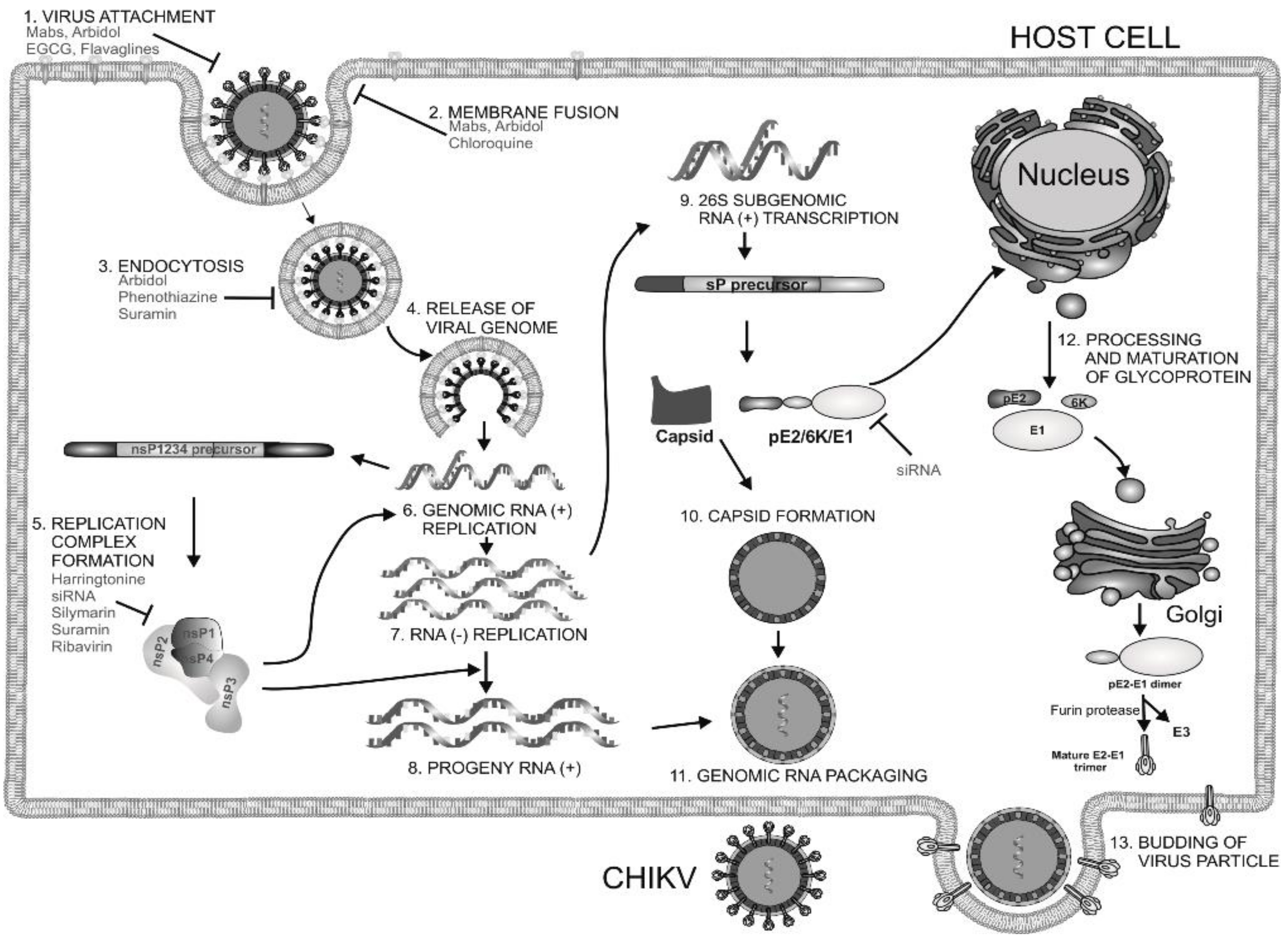
Current Strategies for Inhibition of Chikungunya Infection
Abstract
1. Introduction
2. Current Drug Targets of CHIKV
2.1. Structural Proteins
2.2. Receptors for CHIKV Entry
2.3. Non-Structural Protein 1(nsP1)
2.4. Non-Structural Protein 2(nsP2)
2.5. Non-Structural Protein 3(nsP3)
2.6. Non-Structural Protein 4 (nsP4)
3. Hits Identified by In Silico Approaches
4. Drugs Inhibiting CHIKV Entry
4.1. Chloroquine
4.2. Arbidol and Its Derivatives
4.3. Phenothiazines
4.4. Epigallocatechin Gallate (EGCG)
4.5. Flavaglines
4.6. NSAIDs
4.7. Imipramine
4.8. Monoclonal Antibodies
4.9. Curcumin
5. Inhibitors of Viral Genome Replication
5.1. Andrographolide
5.2. Ribavirin
5.3. Mycophenolic Acid (MPA)
5.5. Favipiravir (T-705)
5.6. RNA Interference Targeting CHIKV Genes
5.7. Silymarin
5.8. Suramin
6. Inhibitors of Viral Protein Translation
Harringtonine and Homoharringtonine
7. Host-Targeting Antivirals
7.1. Furin Inhibitors
7.2. Protein Kinase C (PKC) Modulators
7.3. Kinase Inhibitors
7.4. HSP-90 Inhibitors
7.5. Interferon
7.6. Viperin
7.7. Polyinosinicacid:Polycytidylic Acid (poly I:C)
7.8. Retinoic Acid-Inducible Gene-I (RIG-I) Agonists
7.9. Repurposing Drugs for Targeting Host Factors of CHIKV
7.10. Protein Duslfide Isomerase (PDI) Inhibitors
8. Inhibitors with an Unidentified Target
8.1. Lupenone
8.2. Jatrophan Ester
8.3. Trigocherrierin A and B
8.4. Extract of Hyptis Suaveolens
8.6. Triazolopyrimidines
8.7. Benzouracil-Coumarin-Arene-Conjugates
8.8. Thiazolidinone Derivatives
8.9. MBZM-N-IBT
8.10. Berberine, Abamectin, rough a collaborative research effort, Gigante et al. identifieand Ivermectin
8.11. 5-Chloro-N-{4-[(1E)-1-{2-[(2-phenylcyclopropyl) carbonyl] hydrazinylidene} ethyl] phenyl} Thiophene-2-carboxamide
8.12. Compound ID1452-2
9. Pre-Clinical Validation of Molecules
10. CHIKV Coinfection
11. Conclusions
Acknowledgments
Conflicts of Interest
References
- Robinson, M.C. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53. I. Clinical features. Trans. R. Soc. Trop. Med. Hyg. 1955, 49, 28–32. [Google Scholar] [CrossRef]
- Waggoner, J.J.; Pinsky, B.A. How great is the threat of chikungunya virus? Expert Rev. Anti-Infect. Ther. 2015, 13, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Morrison, T.E. Reemergence of chikungunya virus. J. Virol. 2014, 88, 11644–11647. [Google Scholar] [CrossRef]
- Thiberville, S.D.; Moyen, N.; Dupuis-Maguiraga, L.; Nougairede, A.; Gould, E.A.; Roques, P.; de Lamballerie, X. Chikungunya fever: Epidemiology, clinical syndrome, pathogenesis and therapy. Antivir. Res. 2013, 99, 345–370. [Google Scholar] [CrossRef]
- Tsetsarkin, K.A.; Vanlandingham, D.L.; McGee, C.E.; Higgs, S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007, 3, e201. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; McGee, C.E.; Volk, S.M.; Vanlandingham, D.L.; Weaver, S.C.; Higgs, S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS ONE 2009, 4, e6835. [Google Scholar] [CrossRef]
- Vazeille, M.; Moutailler, S.; Coudrier, D.; Rousseaux, C.; Khun, H.; Huerre, M.; Thiria, J.; Dehecq, J.S.; Fontenille, D.; Schuffenecker, I.; et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS ONE 2007, 2, e1168. [Google Scholar] [CrossRef]
- Schuffenecker, I.; Iteman, I.; Michault, A.; Murri, S.; Frangeul, L.; Vaney, M.C.; Lavenir, R.; Pardigon, N.; Reynes, J.M.; Pettinelli, F.; et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006, 3, e263. [Google Scholar] [CrossRef]
- Hanson, S.M.; Craig, G.B., Jr. Aedes albopictus (Diptera: Culicidae) eggs: Field survivorship during northern Indiana winters. J. Med. Entomol. 1995, 32, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.K.; Chua, K.B.; Hooi, P.S.; Rahimah, M.A.; Kumari, S.; Tharmaratnam, M.; Chuah, S.K.; Smith, D.W.; Sampson, I.A. Chikungunya infection—An emerging disease in Malaysia. Southeast Asian J. Trop. Med. Public Health 2001, 32, 447–451. [Google Scholar] [PubMed]
- Mahendradas, P.; Ranganna, S.K.; Shetty, R.; Balu, R.; Narayana, K.M.; Babu, R.B.; Shetty, B.K. Ocular manifestations associated with chikungunya. Ophthalmology 2008, 115, 287–291. [Google Scholar] [CrossRef]
- Chatterjee, S.N.; Chakravarti, S.K.; Mitra, A.C.; Sarkar, J.K. Virological investigation of cases with neurological complications during the outbreak of haemorrhagic fever in Calcutta. J. Indian Med. Assoc. 1965, 45, 314–316. [Google Scholar] [PubMed]
- Chandak, N.H.; Kashyap, R.S.; Kabra, D.; Karandikar, P.; Saha, S.S.; Morey, S.H.; Purohit, H.J.; Taori, G.M.; Daginawala, H.F. Neurological complications of Chikungunya virus infection. Neurol. India 2009, 57, 177–180. [Google Scholar] [PubMed]
- Economopoulou, A.; Dominguez, M.; Helynck, B.; Sissoko, D.; Wichmann, O.; Quenel, P.; Germonneau, P.; Quatresous, I. Atypical Chikungunya virus infections: Clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Reunion. Epidemiol. Infect. 2009, 137, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Saswat, T.; Kumar, A.; Kumar, S.; Mamidi, P.; Muduli, S.; Debata, N.K.; Pal, N.S.; Pratheek, B.M.; Chattopadhyay, S.; Chattopadhyay, S. High rates of co-infection of Dengue and Chikungunya virus in Odisha and Maharashtra, India during 2013. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 35, 134–141. [Google Scholar] [CrossRef]
- Villamil-Gomez, W.E.; Gonzalez-Camargo, O.; Rodriguez-Ayubi, J.; Zapata-Serpa, D.; Rodriguez-Morales, A.J. Dengue, chikungunya and Zika co-infection in a patient from Colombia. J. Infect. Public Health 2016, 9, 684–686. [Google Scholar] [CrossRef]
- Gandhi, B.S.; Kulkarni, K.; Godbole, M.; Dole, S.S.; Kapur, S.; Satpathy, P.; Khatri, A.M.; Deshpande, P.S.; Azad, F.; Gupte, N.; et al. Dengue and Chikungunya coinfection associated with more severe clinical disease than mono-infection. Int. J. Healthc. Biomed. Res. 2015, 3, 117–123. [Google Scholar]
- Schwartz, O.; Albert, M.L. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 2010, 8, 491–500. [Google Scholar] [CrossRef]
- Smith, T.J.; Cheng, R.H.; Olson, N.H.; Peterson, P.; Chase, E.; Kuhn, R.J.; Baker, T.S. Putative receptor binding sites on alphaviruses as visualized by cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 1995, 92, 10648–10652. [Google Scholar] [CrossRef]
- Ashbrook, A.W.; Burrack, K.S.; Silva, L.A.; Montgomery, S.A.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. Residue 82 of the Chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J. Virol. 2014, 88, 12180–12192. [Google Scholar] [CrossRef]
- Voss, J.E.; Vaney, M.C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef]
- Li, L.; Jose, J.; Xiang, Y.; Kuhn, R.J.; Rossmann, M.G. Structural changes of envelope proteins during alphavirus fusion. Nature 2010, 468, 705–708. [Google Scholar] [CrossRef]
- Ye, F.; Zhang, M. Structures and target recognition modes of PDZ domains: Recurring themes and emerging pictures. Biochem. J. 2013, 455, 1–14. [Google Scholar] [CrossRef]
- Kuo, S.C.; Chen, Y.J.; Wang, Y.M.; Tsui, P.Y.; Kuo, M.D.; Wu, T.Y.; Lo, S.J. Cell-based analysis of Chikungunya virus E1 protein in membrane fusion. J. Biomed. Sci. 2012, 19, 44. [Google Scholar] [CrossRef]
- Rashad, A.A.; Mahalingam, S.; Keller, P.A. Chikungunya virus: Emerging targets and new opportunities for medicinal chemistry. J. Med. Chem. 2014, 57, 1147–1166. [Google Scholar] [CrossRef]
- Hong, E.M.; Perera, R.; Kuhn, R.J. Alphavirus capsid protein helix I controls a checkpoint in nucleocapsid core assembly. J. Virol. 2006, 80, 8848–8855. [Google Scholar] [CrossRef]
- Perera, R.; Owen, K.E.; Tellinghuisen, T.L.; Gorbalenya, A.E.; Kuhn, R.J. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 2001, 75, 1–10. [Google Scholar] [CrossRef]
- Jose, J.; Przybyla, L.; Edwards, T.J.; Perera, R.; Burgner, J.W., 2nd; Kuhn, R.J. Interactions of the cytoplasmic domain of Sindbis virus E2 with nucleocapsid cores promote alphavirus budding. J. Virol. 2012, 86, 2585–2599. [Google Scholar] [CrossRef]
- Snyder, J.E.; Kulcsar, K.A.; Schultz, K.L.; Riley, C.P.; Neary, J.T.; Marr, S.; Jose, J.; Griffin, D.E.; Kuhn, R.J. Functional characterization of the alphavirus TF protein. J. Virol. 2013, 87, 8511–8523. [Google Scholar] [CrossRef]
- Wintachai, P.; Wikan, N.; Kuadkitkan, A.; Jaimipuk, T.; Ubol, S.; Pulmanausahakul, R.; Auewarakul, P.; Kasinrerk, W.; Weng, W.Y.; Panyasrivanit, M.; et al. Identification of prohibitin as a Chikungunya virus receptor protein. J. Med. Virol. 2012, 84, 1757–1770. [Google Scholar] [CrossRef]
- Moller-Tank, S.; Kondratowicz, A.S.; Davey, R.A.; Rennert, P.D.; Maury, W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J. Virol. 2013, 87, 8327–8341. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.A.; Khomandiak, S.; Ashbrook, A.W.; Weller, R.; Heise, M.T.; Morrison, T.E.; Dermody, T.S. A single-amino-acid polymorphism in Chikungunya virus E2 glycoprotein influences glycosaminoglycan utilization. J. Virol. 2014, 88, 2385–2397. [Google Scholar] [CrossRef]
- Fraisier, C.; Koraka, P.; Belghazi, M.; Bakli, M.; Granjeaud, S.; Pophillat, M.; Lim, S.M.; Osterhaus, A.; Martina, B.; Camoin, L.; et al. Kinetic analysis of mouse brain proteome alterations following Chikungunya virus infection before and after appearance of clinical symptoms. PLoS ONE 2014, 9, e91397. [Google Scholar] [CrossRef]
- Van Duijl-Richter, M.K.; Hoornweg, T.E.; Rodenhuis-Zybert, I.A.; Smit, J.M. Early Events in Chikungunya Virus Infection-From Virus CellBinding to Membrane Fusion. Viruses 2015, 7, 3647–3674. [Google Scholar] [CrossRef]
- Solignat, M.; Gay, B.; Higgs, S.; Briant, L.; Devaux, C. Replication cycle of chikungunya: A re-emerging arbovirus. Virology 2009, 393, 183–197. [Google Scholar] [CrossRef] [PubMed]
- Blaising, J.; Polyak, S.J.; Pecheur, E.I. Arbidol as a broad-spectrum antiviral: An update. Antivir. Res. 2014, 107, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.H.; Maric, M.; Madison, M.N.; Maury, W.; Roller, R.J.; Okeoma, C.M. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology 2013, 438, 37–49. [Google Scholar] [CrossRef]
- Gigante, A.; Gomez-SanJuan, A.; Delang, L.; Li, C.; Bueno, O.; Gamo, A.M.; Priego, E.M.; Camarasa, M.J.; Jochmans, D.; Leyssen, P.; et al. Antiviral activity of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones against chikungunya virus targeting the viral capping nsP1. Antivir. Res. 2017, 144, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar]
- Grudkowska, M.; Zagdanska, B. Multifunctional role of plant cysteine proteinases. Acta Biochim. Pol. 2004, 51, 609–624. [Google Scholar]
- Saisawang, C.; Saitornuang, S.; Sillapee, P.; Ubol, S.; Smith, D.R.; Ketterman, A.J. Chikungunya nsP2 protease is not a papain-like cysteine protease and the catalytic dyad cysteine is interchangeable with a proximal serine. Sci. Rep. 2015, 5, 17125. [Google Scholar] [CrossRef]
- Karpe, Y.A.; Aher, P.P.; Lole, K.S. NTPase and 5′-RNA triphosphatase activities of Chikungunya virus nsP2 protein. PLoS ONE 2011, 6, e22336. [Google Scholar] [CrossRef]
- Rathore, A.P.; Haystead, T.; Das, P.K.; Merits, A.; Ng, M.L.; Vasudevan, S.G. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antivir. Res. 2014, 103, 7–16. [Google Scholar]
- Fros, J.J.; Domeradzka, N.E.; Baggen, J.; Geertsema, C.; Flipse, J.; Vlak, J.M.; Pijlman, G.P. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J. Virol. 2012, 86, 10873–10879. [Google Scholar] [CrossRef]
- De, I.; Fata-Hartley, C.; Sawicki, S.G.; Sawicki, D.L. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 2003, 77, 13106–13116. [Google Scholar] [CrossRef]
- Rungrotmongkol, T.; Nunthaboot, N.; Malaisree, M.; Kaiyawet, N.; Yotmanee, P.; Meeprasert, A.; Hannongbua, S. Molecular insight into the specific binding of ADP-ribose to the nsP3 macro domains of chikungunya and Venezuelan equine encephalitis viruses: Molecular dynamics simulations and free energy calculations. J. Mol. Graph. Model. 2010, 29, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Shin, G.; Yost, S.A.; Miller, M.T.; Elrod, E.J.; Grakoui, A.; Marcotrigiano, J. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc. Natl. Acad. Sci. USA 2012, 109, 16534–16539. [Google Scholar] [CrossRef]
- Wada, Y.; Orba, Y.; Sasaki, M.; Kobayashi, S.; Carr, M.J.; Nobori, H.; Sato, A.; Hall, W.W.; Sawa, H. Discovery of a novel antiviral agent targeting the nonstructural protein 4 (nsP4) of chikungunya virus. Virology 2017, 505, 102–112. [Google Scholar] [CrossRef]
- Tardif, K.D.; Waris, G.; Siddiqui, A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005, 13, 159–163. [Google Scholar] [CrossRef]
- Rathore, A.P.; Ng, M.L.; Vasudevan, S.G. Differential unfolded protein response during Chikungunya and Sindbis virus infection: CHIKV nsP4 suppresses eIF2alpha phosphorylation. Virol. J. 2013, 10, 36. [Google Scholar] [CrossRef]
- Kumar, S.; Mamidi, P.; Kumar, A.; Basantray, I.; Bramha, U.; Dixit, A.; Maiti, P.K.; Singh, S.; Suryawanshi, A.R.; Chattopadhyay, S.; et al. Development of novel antibodies against non-structural proteins nsP1, nsP3 and nsP4 of chikungunya virus: Potential use in basic research. Arch. Virol. 2015, 160, 2749–2761. [Google Scholar] [CrossRef]
- Mishra, P.; Kumar, A.; Mamidi, P.; Kumar, S.; Basantray, I.; Saswat, T.; Das, I.; Nayak, T.K.; Chattopadhyay, S.; Subudhi, B.B.; et al. Inhibition of Chikungunya Virus Replication by 1-[(2-Methylbenzimidazol-1-yl) Methyl]-2-Oxo-Indolin-3-ylidene] Amino] Thiourea(MBZM-N-IBT). Sci. Rep. 2016, 6, 20122. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Jadav, S.S.; Sinha, B.N.; Pastorino, B.; de Lamballerie, X.; Hilgenfeld, R.; Jayaprakash, V. Identification of Pyrazole Derivative as an Antiviral Agent Against Chikungunya Through HTVS. Lett. Drug Des. Discov. 2015, 12, 292–301. [Google Scholar] [CrossRef]
- Stahura, F.L.; Bajorath, J. Virtual screening methods that complement HTS. Comb. Chem. High Throughput Screen. 2004, 7, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Ekins, S.; Mestres, J.; Testa, B. In silico pharmacology for drug discovery: Methods for virtual ligand screening and profiling. Br. J. Pharmacol. 2007, 152, 9–20. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Dash, P.K.; Tiwari, M.; Santhosh, S.R.; Parida, M.; Lakshmana Rao, P.V. RNA interference mediated inhibition of Chikungunya virus replication in mammalian cells. Biochem. Biophys. Res. Commun. 2008, 376, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Singh Kh, D.; Kirubakaran, P.; Nagarajan, S.; Sakkiah, S.; Muthusamy, K.; Velmurgan, D.; Jeyakanthan, J. Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J. Mol. Model. 2012, 18, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Bassetto, M.; De Burghgraeve, T.; Delang, L.; Massarotti, A.; Coluccia, A.; Zonta, N.; Gatti, V.; Colombano, G.; Sorba, G.; Silvestri, R.; et al. Computer-aided identification, design and synthesis of a novel series of compounds with selective antiviral activity against chikungunya virus. Antivir. Res. 2013, 98, 12–18. [Google Scholar] [CrossRef]
- Bhat, A.S.; Ahangar, A.A. Methods for detecting chemical-chemical interaction in toxicology. Toxicol. Mech. Methods 2007, 17, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Ozden, S.; Lucas-Hourani, M.; Ceccaldi, P.E.; Basak, A.; Valentine, M.; Benjannet, S.; Hamelin, J.; Jacob, Y.; Mamchaoui, K.; Mouly, V.; et al. Inhibition of Chikungunya virus infection in cultured human muscle cells by furin inhibitors: Impairment of the maturation of the E2 surface glycoprotein. J. Biol. Chem. 2008, 283, 21899–21908. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Yu, H.; Keller, P.A. Identification of chikungunya virus nsP2 protease inhibitors using structure-base approaches. J. Mol. Graph. Model. 2015, 57, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, T.; Asthana, S.; Bissoyi, A. Molecular Modeling and Docking Study to Elucidate Novel Chikungunya Virus nsP2 Protease Inhibitors. Indian J. Pharm. Sci. 2015, 77, 453–460. [Google Scholar]
- Jadav, S.S.; Jayaprakash, V.; Basu, A.; Sinha, B.N. Chikungunya Protease Domain–High throughput Virtual Screening. Int. J. Med. Health Biomed. Bioeng. Pharm. Eng. 2012, 6, 718–721. [Google Scholar]
- Rothan, H.A.; Bahrani, H.; Abdulrahman, A.Y.; Mohamed, Z.; Teoh, T.C.; Othman, S.; Rashid, N.N.; Rahman, N.A.; Yusof, R. Mefenamic acid in combination with ribavirin shows significant effects in reducing chikungunya virus infection in vitro and in vivo. Antivir. Res. 2016, 127, 50–56. [Google Scholar] [CrossRef]
- Kumar, S.P.; Kapopara, R.G.; Patni, M.I.; Pandya, H.A.; Jasrai, Y.T.; Patel, S.K. Exploring the polymerse activity of Chikungunya viral non-strctural protein-4(nsP4) using molecular modeling, e-pharmacophore and docking studies. Int. J. Pharm. Life Sci. 2012, 3, 1752–1765. [Google Scholar]
- Seyedi, S.S.; Shukri, M.; Hassandarvish, P.; Oo, A.; Muthu, S.E.; Abubakar, S.; Zandi, K. Computational Approach Towards Exploring Potential Anti-Chikungunya Activity of Selected Flavonoids. Sci. Rep. 2016, 6, 24027. [Google Scholar] [CrossRef] [PubMed]
- Pohjala, L.; Utt, A.; Varjak, M.; Lulla, A.; Merits, A.; Ahola, T.; Tammela, P. Inhibitors of alphavirus entry and replication identified with a stable Chikungunya replicon cell line and virus-based assays. PLoS ONE 2011, 6, e28923. [Google Scholar] [CrossRef]
- Sharma, R.; Fatma, B.; Saha, A.; Bajpai, S.; Sistla, S.; Dash, P.K.; Parida, M.; Kumar, P.; Tomar, S. Inhibition of chikungunya virus by picolinate that targets viral capsid protein. Virology 2016, 498, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Santhosh, S.R.; Tiwari, M.; Lakshmana Rao, P.V.; Parida, M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J. Med. Virol. 2010, 82, 817–824. [Google Scholar] [CrossRef]
- Gagarinova, V.M.; Ignat’eva, G.S.; Sinitskaia, L.V.; Ivanova, A.M.; Rodina, M.A.; Tur’eva, A.V. The new chemical preparation arbidol: Its prophylactic efficacy during influenza epidemics. Zhurnal Mikrobiol. Epidemiol. Immunobiol. 1993, 5, 40–43. [Google Scholar]
- Obrosova-Serova, N.P.; Burtseva, E.I.; Nevskii, I.M.; Karmanova, R.I.; Nazarov, V.I.; Pitkenen, A.A.; Slepushkin, A.N. The protective action of arbidol during a rise in respiratory diseases in 1990. Vopr. Virusol. 1991, 36, 380–381. [Google Scholar]
- Di Mola, A.; Peduto, A.; La Gatta, A.; Delang, L.; Pastorino, B.; Neyts, J.; Leyssen, P.; de Rosa, M.; Filosa, R. Structure-activity relationship study of arbidol derivatives as inhibitors of chikungunya virus replication. Bioorg. Med. Chem. 2014, 22, 6014–6025. [Google Scholar] [CrossRef]
- Lin, L.T.; Chen, T.Y.; Lin, S.C.; Chung, C.Y.; Lin, T.C.; Wang, G.H.; Anderson, R.; Lin, C.C.; Richardson, C.D. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol. 2013, 13, 187. [Google Scholar] [CrossRef]
- Wintachai, P.; Thuaud, F.; Basmadjian, C.; Roytrakul, S.; Ubol, S.; Desaubry, L.; Smith, D.R. Assessment of flavaglines as potential chikungunya virus entry inhibitors. Microbiol. Immunol. 2015, 59, 129–141. [Google Scholar] [CrossRef]
- Wichit, S.; Hamel, R.; Bernard, E.; Talignani, L.; Diop, F.; Ferraris, P.; Liegeois, F.; Ekchariyawat, P.; Luplertlop, N.; Surasombatpattana, P.; et al. Imipramine Inhibits Chikungunya Virus Replication in Human Skin Fibroblasts through Interference with Intracellular Cholesterol Trafficking. Sci. Rep. 2017, 7, 3145. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir. Res. 2017, 142, 148–157. [Google Scholar] [CrossRef]
- Wintachai, P.; Kaur, P.; Lee, R.C.; Ramphan, S.; Kuadkitkan, A.; Wikan, N.; Ubol, S.; Roytrakul, S.; Chu, J.J.; Smith, D.R. Activity of andrographolide against chikungunya virus infection. Sci. Rep. 2015, 5, 14179. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Bahrani, H.; Mohamed, Z.; Teoh, T.C.; Shankar, E.M.; Rahman, N.A.; Yusof, R. A combination of doxycycline and ribavirin alleviated chikungunya infection. PLoS ONE 2015, 10, e0126360. [Google Scholar] [CrossRef]
- Khan, M.; Dhanwani, R.; Patro, I.K.; Rao, P.V.; Parida, M.M. Cellular IMPDH enzyme activity is a potential target for the inhibition of Chikungunya virus replication and virus induced apoptosis in cultured mammalian cells. Antivir. Res. 2011, 89, 1–8. [Google Scholar] [CrossRef]
- Briolant, S.; Garin, D.; Scaramozzino, N.; Jouan, A.; Crance, J.M. In vitro inhibition of Chikungunya and Semliki Forest viruses replication by antiviral compounds: Synergistic effect of interferon-alpha and ribavirin combination. Antivir. Res. 2004, 61, 111–117. [Google Scholar] [CrossRef]
- Delang, L.; Segura Guerrero, N.; Tas, A.; Querat, G.; Pastorino, B.; Froeyen, M.; Dallmeier, K.; Jochmans, D.; Herdewijn, P.; Bello, F.; et al. Mutations in the chikungunya virus non-structural proteins cause resistance to favipiravir (T-705), a broad-spectrum antiviral. J. Antimicrob. Chemother. 2014, 69, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Albulescu, I.C.; van Hoolwerff, M.; Wolters, L.A.; Bottaro, E.; Nastruzzi, C.; Yang, S.C.; Tsay, S.C.; Hwu, J.R.; Snijder, E.J.; van Hemert, M.J. Suramin inhibits chikungunya virus replication through multiple mechanisms. Antivir. Res. 2015, 121, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Kaur, P.; Thiruchelvan, M.; Lee, R.C.; Chen, H.; Chen, K.C.; Ng, M.L.; Chu, J.J. Inhibition of chikungunya virus replication by harringtonine, a novel antiviral that suppresses viral protein expression. Antimicrob. Agents Chemother. 2013, 57, 155–167. [Google Scholar] [CrossRef]
- Bourjot, M.; Leyssen, P.; Eydoux, C.; Guillemot, J.C.; Canard, B.; Rasoanaivo, P.; Gueritte, F.; Litaudon, M. Chemical constituents of Anacolosa pervilleana and their antiviral activities. Fitoterapia 2012, 83, 1076–1080. [Google Scholar] [CrossRef]
- Corlay, N.; Delang, L.; Girard-Valenciennes, E.; Neyts, J.; Clerc, P.; Smadja, J.; Gueritte, F.; Leyssen, P.; Litaudon, M. Tigliane diterpenes from Croton mauritianus as inhibitors of chikungunya virus replication. Fitoterapia 2014, 97, 87–91. [Google Scholar] [CrossRef]
- Gupta, D.K.; Kaur, P.; Leong, S.T.; Tan, L.T.; Prinsep, M.R.; Chu, J.J. Anti-Chikungunya viral activities of aplysiatoxin-related compounds from the marine cyanobacterium Trichodesmium erythraeum. Mar. Drugs 2014, 12, 115–127. [Google Scholar] [CrossRef]
- Nothias-Scaglia, L.F.; Pannecouque, C.; Renucci, F.; Delang, L.; Neyts, J.; Roussi, F.; Costa, J.; Leyssen, P.; Litaudon, M.; Paolini, J. Antiviral Activity of Diterpene Esters on Chikungunya Virus and HIV Replication. J. Nat. Prod. 2015, 78, 1277–1283. [Google Scholar] [CrossRef]
- Staveness, D.; Abdelnabi, R.; Near, K.E.; Nakagawa, Y.; Neyts, J.; Delang, L.; Leyssen, P.; Wender, P.A. Inhibition of Chikungunya Virus-Induced Cell Death by Salicylate-Derived Bryostatin Analogues Provides Additional Evidence for a PKC-Independent Pathway. J. Nat. Prod. 2016, 79, 680–684. [Google Scholar] [CrossRef]
- Cruz, D.J.; Bonotto, R.M.; Gomes, R.G.; da Silva, C.T.; Taniguchi, J.B.; No, J.H.; Lombardot, B.; Schwartz, O.; Hansen, M.A.; Freitas-Junior, L.H. Identification of novel compounds inhibiting chikungunya virus-induced cell death by high throughput screening of a kinase inhibitor library. PLoS Negl. Trop. Dis. 2013, 7, e2471. [Google Scholar] [CrossRef]
- Karlas, A.; Berre, S.; Couderc, T.; Varjak, M.; Braun, P.; Meyer, M.; Gangneux, N.; Karo-Astover, L.; Weege, F.; Raftery, M.; et al. A human genome-wide loss-of-function screen identifies effective chikungunya antiviral drugs. Nat. Commun. 2016, 7, 11320. [Google Scholar] [CrossRef]
- Langsjoen, R.M.; Auguste, A.J.; Rossi, S.L.; Roundy, C.M.; Penate, H.N.; Kastis, M.; Schnizlein, M.K.; Le, K.C.; Haller, S.L.; Chen, R.; et al. Host oxidative folding pathways offer novel anti-chikungunya virus drug targets with broad spectrum potential. Antivir. Res. 2017, 143, 246–251. [Google Scholar] [CrossRef]
- Nothias-Scaglia, L.F.; Retailleau, P.; Paolini, J.; Pannecouque, C.; Neyts, J.; Dumontet, V.; Roussi, F.; Leyssen, P.; Costa, J.; Litaudon, M. Jatrophane diterpenes as inhibitors of chikungunya virus replication: Structure-activity relationship and discovery of a potent lead. J. Nat. Prod. 2014, 77, 1505–1512. [Google Scholar] [CrossRef]
- Allard, P.M.; Leyssen, P.; Martin, M.T.; Bourjot, M.; Dumontet, V.; Eydoux, C.; Guillemot, J.C.; Canard, B.; Poullain, C.; Gueritte, F.; et al. Antiviral chlorinated daphnane diterpenoid orthoesters from the bark and wood of Trigonostemon cherrieri. Phytochemistry 2012, 84, 160–168. [Google Scholar] [CrossRef]
- Hwu, J.R.; Kapoor, M.; Tsay, S.C.; Lin, C.C.; Hwang, K.C.; Horng, J.C.; Chen, I.C.; Shieh, F.K.; Leyssen, P.; Neyts, J. Benzouracil-coumarin-arene conjugates as inhibiting agents for chikungunya virus. Antivir. Res. 2015, 118, 103–109. [Google Scholar] [CrossRef]
- Jadav, S.S.; Sinha, B.N.; Hilgenfeld, R.; Pastorino, B.; de Lamballerie, X.; Jayaprakash, V. Thiazolidone derivatives as inhibitors of chikungunya virus. Eur. J. Med. Chem. 2015, 89, 172–178. [Google Scholar] [CrossRef]
- Varghese, F.S.; Kaukinen, P.; Glasker, S.; Bespalov, M.; Hanski, L.; Wennerberg, K.; Kummerer, B.M.; Ahola, T. Discovery of berberine, abamectin and ivermectin as antivirals against chikungunya and other alphaviruses. Antivir. Res. 2016, 126, 117–124. [Google Scholar] [CrossRef]
- Bourai, M.; Lucas-Hourani, M.; Gad, H.H.; Drosten, C.; Jacob, Y.; Tafforeau, L.; Cassonnet, P.; Jones, L.M.; Judith, D.; Couderc, T.; et al. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J. Virol. 2012, 86, 3121–3134. [Google Scholar] [CrossRef]
- Kohli, N.; Westerveld, D.R.; Ayache, A.C.; Verma, A.; Shil, P.; Prasad, T.; Zhu, P.; Chan, S.L.; Li, Q.; Daniell, H. Oral delivery of bioencapsulated proteins across blood-brain and blood-retinal barriers. Mol. Ther. J. Am. Soc. Gene Ther. 2014, 22, 535–546. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant-based vaccines against viruses. Virol. J. 2014, 11, 205. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases? Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Bernard, E.; Solignat, M.; Gay, B.; Chazal, N.; Higgs, S.; Devaux, C.; Briant, L. Endocytosis of chikungunya virus into mammalian cells: Role of clathrin and early endosomal compartments. PLoS ONE 2010, 5, e11479. [Google Scholar] [CrossRef] [PubMed]
- De Lamballerie, X.; Boisson, V.; Reynier, J.C.; Enault, S.; Charrel, R.N.; Flahault, A.; Roques, P.; Le Grand, R. On chikungunya acute infection and chloroquine treatment. Vector Borne Zoonotic Dis. 2008, 8, 837–839. [Google Scholar] [CrossRef]
- Freedman, A.; Steinberg, V.L. Chloroquine in rheumatoid arthritis; a double blindfold trial of treatment for one year. Ann. Rheum. Dis. 1960, 19, 243–250. [Google Scholar] [CrossRef]
- Langridge, W.; Denes, B.; Fodor, I. Cholera toxin B subunit modulation of mucosal vaccines for infectious and autoimmune diseases. Curr. Opin. Investig. Drugs 2010, 11, 919–928. [Google Scholar] [PubMed]
- Pecheur, E.I.; Borisevich, V.; Halfmann, P.; Morrey, J.D.; Smee, D.F.; Prichard, M.; Mire, C.E.; Kawaoka, Y.; Geisbert, T.W.; Polyak, S.J. The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J. Virol. 2016, 90, 3086–3092. [Google Scholar] [CrossRef] [PubMed]
- Delogu, I.; Pastorino, B.; Baronti, C.; Nougairede, A.; Bonnet, E.; de Lamballerie, X. In vitro antiviral activity of arbidol against Chikungunya virus and characteristics of a selected resistant mutant. Antivir. Res. 2011, 90, 99–107. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Gastaminza, P.; Whitten-Bauer, C.; Chisari, F.V. Unbiased probing of the entire hepatitis C virus life cycle identifies clinical compounds that target multiple aspects of the infection. Proc. Natl. Acad. Sci. USA 2010, 107, 291–296. [Google Scholar] [CrossRef]
- Steinmann, J.; Buer, J.; Pietschmann, T.; Steinmann, E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. Br. J. Pharmacol. 2013, 168, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Colpitts, C.C.; Schang, L.M. A small molecule inhibits virion attachment to heparan sulfate- or sialic acid-containing glycans. J. Virol. 2014, 88, 7806–7817. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Sliva, K.; von Rhein, C.; Kummerer, B.M.; Schnierle, B.S. The green tea catechin, epigallocatechin gallate inhibits chikungunya virus infection. Antivir. Res. 2015, 113, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Maliakal, P.; Chen, L.; Meng, X.; Bondoc, F.Y.; Prabhu, S.; Lambert, G.; Mohr, S.; Yang, C.S. Pharmacokinetics of tea catechins after ingestion of green tea and (-)-epigallocatechin-3-gallate by humans: Formation of different metabolites and individual variability. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2002, 11, 1025–1032. [Google Scholar]
- Lu King, M.; Chiang, C.-C.; Ling, H.-C.; Fujita, E.; Ochiai, M.; McPhail, A.T. X-Ray crystal structure of rocaglamide, a novel antileulemic 1H-cyclopenta[b]benzofuran from Aglaia elliptifolia. J. Chem. Soc. Chem. Commun. 1982, 0, 1150–1151. [Google Scholar] [CrossRef]
- Joycharat, N.; Greger, H.; Hofer, O.; Saifah, E. Flavaglines and triterpenoids from the leaves of Aglaia forbesii. Phytochemistry 2008, 69, 206–211. [Google Scholar] [CrossRef]
- Polier, G.; Neumann, J.; Thuaud, F.; Ribeiro, N.; Gelhaus, C.; Schmidt, H.; Giaisi, M.; Kohler, R.; Muller, W.W.; Proksch, P.; et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012, 19, 1093–1104. [Google Scholar] [CrossRef]
- Bernard, Y.; Ribeiro, N.; Thuaud, F.; Turkeri, G.; Dirr, R.; Boulberdaa, M.; Nebigil, C.G.; Desaubry, L. Flavaglines alleviate doxorubicin cardiotoxicity: Implication of Hsp27. PLoS ONE 2011, 6, e25302. [Google Scholar] [CrossRef]
- Ganu, M.A.; Ganu, A.S. Post-chikungunya chronic arthritis—Our experience with DMARDs over two year follow up. J. Assoc. Phys. India 2011, 59, 83–86. [Google Scholar]
- Chopra, A.; Saluja, M.; Venugopalan, A. Effectiveness of chloroquine and inflammatory cytokine response in patients with early persistent musculoskeletal pain and arthritis following chikungunya virus infection. Arthritis Rheumatol. 2014, 66, 319–326. [Google Scholar] [CrossRef]
- Phalen, T.; Kielian, M. Cholesterol is required for infection by Semliki Forest virus. J. Cell Biol. 1991, 112, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.T.; Yu, H.; Keller, P.A. Discovery of in silico hits targeting the nsP3 macro domain of chikungunya virus. J. Mol. Model. 2014, 20, 2216. [Google Scholar] [CrossRef]
- Olagnier, D.; Scholte, F.E.; Chiang, C.; Albulescu, I.C.; Nichols, C.; He, Z.; Lin, R.; Snijder, E.J.; van Hemert, M.J.; Hiscott, J. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J. Virol. 2014, 88, 4180–4194. [Google Scholar] [CrossRef] [PubMed]
- Gigante, A.; Canela, M.D.; Delang, L.; Priego, E.M.; Camarasa, M.J.; Querat, G.; Neyts, J.; Leyssen, P.; Perez-Perez, M.J. Identification of [1,2,3]triazolo[4,5-d]pyrimidin-7(6H)-ones as novel inhibitors of Chikungunya virus replication. J. Med. Chem. 2014, 57, 4000–4008. [Google Scholar] [CrossRef] [PubMed]
- Fong, R.H.; Banik, S.S.; Mattia, K.; Barnes, T.; Tucker, D.; Liss, N.; Lu, K.; Selvarajah, S.; Srinivasan, S.; Mabila, M.; et al. Exposure of epitope residues on the outer face of the chikungunya virus envelope trimer determines antibody neutralizing efficacy. J. Virol. 2014, 88, 14364–14379. [Google Scholar] [CrossRef]
- Pal, P.; Dowd, K.A.; Brien, J.D.; Edeling, M.A.; Gorlatov, S.; Johnson, S.; Lee, I.; Akahata, W.; Nabel, G.J.; Richter, M.K.; et al. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog. 2013, 9, e1003312. [Google Scholar] [CrossRef] [PubMed]
- Mathew, D.; Hsu, W.L. Antiviral potential of curcumin. Sci. Direct 2018, 40, 692–699. [Google Scholar] [CrossRef]
- Von Rhein, C.; Weidner, T.; Henss, L.; Martin, J.; Weber, C.; Sliva, K.; Schnierle, B.S. Curcumin and Boswellia serrata gum resin extract inhibit chikungunya and vesicular stomatitis virus infections in vitro. Antivir. Res. 2016, 125, 51–57. [Google Scholar] [CrossRef]
- Liu, W.; Zhai, Y.; Heng, X.; Che, F.Y.; Chen, W.; Sun, D.; Zhai, G. Oral bioavailability of curcumin: Problems and advancements. J. Drug Target. 2016, 24, 694–702. [Google Scholar] [CrossRef]
- Calabrese, C.; Berman, S.H.; Babish, J.G.; Ma, X.; Shinto, L.; Dorr, M.; Wells, K.; Wenner, C.A.; Standish, L.J. A phase I trial of andrographolide in HIV positive patients and normal volunteers. Phytother. Res. PTR 2000, 14, 333–338. [Google Scholar] [CrossRef]
- Chen, J.X.; Xue, H.J.; Ye, W.C.; Fang, B.H.; Liu, Y.H.; Yuan, S.H.; Yu, P.; Wang, Y.Q. Activity of andrographolide and its derivatives against influenza virus in vivo and in vitro. Biol. Pharm. Bull. 2009, 32, 1385–1391. [Google Scholar] [CrossRef]
- Lee, J.C.; Tseng, C.K.; Young, K.C.; Sun, H.Y.; Wang, S.W.; Chen, W.C.; Lin, C.K.; Wu, Y.H. Andrographolide exerts anti-hepatitis C virus activity by up-regulating haeme oxygenase-1 via the p38 MAPK/Nrf2 pathway in human hepatoma cells. Br. J. Pharmacol. 2014, 171, 237–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, X.F.; Nguyen, V.S.; Yang, P.; Zhou, J.; Gao, M.; Li, Z.; Lim, T.K.; He, Y.; Ong, C.S.; et al. A quantitative chemical proteomics approach to profile the specific cellular targets of andrographolide, a promising anticancer agent that suppresses tumor metastasis. Mol. Cell. Proteom. MCP 2014, 13, 876–886. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Khurana, N.; Pounikar, Y.; Gajbhiye, A.; Kharya, M.D. Enhancement of absorption and hepatoprotective potential through soya-phosphatidylcholine-andrographolide vesicular system. J. Liposome Res. 2013, 23, 110–118. [Google Scholar] [CrossRef]
- Turner, T.L.; Kopp, B.T.; Paul, G.; Landgrave, L.C.; Hayes, D., Jr.; Thompson, R. Respiratory syncytial virus: Current and emerging treatment options. ClinicoEcon. Outcomes Res. CEOR 2014, 6, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Paeshuyse, J.; Dallmeier, K.; Neyts, J. Ribavirin for the treatment of chronic hepatitis C virus infection: A review of the proposed mechanisms of action. Curr. Opin. Virol. 2011, 1, 590–598. [Google Scholar] [CrossRef]
- Cline, J.C.; Nelson, J.D.; Gerzon, K.; Williams, R.H.; Delong, D.C. In vitro antiviral activity of mycophenolic acid and its reversal by guanine-type compounds. Appl. Microbiol. 1969, 18, 14–20. [Google Scholar]
- Smee, D.F.; Bray, M.; Huggins, J.W. Antiviral activity and mode of action studies of ribavirin and mycophenolic acid against orthopoxviruses in vitro. Antivir. Chem. Chemother. 2001, 12, 327–335. [Google Scholar] [CrossRef]
- Diamond, M.S.; Zachariah, M.; Harris, E. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 2002, 304, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Termsarasab, P.; Katirji, B. Opportunistic infections in myasthenia gravis treated with mycophenolate mofetil. J. Neuroimmunol. 2012, 249, 83–85. [Google Scholar] [CrossRef]
- Rashad, A.A.; Neyts, J.; Leyssen, P.; Keller, P.A. A reassessment of mycophenolic acid as a lead compound for the development of inhibitors of chikungunya virus replication. Tetrahedron 2018, 74, 1294–1306. [Google Scholar] [CrossRef]
- Deneau, D.G.; Farber, E.M. The treatment of psoriasis with azaribine. Dermatologica 1975, 151, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Crutcher, W.A.; Moschella, S.L. Double-blind controlled crossover high-dose study of Azaribine in psoriasis. Br. J. Dermatol. 1975, 92, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Rada, B.; Dragun, M. Antiviral action and selectivity of 6-azauridine. Ann. N. Y. Acad. Sci. 1977, 284, 410–417. [Google Scholar] [CrossRef]
- Scholte, F.E.; Tas, A.; Martina, B.E.; Cordioli, P.; Narayanan, K.; Makino, S.; Snijder, E.J.; van Hemert, M.J. Characterization of synthetic Chikungunya viruses based on the consensus sequence of recent E1–226V isolates. PLoS ONE 2013, 8, e71047. [Google Scholar] [CrossRef]
- Plevova, J.; Janku, I.; Seda, M. Toxicity of 6-azauridine triacetate. Toxicol. Appl. Pharmacol. 1970, 17, 511–518. [Google Scholar] [CrossRef]
- Caroline, A.L.; Powell, D.S.; Bethel, L.M.; Oury, T.D.; Reed, D.S.; Hartman, A.L. Broad spectrum antiviral activity of favipiravir (T-705): Protection from highly lethal inhalational Rift Valley Fever. PLoS Negl. Trop. Dis. 2014, 8, e2790. [Google Scholar] [CrossRef]
- Smither, S.J.; Eastaugh, L.S.; Steward, J.A.; Nelson, M.; Lenk, R.P.; Lever, M.S. Post-exposure efficacy of oral T-705 (Favipiravir) against inhalational Ebola virus infection in a mouse model. Antivir. Res. 2014, 104, 153–155. [Google Scholar] [CrossRef]
- Furuta, Y.; Gowen, B.B.; Takahashi, K.; Shiraki, K.; Smee, D.F.; Barnard, D.L. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antivir. Res. 2013, 100, 446–454. [Google Scholar] [CrossRef]
- Oestereich, L.; Rieger, T.; Ludtke, A.; Ruibal, P.; Wurr, S.; Pallasch, E.; Bockholt, S.; Krasemann, S.; Munoz-Fontela, C.; Gunther, S. Efficacy of Favipiravir Alone and in Combination With Ribavirin in a Lethal, Immunocompetent Mouse Model of Lassa Fever. J. Infect. Dis. 2016, 213, 934–938. [Google Scholar] [CrossRef] [PubMed]
- Parashar, D.; Paingankar, M.S.; Kumar, S.; Gokhale, M.D.; Sudeep, A.B.; Shinde, S.B.; Arankalle, V.A. Administration of E2 and NS1 siRNAs inhibit chikungunya virus replication in vitro and protects mice infected with the virus. PLoS Negl. Trop. Dis. 2013, 7, e2405. [Google Scholar] [CrossRef]
- Lam, S.; Chen, K.C.; Ng, M.M.; Chu, J.J. Expression of plasmid-based shRNA against the E1 and nsP1 genes effectively silenced Chikungunya virus replication. PLoS ONE 2012, 7, e46396. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef]
- Lam, S.; Chen, H.; Chen, C.K.; Min, N.; Chu, J.J. Antiviral Phosphorodiamidate Morpholino Oligomers are Protective against Chikungunya Virus Infection on Cell-based and Murine Models. Sci. Rep. 2015, 5, 12727. [Google Scholar] [CrossRef]
- Borah, A.; Paul, R.; Choudhury, S.; Choudhury, A.; Bhuyan, B.; Das Talukdar, A.; Dutta Choudhury, M.; Mohanakumar, K.P. Neuroprotective potential of silymarin against CNS disorders: Insight into the pathways and molecular mechanisms of action. CNS Neurosci. Ther. 2013, 19, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Wagoner, J.; Negash, A.; Kane, O.J.; Martinez, L.E.; Nahmias, Y.; Bourne, N.; Owen, D.M.; Grove, J.; Brimacombe, C.; McKeating, J.A.; et al. Multiple effects of silymarin on the hepatitis C virus lifecycle. Hepatology 2010, 51, 1912–1921. [Google Scholar] [CrossRef]
- Lani, R.; Hassandarvish, P.; Chiam, C.W.; Moghaddam, E.; Chu, J.J.; Rausalu, K.; Merits, A.; Higgs, S.; Vanlandingham, D.; Abu Bakar, S.; et al. Antiviral activity of silymarin against chikungunya virus. Sci. Rep. 2015, 5, 11421. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Q.; Song, D.F.; Li, R.Q.; Yang, H.; Qi, L.W.; Xin, G.Z.; Wang, D.Q.; Song, H.P.; Chen, J.; Hao, H.; et al. Identification of effective combinatorial markers for quality standardization of herbal medicines. J. Chromatogr. A 2014, 1345, 78–85. [Google Scholar] [CrossRef]
- Ho, Y.J.; Wang, Y.M.; Lu, J.W.; Wu, T.Y.; Lin, L.I.; Kuo, S.C.; Lin, C.C. Suramin Inhibits Chikungunya Virus Entry and Transmission. PLoS ONE 2015, 10, e0133511. [Google Scholar] [CrossRef] [PubMed]
- Hwu, J.R.; Gupta, N.K.; Tsay, S.C.; Huang, W.C.; Albulescu, I.C.; Kovacikova, K.; van Hemert, M.J. Bis(benzofuran-thiazolidinone)s and bis(benzofuran-thiazinanone)s as inhibiting agents for chikungunya virus. Antivir. Res. 2017, 146, 96–101. [Google Scholar] [CrossRef]
- Lu, J.W.; Hsieh, P.S.; Lin, C.C.; Hu, M.K.; Huang, S.M.; Wang, Y.M.; Liang, C.Y.; Gong, Z.; Ho, Y.J. Synergistic effects of combination treatment using EGCG and suramin against the chikungunya virus. Biochem. Biophys. Res. Commun. 2017, 491, 595–602. [Google Scholar] [CrossRef]
- Efferth, T.; Sauerbrey, A.; Halatsch, M.E.; Ross, D.D.; Gebhart, E. Molecular modes of action of cephalotaxine and homoharringtonine from the coniferous tree Cephalotaxus hainanensis in human tumor cell lines. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2003, 367, 56–67. [Google Scholar] [CrossRef]
- Zhang, X.; Fugere, M.; Day, R.; Kielian, M. Furin processing and proteolytic activation of Semliki Forest virus. J. Virol. 2003, 77, 2981–2989. [Google Scholar] [CrossRef]
- Craig, J.; Galban, S.G.; Van Dort, M.E.; Lucker, G.D.; Bhojani, M.S.; Rehemtulla, A.; Ross, B.D. Progress in Molecular Biology and Translational Science. Molecular Biology of Cancer: Translation to the Clinic, 1 ed.; Ruddon, R.W., Ed.; Elsevier Science Publishing Co. Inc.: Atlanta, GA, USA, 2010; Volume 95. [Google Scholar]
- Mehla, R.; Bivalkar-Mehla, S.; Zhang, R.; Handy, I.; Albrecht, H.; Giri, S.; Nagarkatti, P.; Nagarkatti, M.; Chauhan, A. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS ONE 2010, 5, e11160. [Google Scholar] [CrossRef]
- Van Apeldoorn, M.E.; van Egmond, H.P.; Speijers, G.J.; Bakker, G.J. Toxins of cyanobacteria. Mol. Nutr. Food Res. 2007, 51, 7–60. [Google Scholar] [CrossRef] [PubMed]
- Staveness, D.; Abdelnabi, R.; Schrier, A.J.; Loy, B.A.; Verma, V.A.; DeChristopher, B.A.; Near, K.E.; Neyts, J.; Delang, L.; Leyssen, P.; et al. Simplified Bryostatin Analogues Protect Cells from Chikungunya Virus-Induced Cell Death. J. Nat. Prod. 2016, 79, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Mochly-Rosen, D.; Das, K.; Grimes, K.V. Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug Discov. 2012, 11, 937–957. [Google Scholar] [CrossRef]
- Reid, S.P.; Tritsch, S.R.; Kota, K.; Chiang, C.Y.; Dong, L.; Kenny, T.; Brueggemann, E.E.; Ward, M.D.; Cazares, L.H.; Bavari, S. Sphingosine kinase 2 is a chikungunya virus host factor co-localized with the viral replication complex. Emerg. Microbes Infect. 2015, 4, e61. [Google Scholar] [CrossRef]
- Geller, R.; Taguwa, S.; Frydman, J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim. Biophys. Acta 2012, 1823, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.G.; Siripanyaphinyo, U.; Tumkosit, U.; Noranate, N.; A-nuegoonpipat, A.; Pan, Y.; Kameoka, M.; Kurosu, T.; Ikuta, K.; Takeda, N.; et al. Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol. J. 2012, 9, 114. [Google Scholar] [CrossRef]
- Das, I.; Basantray, I.; Mamidi, P.; Nayak, T.K.; Pratheek, B.M.; Chattopadhyay, S.; Chattopadhyay, S. Heat shock protein 90 positively regulates Chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS ONE 2014, 9, e100531. [Google Scholar] [CrossRef]
- Jhaveri, K.; Modi, S. Ganetespib: Research and clinical development. OncoTargets Ther. 2015, 8, 1849–1858. [Google Scholar]
- Belardelli, F. Role of interferons and other cytokines in the regulation of the immune response. APMIS Acta Pathol. Microbiol. Immunol. Scand. 1995, 103, 161–179. [Google Scholar] [CrossRef]
- Friedman, R.M. Clinical uses of interferons. Br. J. Clin. Pharmacol. 2008, 65, 158–162. [Google Scholar] [CrossRef]
- Brehin, A.C.; Casademont, I.; Frenkiel, M.P.; Julier, C.; Sakuntabhai, A.; Despres, P. The large form of human 2’,5’-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology 2009, 384, 216–222. [Google Scholar] [CrossRef]
- Schilte, C.; Couderc, T.; Chretien, F.; Sourisseau, M.; Gangneux, N.; Guivel-Benhassine, F.; Kraxner, A.; Tschopp, J.; Higgs, S.; Michault, A.; et al. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J. Exp. Med. 2010, 207, 429–442. [Google Scholar] [CrossRef]
- Rudd, P.A.; Wilson, J.; Gardner, J.; Larcher, T.; Babarit, C.; Le, T.T.; Anraku, I.; Kumagai, Y.; Loo, Y.M.; Gale, M., Jr.; et al. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J. Virol. 2012, 86, 9888–9898. [Google Scholar] [CrossRef]
- Gallegos, K.M.; Drusano, G.L.; DZ, D.A.; Brown, A.N. Chikungunya Virus: In Vitro Response to Combination Therapy With Ribavirin and Interferon Alfa 2a. J. Infect. Dis. 2016, 214, 1192–1197. [Google Scholar] [CrossRef]
- Seo, J.Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 2011, 10, 534–539. [Google Scholar] [CrossRef]
- Teng, T.S.; Foo, S.S.; Simamarta, D.; Lum, F.M.; Teo, T.H.; Lulla, A.; Yeo, N.K.; Koh, E.G.; Chow, A.; Leo, Y.S.; et al. Viperin restricts chikungunya virus replication and pathology. J. Clin. Investig. 2012, 122, 4447–4460. [Google Scholar] [CrossRef] [PubMed]
- Phillips, B.M.; Hartnagel, R.E.; Kraus, P.J.; Tamayo, R.P.; Fonseca, E.H.; Kowalski, R.L. Systemic toxicity of polyinosinic acid: Polycytidylic acid in rodents and dogs. Toxicol. Appl. Pharmacol. 1971, 18, 220–230. [Google Scholar] [CrossRef]
- Priya, R.; Patro, I.K.; Parida, M.M. TLR3 mediated innate immune response in mice brain following infection with Chikungunya virus. Virus Res. 2014, 189, 194–205. [Google Scholar] [CrossRef]
- Sumpter, R., Jr.; Loo, Y.M.; Foy, E.; Li, K.; Yoneyama, M.; Fujita, T.; Lemon, S.M.; Gale, M., Jr. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 2005, 79, 2689–2699. [Google Scholar] [CrossRef]
- Matsumiya, T.; Stafforini, D.M. Function and regulation of retinoic acid-inducible gene-I. Crit. Rev. Immunol. 2010, 30, 489–513. [Google Scholar] [CrossRef]
- Geller, R.; Vignuzzi, M.; Andino, R.; Frydman, J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 2007, 21, 195–205. [Google Scholar] [CrossRef]
- Madureira, A.M.; Ascenso, J.R.; Valdeira, L.; Duarte, A.; Frade, J.P.; Freitas, G.; Ferreira, M.J. Evaluation of the antiviral and antimicrobial activities of triterpenes isolated from Euphorbia segetalis. Nat. Prod. Res. 2003, 17, 375–380. [Google Scholar] [CrossRef]
- Devappa, R.K.; Makkar, H.P.S.; Becker, K. Jatropha Diterpenes: A Review. J. Am. Oil Chem. Soc. 2011, 88, 301–322. [Google Scholar] [CrossRef]
- Patil, D.; Roy, S.; Dahake, R.; Rajopadhye, S.; Kothari, S.; Deshmukh, R.; Chowdhary, A. Evaluation of Jatropha curcas Linn. leaf extracts for its cytotoxicity and potential to inhibit hemagglutinin protein of influenza virus. Indian J. Virol. Off. Organ Indian Virol. Soc. 2013, 24, 220–226. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, R.H.; Wang, F.; Dong, Z.J.; Yang, L.M.; Zheng, Y.T.; Liu, J.K. Daphnane diterpenoids isolated from Trigonostemon thyrsoideum as HIV-1 antivirals. Phytochemistry 2010, 71, 1879–1883. [Google Scholar] [CrossRef]
- Kothandan, S.; Swaminathan, R. Evaluation of in vitro antiviral activity of Vitex Negundo L., Hyptis suaveolens (L) poit., Decalepis hamiltonii Wight & Arn., to Chikungunya virus. Asian Pac. J. Trop. Dis. 2014, 4, S111–S115. [Google Scholar]
- Murali, K.S.; Sivasubramanian, S.; Vincent, S.; Murugan, S.B.; Giridaran, B.; Dinesh, S.; Gunasekaran, P.; Krishnasamy, K.; Sathishkumar, R. Anti-chikungunya activity of luteolin and apigenin rich fraction from Cynodon dactylon. Asian Pac. J. Trop. Med. 2015, 8, 352–358. [Google Scholar] [CrossRef]
- Abdel-Aal, M.T. Synthesis and anti-hepatitis B activity of new substituted uracil and thiouracil glycosides. Arch. Pharmacal Res. 2010, 33, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Ordonez, P.; Hamasaki, T.; Isono, Y.; Sakakibara, N.; Ikejiri, M.; Maruyama, T.; Baba, M. Anti-human immunodeficiency virus type 1 activity of novel 6-substituted 1-benzyl-3-(3,5-dimethylbenzyl)uracil derivatives. Antimicrob. Agents Chemother. 2012, 56, 2581–2589. [Google Scholar] [CrossRef]
- Kostova, I.; Raleva, S.; Genova, P.; Argirova, R. Structure-Activity Relationships of Synthetic Coumarins as HIV-1 Inhibitors. Bioinorg. Chem. Appl. 2006. [Google Scholar] [CrossRef] [PubMed]
- Gomord, V.; Fitchette, A.C.; Menu-Bouaouiche, L.; Saint-Jore-Dupas, C.; Plasson, C.; Michaud, D.; Faye, L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 2010, 8, 564–587. [Google Scholar] [CrossRef] [PubMed]
- Tillhon, M.; Guaman Ortiz, L.M.; Lombardi, P.; Scovassi, A.I. Berberine: New perspectives for old remedies. Biochem. Pharmacol. 2012, 84, 1260–1267. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Hourani, M.; Lupan, A.; Despres, P.; Thoret, S.; Pamlard, O.; Dubois, J.; Guillou, C.; Tangy, F.; Vidalain, P.O.; Munier-Lehmann, H. A phenotypic assay to identify Chikungunya virus inhibitors targeting the nonstructural protein nsP2. J. Biomol. Screen. 2013, 18, 172–179. [Google Scholar] [CrossRef]
- Chan, Y.-H.; Lum, F.-M.; Ng, L. Limitations of Current in Vivo Mouse Models for the Study of Chikungunya Virus Pathogenesis. Med. Sci. 2015, 3, 64–77. [Google Scholar] [CrossRef]
- Labadie, K.; Larcher, T.; Joubert, C.; Mannioui, A.; Delache, B.; Brochard, P.; Guigand, L.; Dubreil, L.; Lebon, P.; Verrier, B.; et al. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J. Clin. Investig. 2010, 120, 894–906. [Google Scholar] [CrossRef]
- Akkina, R. New generation humanized mice for virus research: Comparative aspects and future prospects. Virology 2013, 435, 14–28. [Google Scholar] [CrossRef]
- Goertz, G.P.; Vogels, C.B.F.; Geertsema, C.; Koenraadt, C.J.M.; Pijlman, G.P. Mosquito co-infection with Zika and chikungunya virus allows simultaneous transmission without affecting vector competence of Aedes aegypti. PLoS Negl. Trop. Dis. 2017, 11, e0005654. [Google Scholar] [CrossRef] [PubMed]
- Le Coupanec, A.; Tchankouo-Nguetcheu, S.; Roux, P.; Khun, H.; Huerre, M.; Morales-Vargas, R.; Enguehard, M.; Lavillette, D.; Misse, D.; Choumet, V. Co-Infection of Mosquitoes with Chikungunya and Dengue Viruses Reveals Modulation of the Replication of Both Viruses in Midguts and Salivary Glands of Aedes aegypti Mosquitoes. Int. J. Mol. Sci. 2017, 18, 1708. [Google Scholar] [CrossRef]
- Farias, K.J.; Machado, P.R.; Muniz, J.A.; Imbeloni, A.A.; da Fonseca, B.A. Antiviral activity of chloroquine against dengue virus type 2 replication in Aotus monkeys. Viral Immunol. 2015, 28, 161–169. [Google Scholar] [CrossRef]
- Crance, J.M.; Scaramozzino, N.; Jouan, A.; Garin, D. Interferon, ribavirin, 6-azauridine and glycyrrhizin: Antiviral compounds active against pathogenic flaviviruses. Antivir. Res. 2003, 58, 73–79. [Google Scholar] [CrossRef]
- Zandi, K.; Teoh, B.T.; Sam, S.S.; Wong, P.F.; Mustafa, M.R.; Abubakar, S. Antiviral activity of four types of bioflavonoid against dengue virus type-2. Virol. J. 2011, 8, 560. [Google Scholar] [CrossRef]
- Basavannacharya, C.; Vasudevan, S.G. Suramin inhibits helicase activity of NS3 protein of dengue virus in a fluorescence-based high throughput assay format. Biochem. Biophys. Res. Commun. 2014, 453, 539–544. [Google Scholar] [CrossRef]
- Carneiro, B.M.; Batista, M.N.; Braga, A.C.; Nogueira, M.L.; Rahal, P. The green tea molecule EGCG inhibits Zika virus entry. Virology 2016, 496, 215–218. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Osman, H. Novel targets for anti-retroviral therapy. J. Infect. 2009, 59, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Schito, M.L.; Goel, A.; Song, Y.; Inman, J.K.; Fattah, R.J.; Rice, W.G.; Turpin, J.A.; Sher, A.; Appella, E. In vivo antiviral activity of novel human immunodeficiency virus type 1 nucleocapsid p7 zinc finger inhibitors in a transgenic murine model. AIDS Res. Hum. Retrovir. 2003, 19, 91–101. [Google Scholar] [CrossRef]
| Sl No. | Compound | Structure | Target | References |
|---|---|---|---|---|
| 1 | N-butyl-9-[3,4-dipropoxy-5-(propoxymethyl) oxolan-2-yl]purin-6-amine | 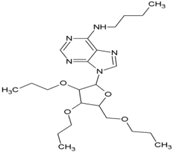 | nsP2 Protease | [56] |
| 2 | ASN 01541696 | 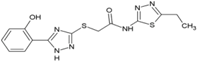 | nsP2 Protease | [59] |
| 3 | (2E)-3-(4-tert-butylphenyl) methylidene]prop-2-Enehydrazide | 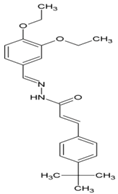 | nsP2 Protease | [60] |
| 4 | NCL 61610 | 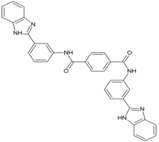 | nsP2 Protease | [63] |
| 5 | CID_5808891 |  | nsP2 Protease | [64] |
| 6 | ZINC67680487 | 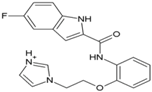 | nsP2 Protease | [65] |
| 7 | ZINC04725220 | 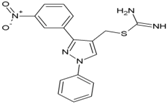 | nsP2 Protease | [54] |
| 8 | Doxycycline |  | nsP2, E2 | [66] |
| 9 | BILN2106 | 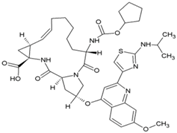 | nsP4 | [67] |
| 10 | JTK 109 | 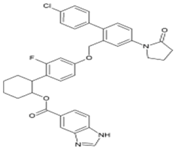 | nsP4 | [67] |
| 11 | Baicalin | 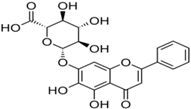 | nsP3 | [68] |
| 12 | Quercetagetin |  | nsP3 | [68] |
| 13 | Naringenin | 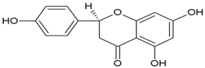 | nsP3 | [69] |
| 14 | Picolinic acid | 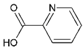 | Capsid protein | [70] |
| Sl No. | Compound | Structure | EC50 | CC50 | Reference |
|---|---|---|---|---|---|
| Compounds interfering with CHIKV internalization | |||||
| 1 | Chloroquine | 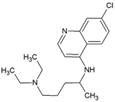 | 17.2 µM | 260 µM | [71] |
| 2 | Arbidol/Umifenovir | 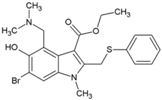 | 12.2 µM | 376 µM | [72,73] |
| 3 | tert-Butyl 5-(hydroxymethyl)-1-methyl-2-((2,6-dichloro phenyl sulfynyl)methyl)-1H-indol-3-carboxylate | 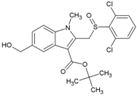 | 30 ± 4 µM | 397 ± 24 µM | [74] |
| 4 | tert-Butyl 5-(hydroxyl methyl)-1-methyl-2-((2-trifluoromethyl phenyl sulfynyl) methyl)-1H-indol-3-carboxylate | 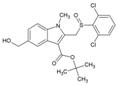 | 32 ± 1.1 µM (Vero cells) | >468 µM (MTS/PMS) | [74] |
| 5 | Chlorpromazine | 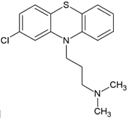 | NR | NR | [69] |
| 6 | EGCG | 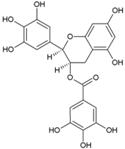 | NR | NR | [75] |
| 7 | FL3(Flavagline) | 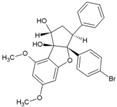 | 22.4 nM(HEK293T) | 118.77nM(MTT) | [76] |
| 8 | Mefenamic acid | 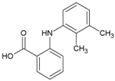 | 13μM(VeroE6) | >100 μM(MTT) | [66] |
| 9 | Meclofenamic acid | 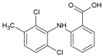 | 18 μM(VeroE6) | >100 μM(MTT) | [66] |
| 10 | U18666A | 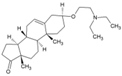 | NR | NR | [77] |
| 11 | Imipramine |  | NR | NR | [77] |
| 12 | Curcumin |  | 3.89 μM(Hela, BHK21, VeroE6) | 11.6 μM(Trypan blue) | [78] |
| 13 | Demethoxycurcumin |  | 0.89μM (Hela, BHK21, VeroE6) | 13.2 μM(Trypan blue) | [78] |
| Compounds inhibiting CHIKV genome replication | |||||
| 14 | Andrographolide | 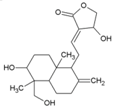 | 77 µM | 1098 µM (MTT Almarblue assay), | [79] |
| 15 | Ribavirin | 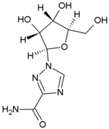 | 341.1 µM | 30.7 mM | [80] |
| 16 | Mycophenolic acid |  | 0.1 µM (VeroE6) | 30 µM (MTT) | [81] |
| 17 | 6-Azauridine |  | 0.2 μg/mL (VeroE6) | 51 μg/mL (Trypan blue) | [82] |
| 18 | Favipiravir | 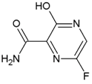 | 5.9 ± 3.3 µM (VeroE6) | NR | [83] |
| 19 | T-1105 | 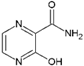 | 2.8 ± 0.3 (Vero E6) | NR | [83] |
| 20 | Suramin | 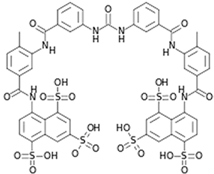 | 8.8–28.9 µM (VeroE6, BHK21) | 700 µM (MTS) | [84] |
| Compounds inhibiting CHIKV protein translation | |||||
| 21 | Harringtonine | 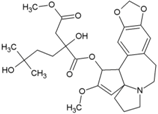 | 0.24 µM (BHK21) | NR | [85] |
| Compounds targeting host factors to inhibit CHIKV | |||||
| 22 | dec-RVKR-cmk | 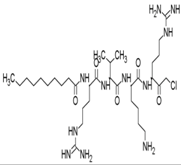 | NR | NR | [62] |
| 23 | Prostratin | 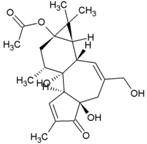 | 5.7µM (VeroE6) | NR | [86] |
| 24 | 12-O-tetradecanoylphorbol 13-acetate | 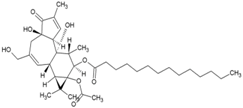 | 2.9 nM (Vero Cells) | 5.7 µM (MTT) | [86] |
| 25 | 12-O-decanoyl-7-hydroperoxy-phorbol-5-ene-13-acetate | 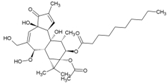 | NR | NR | [87] |
| 26 | Debromoaplysiatoxin |  | 1.3 µM (STCRH30) | 13.9 µM (Almar Blue) | [88] |
| 27 | 3-methoxy debromoaplysiatoxin | 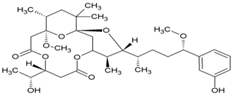 | 2.7 µM (STCRH30) | 24.8 µM (AlmarBlue) | [88] |
| 28 | Phorbol-12, 13-didecanoate | 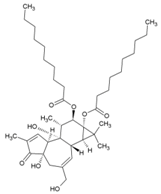 | 6.0 ± 0.9 μM | NR | [89] |
| 29 | Bryostatin-21 |  | 2.2 μM | >50 μM | [90] |
| 30 | 2-(1-hydroxy-2-methylpropyl)-N-[4-(propan-2-yl)phenyl]-1,3-thiazole-4-carboxamide |  | 2.2 µM (HuH7) | >50 µM (Resazurin) | [91] |
| 31 | Geldanamycin | 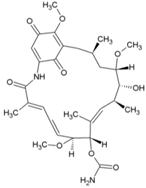 | NR | NR | [43] |
| 32 | Bafilomycin | 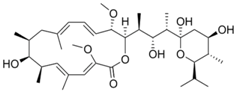 | 0.33 nM (HEK293T | 0.003 µM (WST-1 assay) | [92] |
| 33 | Pimozide |  | 0.28 μM(HEK293T) | 19.18 μM (WST-1 assay) | [92] |
| 34 | 5-tetradecyloxy-2-furoic acid |  | 0.15 μM (HEK293T) | >60 μM(WST-1 assay) | [92] |
| 35 | Cerulenin | 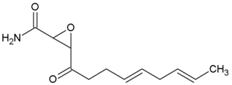 | 3 µM (HEK293T) | 7.57 µM (WST-1 assay) | [92] |
| 36 | Tivozanib |  | 0.8 μM (HEK293T) | 8.34 μM(WST-1 assay) | [92] |
| 37 | Anacardic acid | 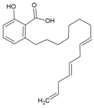 | 0.58 μM (HEK293T) | 2.68 μM(WST-1 assay) | [92] |
| 38 | 16F16 | 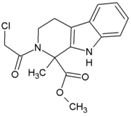 | 6.6 μM (HEK293T) | 8.9 μM (Almarblue assay) | [93] |
| 39 | PACMA31 | 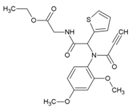 | 12.1 μM (HEK293T) | 12.2 μM (Almarblue assay) | [93] |
| 40 | Auranofin |  | 27.0 μM (HEK293T) | 31.1 μM (Almarblue assay) | [93] |
| 41 | EN460 | 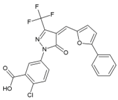 | 1.0 μM (HEK293T) | 1.6 μM (Almarblue assay) | [93] |
| Compound with unknown CHIKV target | |||||
| 42 | Lupenone |  | 77 µM (Vero) | >235 µM | [86] |
| 43 | β-amyrone | 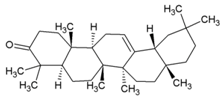 | 86 (Vero) | [86] | |
| 44 | Jatropha ester | 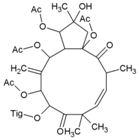 | 0.76 ± 0.14 µM | 159 µM | [94] |
| 45 | Trigocherrin A |  | 1.5 ± 0.6 µM (VeroE6) | 35 ± 8 µM | [95] |
| 46 | Trigocherrin B | 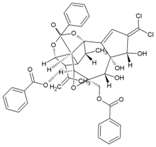 | 2.6 ± 0.7 µM (VeroE6) | 93 ± 3 µM | [95] |
| 47 | Apigenin |  | 70.8 µM | >200 µM | [69] |
| 48 | 5-Ethyl-3-(3′-isopropoxyphenyl)-3H-[1,2,3] triazolo [4,5-d]-pyrimidin-7(6H)-one |  | 3 ± 1 µM (VeroE6) | >668 µM | [38] |
| 49 | 2-Oxo-4-([(4-oxo-3,4-dihydroquinazolin-2-yl)thio]methyl)-2H-chromen-7-yl4-methylbenzenesulfonate |  | 10.2 µM (VeroE6) | 117 µM | [96] |
| 50 | 5-[(2-Methylphenyl)-methylidene]-2-sulfanylidene-1,3-thiazolidin-4-one |  | 0.42 μM(VeroE6) | >100 μM | [97] |
| 51 | MBZM-N-IBT | 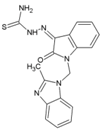 | 38.68 µM (S27),58.33 µM (DRDE-06) (VeroE6) | >800 μM | [52] |
| 52 | Abamectin |  | 1.5 ± 0.6 µM (BHK21) | 28.2 ± 1.1 µM | [98] |
| 53 | Ivermectin |  | 0.6 ± 0.1 µM (BHK21) | 37.9 ± 7.6 µM | [98] |
| 54 | Berberin | 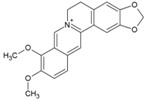 | 1.8 ± 0.5 µM (BHK21) | >100 µM | [98] |
| 55 | 5-chloro-N-{4-[(1E)-1-{2-[(2-phenylcyclopropyl) carbonyl] hydrazinylidene} ethyl] phenyl} thiophene-2-carboxamide | 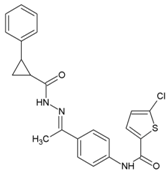 | 1.5 μM | >200 μM | [62] |
| 56 | ID1452-2 | 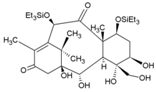 | NR | NR | [99] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subudhi, B.B.; Chattopadhyay, S.; Mishra, P.; Kumar, A. Current Strategies for Inhibition of Chikungunya Infection. Viruses 2018, 10, 235. https://doi.org/10.3390/v10050235
Subudhi BB, Chattopadhyay S, Mishra P, Kumar A. Current Strategies for Inhibition of Chikungunya Infection. Viruses. 2018; 10(5):235. https://doi.org/10.3390/v10050235
Chicago/Turabian StyleSubudhi, Bharat Bhusan, Soma Chattopadhyay, Priyadarsee Mishra, and Abhishek Kumar. 2018. "Current Strategies for Inhibition of Chikungunya Infection" Viruses 10, no. 5: 235. https://doi.org/10.3390/v10050235
APA StyleSubudhi, B. B., Chattopadhyay, S., Mishra, P., & Kumar, A. (2018). Current Strategies for Inhibition of Chikungunya Infection. Viruses, 10(5), 235. https://doi.org/10.3390/v10050235





