Abstract
Clostridium perfringens is one of the most common causes of food-borne illness. The increasing prevalence of multidrug-resistant bacteria requires the development of alternatives to typical antimicrobial treatments. Here, we isolated and characterized a C. perfringens-specific virulent bacteriophage CPS2 from chicken feces. The CPS2 phage contains a 17,961 bp double-stranded DNA genome with 25 putative ORFs, and belongs to the Picovirinae, subfamily of Podoviridae. Bioinformatic analysis of the CPS2 genome revealed a putative endolysin, LysCPS2, which is homologous to the endolysin of Clostridium phage phiZP2 and phiCP7R. The enzyme showed strong lytic activity against C. perfringens with optimum conditions at pH 7.5–10, 25–65 °C, and over a broad range of NaCl concentrations. Interestingly, LysCPS2 was found to be highly thermostable, with up to 30% of its lytic activity remaining after 10 min of incubation at 95 °C. The cell wall binding domain in the C-terminal region of LysCPS2 showed a binding spectrum specific to C. perfringens strains. This is the first report to characterize highly thermostable endolysin isolated from virulent C. perfringens bacteriophage. The enzyme can be used as an alternative biocontrol and detection agent against C. perfringens.
1. Introduction
Clostridium perfringens is a Gram-positive anaerobic bacterium that has the ability to form spores [1]. C. perfringens is responsible for a wide range of diseases: including gas gangrene (clostridial myonecrosis), necrotic enteritis, and non-foodborne gastrointestinal infections [2]. Approximately 5% of all C. perfringens strains produce C. perfringens enterotoxin (CPE), which causes diarrhea and abdominal cramping symptoms. Most CPE-positive stains are classified as type A, and food poisoning by cpe-producing C. perfringens type A is the second most common foodborne illness in developed countries [3]. In addition, C. perfringens has become a significant problem in the poultry industry because it is a causative agent of necrotic enteritis, characterized by outbreaks with high mortality and small intestinal mucosal necrosis [4]. Increased mortality, economic loss, and contamination of poultry products for human consumption are important concerns regarding C. perfringens in the poultry industry.
Furthermore, the increasing incidence of antibiotic resistance of bacterial pathogens and the lack of novel antibiotics have become serious worldwide problems. Bacteriophages (phages) and gene products such as endolysins have been attracting considerable attention as alternatives to antibiotics. Endolysins are phage-encoded peptidoglycan hydrolases that break down the bacterial peptidoglycan at the end of their reproduction cycles to release the viral progeny [5]. The purified endolysin protein has potent hydrolytic activity against Gram-positive bacteria when applied exogenously. In addition, endolysins have significant advantages over classic antibiotics, such as narrow host specificity, high sensitivity, and low probability for development of resistant bacteria [6].
To date, there have been few reports on C. perfringens phage endolysins. Most studies have attempted to isolate endolysin from the prophage because of the relative difficulties in isolating the phage from anaerobic C. perfringens. N-acetylmuramidases of C. perfringens phage phiSM101 and N-acetylmuramoyl-l-alanine amidase of φ3626 phage were isolated from lysogenic C. perfringens strains and characterized [7,8]. CP25L endolysin was examined for the activity to kill C. perfringens, and endolysin delivery by Lactobacillus johnsonii was reported [9]. Two endolysins from the clostridial phages ΦCP39O and ΦCP26F have been reported to have lytic activity against C. perfringens stains [10]. However, detailed information about these endolysins have not been reported.
Here we isolated novel virulent C. perfringens bacteriophage CPS2 from chicken feces, and its predicted endolysin LysCPS2 was identified in the genome of bacteriophage CPS2. The gene was cloned and expressed in Escherichia coli, and the purified endolysin was biochemically characterized for its potential as an antimicrobial and a detection agent.
2. Materials and Methods
2.1. Bacterial Strains, and Growth Conditions
C. perfringens ATCC 13124 was used as a host strain for isolation and propagation of the bacteriophage CPS2. E. coli BL21 was grown in Luria-Bertani (LB) broth (Difco, Detroit, MI, USA) at 37 °C and used as the host for expression of the recombinant LysCPS2. Bacterial strains that were used for antimicrobial spectrum determination are listed in Table 1, along with the results. All of the bacterial strains were routinely grown at 37 °C in Brain Heart Infusion (BHI) broth medium (Difco) under anaerobic conditions.

Table 1.
Antimicrobial spectra of the CPS2 and LysCPS2, and binding spectrum of LysCPS2_CBD.
2.2. Isolation and Propagation of Bacteriophage CPS2
To isolate a bacteriophage, we applied the same method as described in the previous study [11]. Isolated phages were amplified by serial propagation and concentrated by polyethylene glycol precipitation and subsequent CsCl density gradient ultracentrifugation (78,500× g at 4 °C for 2 h). The concentrated phages were dialyzed using 2 L of standard dialysis buffer (10 mM NaCl, 10 mM MgSO4 and 1 M Tris-HCl; pH 8.0) for 2 h. The phage stock obtained was stored in glass vials at 4 °C.
2.3. Transmission Electron Microscopy (TEM) Analysis
Purified CPS2 (1 × 109 PFU/mL) was placed on carbon-coated copper grids and negatively stained with 2% aqueous uranyl acetate (pH 4.0) for 20 s. The morphology of CPS2 was analyzed by TEM (LEO 912AB transmission electron microscope; Carl Zeiss, Wezlar, Germany). Images were scanned at the National Academy of Agricultural Science (Jeonju, South Korea).
2.4. DNA Purification and Whole Genome Sequencing of Bacteriophage CPS2
To extract genomic DNA from CPS2, host DNA was removed by treatment of the virions with DNaseI and RNaseA (1 μg/mL each) at room temperature for 30 min. The virions were then lysed by reacting with proteinase K mixture (50 μg/mL proteinase K, 20 mM ethylenediaminetetraacetic acid (EDTA), 0.5% sodium dodecyl sulfate (SDS)) at 56 °C for 1 h. After lysis, the DNA was purified by the phenol-chloroform extraction [12] and concentrated by ethanol precipitation [13]. The purified genomic DNA of CPS2 was sequenced using the GS-FLX Titanium sequencer (Roche Holding AG, Basel, Switzerland). Total 23,050 sequencing reads obtained were assembled using the GS De Novo Assembler version 2.9 (Roche Holding AG, Basel, Switzerland) with default parameters. Additional DNA sequencing of phage was performed to identify end regions of genomic DNA sequence using primers (CPS2endF, 5’-CAC CCT GGA GCA TTT ACA C-3’; CPS2endR, 5’-TCC ATA ACA GAC AAT AAA AAT TTT AAA T-3’) in Macrogen inc. (Seoul, South Korea). The position of open reading frames (ORFs) was predicted by bioinformatics tools, including Glimmer 3.02 [14] and Rapid Annotation using Subsystem Technology (RAST) software [15]. The function of each ORF was predicted using NCBI BLASTP and InterProScan [14] databases. Based on the information, each ORF’s name was annotated manually. The gene encoding endolysin was identified, and its domain structure was investigated using InterProScan databases. The complete genome sequence of CPS2 phage was deposited in GenBank under accession number MH248069.
2.5. Cloning, Expression, and Purification of LysCPS2
The endolysin gene (lysCPS2) was amplified from the genomic DNA of the bacteriophage CPS2 by polymerase chain reaction (PCR) using primers lysCPS2F (5’-GCG GGA TCC ATG AAA ATA ATA CAA TCA AAT ATC CAT TTT-3’) and lysCPS2R (5’-CGC AAG CTT TTA GTC TTT TTT AAT ATA TTT TGC GGA-3’). The PCR product was cloned into pET28a (Novagen, Madison, WI, USA), which has an N-terminal hexahistidine (His)-tag sequence. Plasmid with a correct insert was transformed into competent E. coli BL21 (DE3). Expression of the recombinant LysCPS2 was induced with 0.5 mM isopropyl-b-d-thiogalactopyranoside, with adjusted OD600 to 0.6–0.8, followed by incubation for an additional 15 h at 30 °C. Bacterial cells were suspended in lysis buffer (50 mM Tris-HCl, 100 mM sodium chloride, pH 7.5) and disrupted by sonication (Branson Ultrasonics, Shanghai, China). After centrifugation at 15,000× g for 40 min, the supernatant was collected, mixed with 500 μL of nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Hilden, Germany) and incubated at 4 ℃ for 1 h with gentle shaking. The flow-through was discarded and the resin was serially washed with 10 mL of 10 mM imidazole and 5 mL of 20 mM imidazole. An elution buffer (50 mM Tris-HCl, 100 mM sodium chloride, and 250 mM imidazole; pH 7.5) was used to elute the protein. The purified protein was stored at −4 °C until use, after the buffer was changed to the storage buffer (50 mM Tris-HCl, 200 mM sodium chloride, pH 7.5, 30% glycerol) using PD Miditrap G-25 (GE Healthcare, Little Chalfont, UK).
2.6. Lytic Activity Assay
The lytic activity of the endolysin against bacterial cells was assayed by monitoring the decrease in OD600. All tested bacteria were cultivated to the exponential phase. Cells were harvested and resuspended with reaction buffer (50 mM Tris-HCl, 200 mM NaCl, pH 7.5) to OD600 to 0.8–1.0. The endolysin (50 μL) was added to the cell suspension (950 μL), followed by incubation at room temperature, unless indicated otherwise. OD600 values were monitored over time. The lytic activity was calculated after 40 min as follows: {ΔOD600 test (endolysin added)-ΔOD600 control (buffer only)}/initial OD600. Antimicrobial spectrum was tested by plate lysis assay as previously described [16]. In brief, 10 μL of diluted endolysin in reaction buffer (32 μM; final concentration) was spotted onto a freshly prepared bacterial lawn on BHI agar plates. Spotted plates were air-dried in a laminar flow hood for 15 min and incubated overnight at 37 °C. To evaluate the effect of pH on LysCPS2 enzymatic activity, the endolysin (162 nM) was added to C. perfringens cells suspended with a universal pH buffer [17]. The universal buffer consists of 50 mM KCl, 10 mM KH2PO4, 10 mM Na-citrate, and 10 mM H3BO4, and was adjusted to different pH values—between 4 and 10—using 5 M NaOH or 5 M HCl. Different temperatures (25–95 °C) were applied to test the effect of temperature on LysCPS2 (162 nM) enzymatic activity. To evaluate the stability of the endolysin, the lysis assays were performed against C. perfringens ATCC 13124 at room temperature and pH 7.5 after the enzyme was incubated for 10 min at different temperatures. The influence of NaCl on lytic activity of LysCPS2 (162 nM) was tested with the addition of concentrations of 0, 100, 200, 300, 400, and 500 mM NaCl. The effects of metal ions on lysis activity were determined as previously reported [6]. To chelate metal ions attached to the endolysin, thereby inhibiting its catalytic function, EDTA (100 mM; final concentration) was added to the endolysin (4.86 μM) and incubated at 37 °C for 1 h. The EDTA was removed by exchanging the endolysin into the reaction buffer using a PD trap G-25 column. The EDTA-treated enzyme was added to cell suspensions with metal ions (MgCl2, CaCl2, MnCl2, ZnCl2, CuCl2; 1.0 mM final concentration), and the lysis activity was assayed in the reaction buffer.
2.7. CBD Binding and Fluorescence Microscopy
The gene encoding the putative CBD (between bases 457–681 of lysCPS2) was amplified from the genomic DNA of CPS2 by PCR using primers lysCPS2_CBDF (5’-GCG GGA TCC GGA AAC TTA GAT TTA AAC AAA TTA AGA ACA GAT GTA AAC-3’) and lysCPS2_CBDR (5’-CGC AAG CTT TTA GTC TTT TTT AAT ATA TTT TGC GGA-3’). The resulting PCR product was cloned using the BamHI and SalI restriction sites into pET28a-mCherry (mCherry gene inserted between NdeI and BamHI sites into pET28a) [18]. mCherry-tagged LysCPS2_CBD protein was produced in E. coli BL21 and purified by affinity chromatography, as described earlier [19]. Purified protein at a final concentration of 100 μM was added to C. perfringens cells, and the mixture was then incubated for 10 min at 25 °C. Subsequently, cells were collected by centrifugation and washed twice with Dulbecco’s phosphate-buffered saline (PBS). For fluorescence microscopy, images were captured on a DE/Axio Imager A1 microscope (Carl Zeiss) with a charge-coupled-device camera using AxioVision release 4.7 (Carl Zeiss, Oberkochen, Germany).
3. Results and Discussion
3.1. Morphology of Phage CPS2
The C. perfringens phage CPS2 was isolated from chicken feces using C. perfringens isolate as a host stain. Morphological observation of phage CPS2 revealed that it belongs to the family Podoviridae due to the presence of short and non-contractile tails (Figure 1A). The diameter of the icosahedral head was approximately 40 nm and the length of non-contractile tails (from baseplate of virion to tail fiber) was 15 nm.
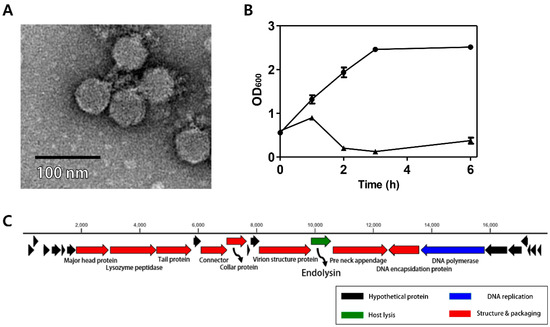
Figure 1.
Characterization of C. perfringens virulent phage CPS2. (A) TEM image of CPS2; (B) bacterial challenge assay of CPS2 against C. perfringens ATCC 13124. The closed circle indicates non-phage-treated C. perfringens ATCC 13124, and the closed triangle indicates phage-treated C. perfringens ATCC 13124; (C) genome map of CPS2. Red: structure and packaging, blue: DNA replication, green: host lysis, black: hypothetical protein. Twenty-five putative ORFs were predicted in the CPS2 genome.
3.2. Antibacterial Properties of Phage CPS2
The bacterial challenge assay was performed to evaluate the growth inhibition ability of the CPS2 phage. When phage CPS2 was added to exponentially growing C. perfringens at a multiplicity of infection (MOI) of 1.0, complete inhibition of host cells was observed 1 h after phage infection, and growth inhibition was maintained for up to 6 h (Figure 1B). A host range test showed that CPS2 can inhibit the growth of only C. perfringens ATCC 13124, and C. perfringens human stool isolate 3 out of the 10 tested C. perfringens strains. CPS2 phage did not show lytic activity against other clostridial bacteria or other Gram-positive bacteria, indicating very narrow host specificity (Table 1).
3.3. Genomic Analysis of CPS2
The complete genome of C. perfringens bacteriophage CPS2 comprises 17,961-bp with an overall G + C content of 33.30%. Twenty-five putative ORFs were identified, and no tRNA genes were detected. The functional ORFs were clustered into 4 functional groups of DNA replication, host lysis, structure plus packaging, and additional function (Figure 1C). The sequence analysis showed the linear structure of CPS2 double-stranded DNA, as well as the inverted terminal repeats (ITR) of 366 nucleotide pairs on the phage genomic termini. Furthermore, the CPS2 genome encoded a putative Type-B DNA polymerase that is utilized in a protein-primed replication mechanism, implying the presence of terminal proteins for DNA replication [22]. These findings suggest that CPS2 belong to the Podoviridae sub-family Picovirinae [23]. Importantly, the toxin production- and bacterial virulence-associated genes were not identified in the CPS2 genome. Genes associated with lysogenization were not detected, suggesting that CPS2 is a virulent phage.
3.4. Identification and Expression of the LysCPS2 Endolysin
The putative endolysin gene was identified from the CPS2 genome and designated as LysCPS2. Pfam and Conserved Domain Database analysis revealed that LysCPS2 is a putative N-acetylmuramoyl-l-alanine amidase that consists of N-terminal amidase_2 domain (PF01520) as an enzymatically active domain (EAD) and a C-terminal SH3_3 (PF08239) as a cell wall binding domain (CBD). Amino acid sequence alignment revealed that LysCPS2 was highly homologous to endolysins of Clostridium phage phiZP2 and phiCP7R at the amino acid sequence level (Figure 2B) [22] The N-terminal region from LysCPS2 showed 51% sequence identity with previously reported Clostridium phage vB_CpeS-CP51 endolysin. Although BLAST analysis revealed many proteins homologous to LysCPS2, most of them were autolysins. The LysCPS2 was cloned and expressed in E. coli with an N-terminal His-tag. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) showed a single band of the purified endolysin (Figure 2C). Turbidity reduction assay revealed that 32 nM of the purified LysCPS2 showed saturated lysis activity against C. perfringens ATCC 13124 cells (Figure 2D). These results indicate that LysCPS2 is highly active compared to endolysin from C. difficile phage ΦCD27, in which a similar amount of endolysin had barely exhibited lytic activity against C. difficile [24].
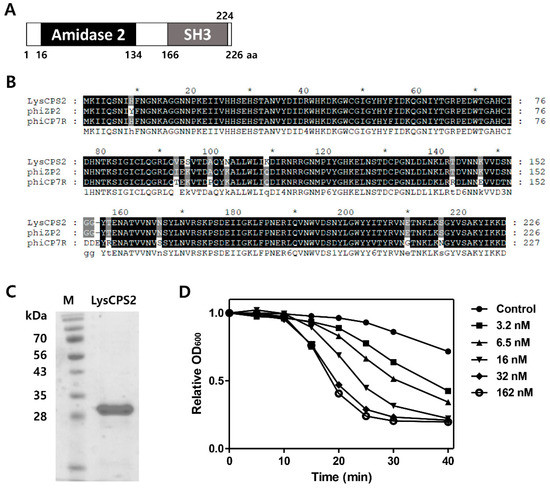
Figure 2.
Modular structure and lytic activities of the LysCPS2 endolysin from CPS2. (A) Schematic representation of LysCPS2. The conserved amidase2 domain is shown; (B) Sequence alignment of various Clostridial phage endolysins; CPS2 phage endolysin, phiZP2 phage endolysin, phiCP7R phage endolysin; (C) SDS-PAGE analysis of purified LysCPS2. M: standard molecular weight marker, LysCPS2: purified LysCPS2 fraction; (D) Lysis of C. perfringens ATCC 13124 cells treated externally with various concentrations of recombinant LysCPS2. Optical density was measured periodically after LysCPS2 treatment.
3.5. Antimicrobial Spectrum of LysCPS2
Antimicrobial activity against Clostridium and other Gram-positive bacterial strains was examined (Table 1). All of the tested C. perfringens strains were susceptible to LysCPS2, indicating a much broader antimicrobial spectrum of LysCPS2, considering the narrow host range of the CPS2 phage. However, other Clostridium species such as C. histolyticum and C. indolis were not lysed by treatment of LysCPS2, meaning that its enzymatic activity requires species-specific moieties of the cell wall component. This enzyme did not show lytic activity against other Gram-positive bacteria, such as Bacillus cereus, Bacillus subtilis, Listeria monocytogenes, and Staphylococcus aureus. B. cereus, B. subtilis, and L. monocytogenes have an A1γ type peptidoglycan directly cross-linked with meso-diaminopimelic acid (m-DAP), and S. aureus has an A3α type cross-linked by penta-glycine bridges in its peptidoglycan. C. perfringens has a different type of peptidoglycan structure containing L,L-DAP and glycine instead of m-DAP in the A1γ type peptidoglycan [25].
3.6. pH, Temperature, NaCl, and Metal Effects on LysCPS2 Activity
To determine optimum conditions for the endolysin activity, the biochemical properties of LysCPS2 were evaluated. Analysis of lytic activity at different pH levels showed that LysCPS2 had the highest lytic activity at pH 7.5–10 (Figure 3A). More than 60% residual lytic activity was observed in the temperature range from 25 °C to 65 °C (Figure 3B). The effect of ionic strength on the lytic activity of LysCPS2 was assessed at various concentrations of NaCl, ranging from 0 to 500 mM (Figure 3C). LysCPS2 retained its lytic activity even at 500 mM NaCl, suggesting that LysCPS2 is highly stable in a broad range of NaCl concentrations in contrast to the C. perfringens phage phiSM101 endolysin that showed reduced activity in the presence of more than 200 mM of NaCl [7]. Even though phiSM101 endolysin showed 36% amino acid sequence identity with LysCPS2, it was the only C. perfringens endolysin that could be compared with the influence of NaCl on the lytic activity of LysCPS2. To examine the effect of metal ions on the enzyme activity of LysCPS2, metal ions were initially removed from the endolysin with 100 mM EDTA. Addition of EDTA showed little effect on the LysCPS2 activity, indicating that LysCPS2 activity is independent of the presence of divalent cations. All tested metal ions except Ca2+ decreased enzymatic activity of LysCPS2, and the enzyme was completely inactivated in the presence of Zn2+ (Figure 3D). Although many divalent ions have been reported to enhance the enzymatic activity of endolysin, the inhibition effect by certain metal ions on endolysin activity has also been reported in several studies [26,27]. The peptidoglycan hydrolase of Burkholderia pseudomallei phage ST79 showed reduced lytic activity in the presence of Zn2+, Mg2+, or Mn2+ [28]. The inhibition effect of Zn2+ is also found in the T5 phage endolysin [29].
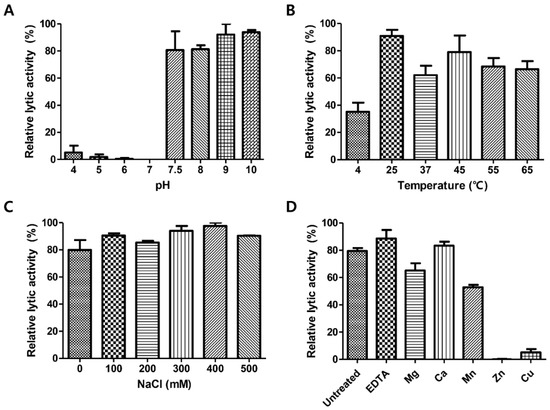
Figure 3.
Effects of pH, temperature, NaCl and metal ions on the lytic activity of LysCPS2. Effects of (A) pH, (B) temperatures, (C) NaCl, and (D) metal ions on the lytic activity of LysCPS2 against C. perfringens ATCC 13124 cells. Each column represents the mean of triplicate experiments, and error bars indicate the standard deviation.
3.7. Determination of Thermal Stability
Generally, endolysins are not heat stable, and most lose their hydrolase activity above 50–60 °C [30]. However, few endolysins such as listerial phage endolysins HPL511, HPL118, and HPLP35 have been reported to be thermostable, showing residual activity after 10 min incubation at 90 °C [31]. Stenotrophomonas maltophilia phage endolysin P28 retained 55% of its activity after treatment at 70 °C for 30 min [32], and a thermostable chimeric endolysin, PlyGVE2CpCWB, targeting C. perfringens has exhibited 57% residual activity at 55 °C [33]. LysCPS2 showed remarkable heat stability. The lytic activity after pre-incubation of the enzyme at temperatures between 4 °C and 65 °C was not reduced compared with the heat-untreated control. LysCPS2 retained more than 30% of its activity after heating at 95 °C for 10 min and 60% after heating at 75 °C for 10 min (Figure 4). This is the first report to characterize highly thermostable endolysin isolated from virulent C. perfringens bacteriophage. This enzyme will provide powerful tools for many applications in molecular biology, biotechnology, and medicine [34].
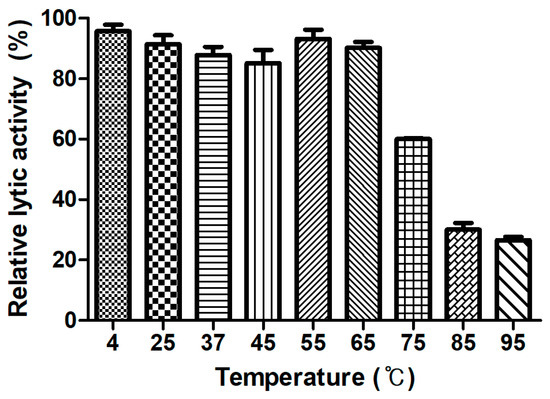
Figure 4.
LysCPS2 thermal stability. LysCPS2 was incubated at different temperatures for 10 min, then cooled on ice before enzyme assay at 25 °C. Relative lytic activities are calculated using the activity of enzyme stored at 4 °C, which showed the maximal activity.
3.8. Binding Activity of LysCPS2_CBD
The specific bacterial binding activity of the putative LysCPS2_CBD was identified with an mCherry (red fluorescent protein) fusion protein (Figure 5). As shown in Table 1, all tested C. perfringens cells were decorated by mCherry_LysCPS2_CBD, whereas other Clostridium species and other Gram-positive bacteria could not be labeled by the fusion protein (Table 1). These results demonstrated that LysCPS2_CBD specifically binds to C. perfringens cells, which is consistent with the antimicrobial spectrum of the LysCPS2. The specificity and affinity of LysCPS2_CBD could be useful for developing an efficient bio-probe against C. perfringens.
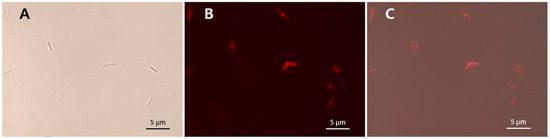
Figure 5.
mCherry_LysCPS2_CBD binding activity. LysCPS2_CBD binds to C. perfringens human isolate cells. Bright field (A), fluorescence (B) and merged (C) images are shown.
4. Conclusions
In this study, we isolated a C. perfringens virulent phage CPS2 and identified a gene encoding LysCPS2 endolysin from CPS2 genome. LysCPS2 is a highly thermostable endolysin showing 30% of its lytic activity against C. perfringens even after 10 min of incubation at 95 °C. Optimum conditions for LysCPS2 were at pH 7.5–10, 25–65 °C, and over a broad range of NaCl concentrations, suggesting that LysCPS2 has high potential as an effective antibacterial agent to control C. perfringens. The cell wall binding domain in the C-terminal region of LysCPS2 showed a binding spectrum specific to C. perfringens strains. Taken together, LysCPS2 would be useful for the development of a powerful biocontrol and detection agent against C. perfringens.
Author Contributions
E.H. and B.S. carried out all the experiments. E.S., B.S. and S.R. conceived of the study, participated in its design and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by Basic Science Research Programs (NRF-2017R1A2A1A17069378) through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET) through Agriculture, Food and Rural Affairs Research Center Support Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) (710012-03-1-SB110).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Canard, B.; Saint-Joanis, B.; Cole, S. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol. Microbiol. 1992, 6, 1421–1429. [Google Scholar] [CrossRef] [PubMed]
- Myers, G.S.; Rasko, D.A.; Cheung, J.K.; Ravel, J.; Seshadri, R.; DeBoy, R.T.; Ren, Q.; Varga, J.; Awad, M.M.; Brinkac, L.M. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006, 16, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Freedman, J.C.; Shrestha, A.; McClane, B.A. Clostridium perfringens enterotoxin: Action, genetics, and translational applications. Toxins 2016, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Timbermont, L.; Haesebrouck, F.; Ducatelle, R.; van Immerseel, F. Necrotic enteritis in broilers: An updated review on the pathogenesis. Avian Pathol. 2011, 40, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Borysowski, J.; Weber-Dąbrowska, B.; Górski, A. Bacteriophage endolysins as a novel class of antibacterial agents. Exp. Biol. Med. 2006, 231, 366–377. [Google Scholar] [CrossRef]
- Nariya, H.; Miyata, S.; Tamai, E.; Sekiya, H.; Maki, J.; Okabe, A. Identification and characterization of a putative endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 90, 1973–1979. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M.; Vukov, N.; Scherer, S.; Loessner, M.J. The murein hydrolase of the bacteriophage φ3626 dual lysis system is active against all tested Clostridium perfringens strains. Appl. Environ. Microbiol. 2002, 68, 5311–5317. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, T.; Horn, N.; Wegmann, U.; Dugo, G.; Narbad, A.; Mayer, M.J. Expression and delivery of an endolysin to combat Clostridium perfringens. Appl. Microbiol. Biotechnol. 2014, 98, 2495–2505. [Google Scholar] [CrossRef] [PubMed]
- Seal, B.S. Characterization of bacteriophages virulent for Clostridium perfringens and identification of phage lytic enzymes as alternatives to antibiotics for potential control of the bacterium1. Poult. Sci. 2013, 92, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Seal, B.S.; Fouts, D.E.; Simmons, M.; Garrish, J.K.; Kuntz, R.L.; Woolsey, R.; Schegg, K.M.; Kropinski, A.M.; Ackermann, H.-W.; Siragusa, G.R. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: Genomic organization and proteomic analysis of the virions. Arch. Virol. 2011, 156, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Kirby, K. A new method for the isolation of ribonucleic acids from mammalian tissues. Biochem. J. 1956, 64, 405. [Google Scholar] [CrossRef] [PubMed]
- Zeugin, J.A.; Hartley, J.L. Ethanol precipitation of DNA. Focus 1985, 7, 1–2. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Kim, M.; Ryu, S. Characterization of a novel endolysin LysSA11 and its utility as a potent biocontrol agent against Staphylococcus aureus on food and utensils. Food Microbiol. 2017, 68, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Walmagh, M.; Boczkowska, B.; Grymonprez, B.; Briers, Y.; Drulis-Kawa, Z.; Lavigne, R. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl. Microbiol. Biotechnol. 2013, 97, 4369–4375. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Shin, J.H.; Heu, S.; Park, J.-K.; Ryu, S. Lateral flow assay-based bacterial detection using engineered cell wall binding domains of a phage endolysin. Biosens. Bioelectron. 2017, 96, 173–177. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Sim, J.; Kang, T.; Nguyen, H.H.; Park, H.K.; Chung, B.H.; Ryu, S. A novel and highly specific phage endolysin cell wall binding domain for detection of Bacillus cereus. Eur. Biophys. J. 2015, 44, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Cherniak, R. Structure of the capsular polysaccharide of Clostridium perfringens Hobbs 10 determined by NMR spectroscopy. Carbohydr. Res. 1997, 305, 65–72. [Google Scholar] [CrossRef]
- Kretzer, J.W.; Lehmann, R.; Schmelcher, M.; Banz, M.; Kim, K.-P.; Korn, C.; Loessner, M.J. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl. Environ. Microbiol. 2007, 73, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Volozhantsev, N.V.; Oakley, B.B.; Morales, C.A.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Popova, A.V.; Zhilenkov, E.L.; Garrish, J.K.; Schegg, K.M. Molecular characterization of podoviral bacteriophages virulent for Clostridium perfringens and their comparison with members of the Picovirinae. PLoS ONE 2012, 7, e38283. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Schuch, R.; Zhu, S.; Tscherne, D.M.; Fischetti, V.A. Genomic sequence of C1, the first streptococcal phage. J. Bacteriol. 2003, 185, 3325–3332. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.J.; Narbad, A.; Gasson, M.J. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J. Bacteriol. 2008, 190, 6734–6740. [Google Scholar] [CrossRef] [PubMed]
- Schleifer, K.H.; Kandler, O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [PubMed]
- Son, B.; Yun, J.; Lim, J.-A.; Shin, H.; Heu, S.; Ryu, S. Characterization of LysB4, an endolysin from the Bacillus cereus-infecting bacteriophage B4. BMC Microbiol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, D.G.; Dong, S.; Baker, J.R.; Engler, J.A. The bifunctional peptidoglycan lysin of Streptococcus agalactiae bacteriophage B30. Microbiology 2004, 150, 2079–2087. [Google Scholar] [CrossRef] [PubMed]
- Khakhum, N.; Yordpratum, U.; Boonmee, A.; Tattawasart, U.; Rodrigues, J.L.; Sermswan, R.W. Cloning, expression, and characterization of a peptidoglycan hydrolase from the Burkholderia pseudomallei phage ST79. AMB Express 2016, 6, 77. [Google Scholar] [CrossRef] [PubMed]
- Mikoulinskaia, G.V.; Odinokova, I.V.; Zimin, A.A.; Lysanskaya, V.Y.; Feofanov, S.A.; Stepnaya, O.A. Identification and characterization of the metal ion-dependent l-alanoyl-d-glutamate peptidase encoded by bacteriophage T5. FEBS J. 2009, 276, 7329–7342. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Briers, Y.; Hertveldt, K.; Robben, J.; Volckaert, G. Identification and characterization of a highly thermostable bacteriophage lysozyme. Cell. Mol. Life Sci. CMLS 2004, 61, 2753–2759. [Google Scholar] [CrossRef] [PubMed]
- Schmelcher, M.; Waldherr, F.; Loessner, M.J. Listeria bacteriophage peptidoglycan hydrolases feature high thermoresistance and reveal increased activity after divalent metal cation substitution. Appl. Microbiol. Biotechnol. 2012, 93, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhu, C.; Chen, J.; Ye, X.; Huang, Y.-P. Antibacterial Activity of Stenotrophomonas maltophilia Endolysin P28 against both Gram-positive and Gram-negative Bacteria. Front. Microbiol. 2015, 6, 1299. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Seal, B.S.; Garrish, J.K.; Oakley, B.B.; Hiett, K.; Yeh, H.-Y.; Woolsey, R.; Schegg, K.M.; Line, J.E.; Donovan, D.M. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-Clostridium antimicrobial with improved thermostability. Viruses 2015, 7, 3019–3034. [Google Scholar] [CrossRef] [PubMed]
- Loessner, M.J. Bacteriophage endolysins—Current state of research and applications. Curr. Opin. Microbiol. 2005, 8, 480–487. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).