| NS5B-WT | 5′UTR-SM | 314.5 ± 120.8 | 49.64 ± 7.55 | 4.5 | 58.3 | −13.9 |
| 5′UTR-WT | NS5B-LM | 551.8 ± 151.1 | 54.77 ± 6.40 | 6.6 | 95.6 | −6.9 |
| 5′UTR-WT | NS5B-SM | 489.8 ± 129.4 | 43.77 ± 5.14 | 5.8 | 76.4 | −14.2 |
| NS5B-LM | 5′UTR-LM | 74.84 ± 7.28 | 83.35 ± 2.15 | 1.1 | 97.8 | −24.6 |
| NS5B-SM | 5′UTR-SM | 72.35 ± 8.04 | 67.45 ± 1.85 | 1.3 | 118 | −22.6 |
| Virus Construct | HCV RNA Copies 1 | MT WT | Progeny Virus 1 | MT WT | Progeny HCV RNA Copies 2 | MT WT |
|---|---|---|---|---|---|---|
| WT genome | 1.7 × 106 ± 3.5 × 105 | 6.9 × 103 ± 2.1 × 103 | 1.9 × 104 ± 3.8 × 103 | |||
| 5′UTRLM | 3.2 × 105 ± 1.2 × 105 | −5.3 | 1.4 × 103 ± 7.6 × 102 | −4.8 | 6.3 × 103 ± 1.8 × 103 | −2.9 |
| NS5BLM | 2.9 × 105 ± 1.1 × 105 | −6.0 | 2.1 × 103 ± 1.0 × 103 | −3.2 | 1.4 × 103 ± 1.4 × 102 | −13.1 |
| 5′UTRSM | 6.7 × 105 ± 2.9 × 105 | −2.6 | 6.9 × 102 ± 2.4 × 102 | −10 | 4.4 × 103 ± 1.3 × 103 | −4.3 |
| NS5BSM | 4.0 × 105 ± 1.6 × 105 | −4.2 | 9.9 × 102 ± 4.0 × 102 | −7 | 3.3 × 103 ± 5.9 × 102 | −5.7 |
| DblLM | 4.7 × 106 ± 2.9 × 106 | 1.9 | 7.2 × 103 ± 1.9 × 103 | 2 | 2.8 × 104 ± 5.3 × 103 | 1.7 |
| DblSM | 4.1 × 106 ± 2.7 × 106 | 1.7 | 9.4 × 103 ± 2.3 × 104 | 1.7 | 5.4 × 104 ± 2.2 × 104 | 1.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rance, E.; Tanner, J.E.; Alfieri, C. Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5′UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication. Viruses 2019, 11, 17. https://doi.org/10.3390/v11010017
Rance E, Tanner JE, Alfieri C. Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5′UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication. Viruses. 2019; 11(1):17. https://doi.org/10.3390/v11010017
Chicago/Turabian StyleRance, Elodie, Jerome E. Tanner, and Caroline Alfieri. 2019. "Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5′UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication" Viruses 11, no. 1: 17. https://doi.org/10.3390/v11010017
APA StyleRance, E., Tanner, J. E., & Alfieri, C. (2019). Genomic-Scale Interaction Involving Complementary Sequences in the Hepatitis C Virus 5′UTR Domain IIa and the RNA-Dependent RNA Polymerase Coding Region Promotes Efficient Virus Replication. Viruses, 11(1), 17. https://doi.org/10.3390/v11010017




