T = 4 Icosahedral HIV-1 Capsid As an Immunogenic Vector for HIV-1 V3 Loop Epitope Display
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and Protein Purification
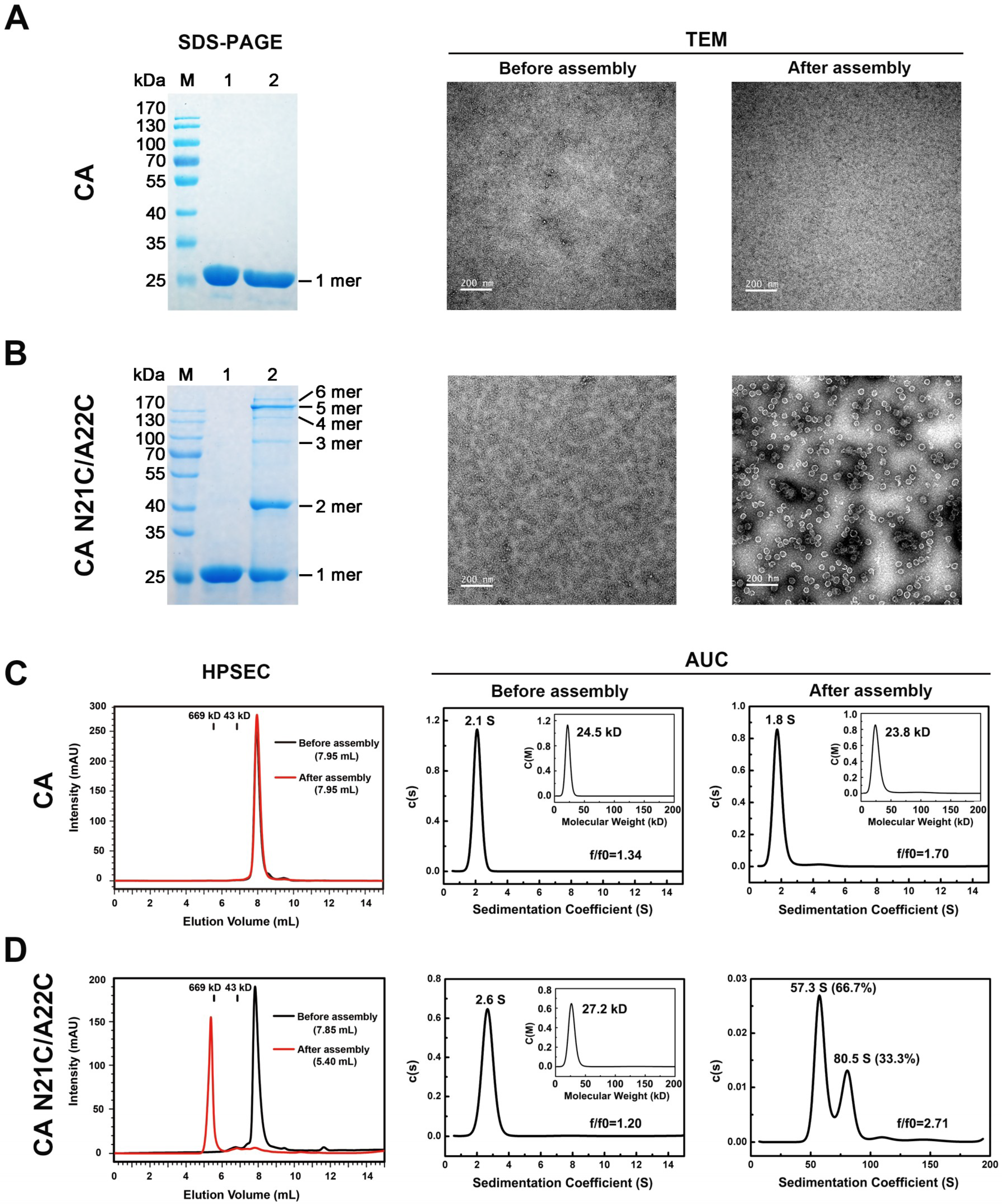
2.2. Protein Assembly In Vitro, SDS–PAGE and Western Blotting (WB)
2.3. Size-exclusion Chromatography, Analytical Ultracentrifugation (AUC), and Transmission Electron Microscopy (TEM)
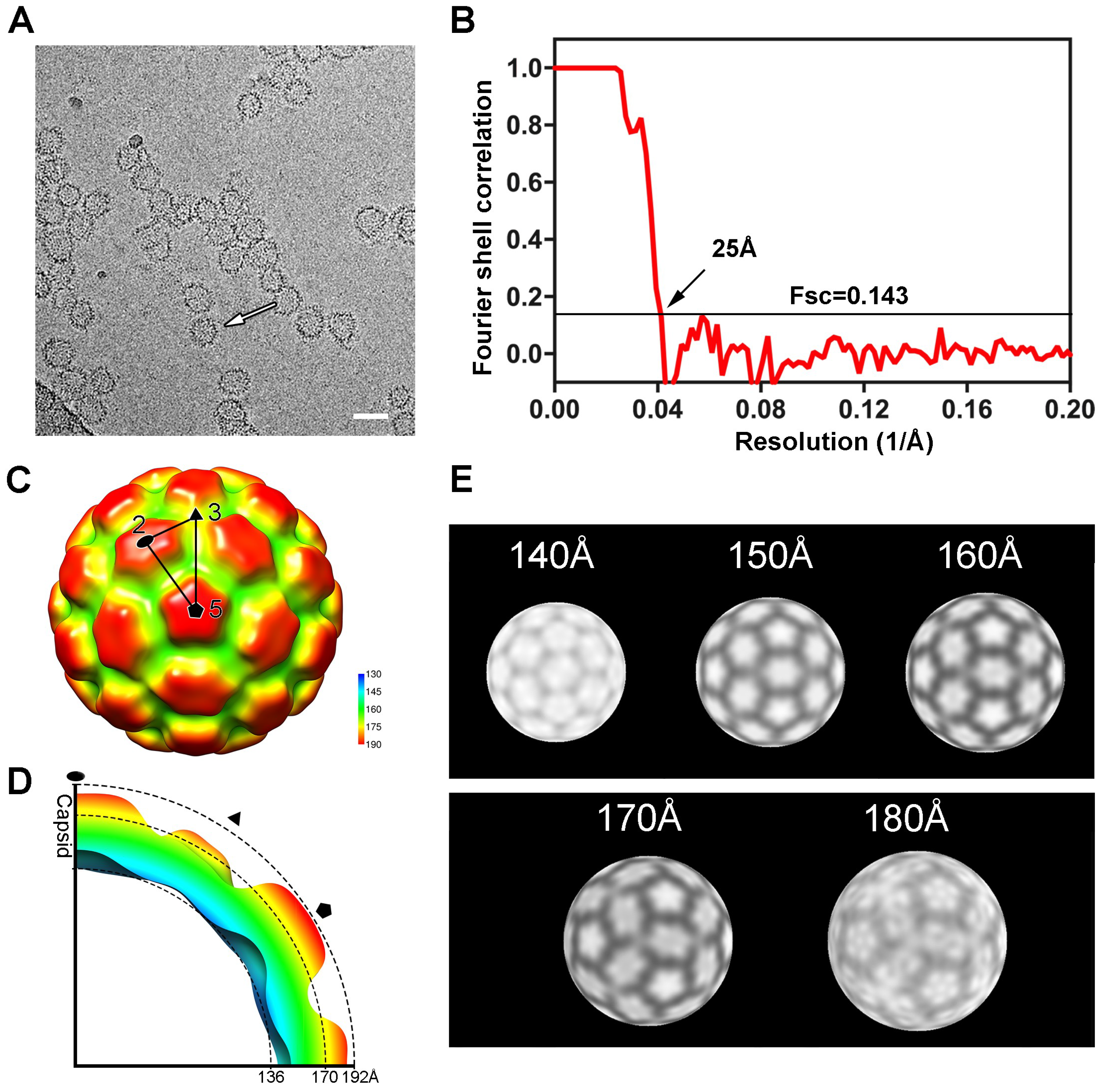
2.4. CryoEM Data Collection and Structure Determination
2.5. Ethics Statement and Mice Immunization
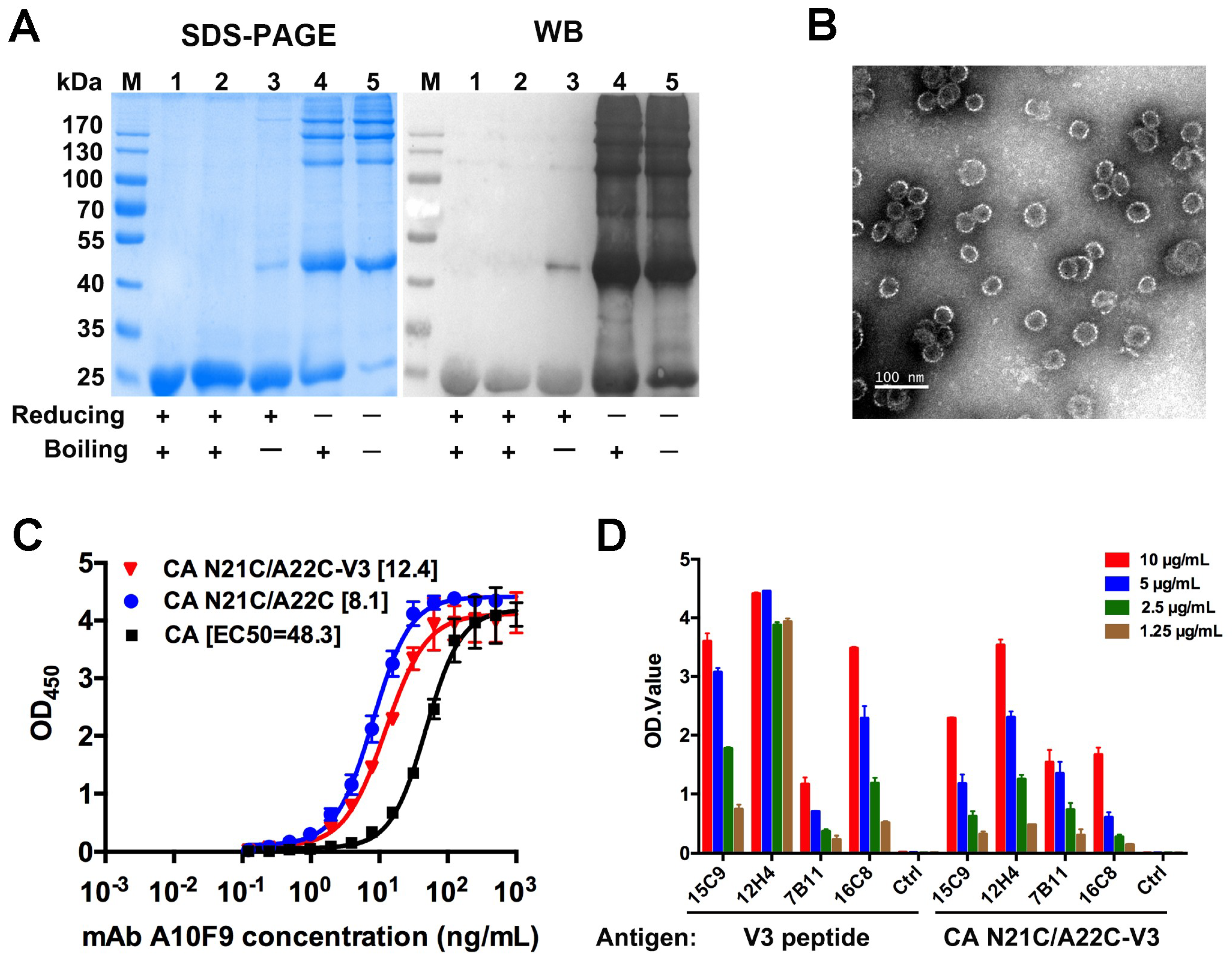
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. HIV-1 Neutralization Assay
3. Results
3.1. Purification and Characterization of HIV-1 CA N21C/A22C
3.2. Three-Dimensional Reconstruction of HIV-1 CA N21C/A22C Particles
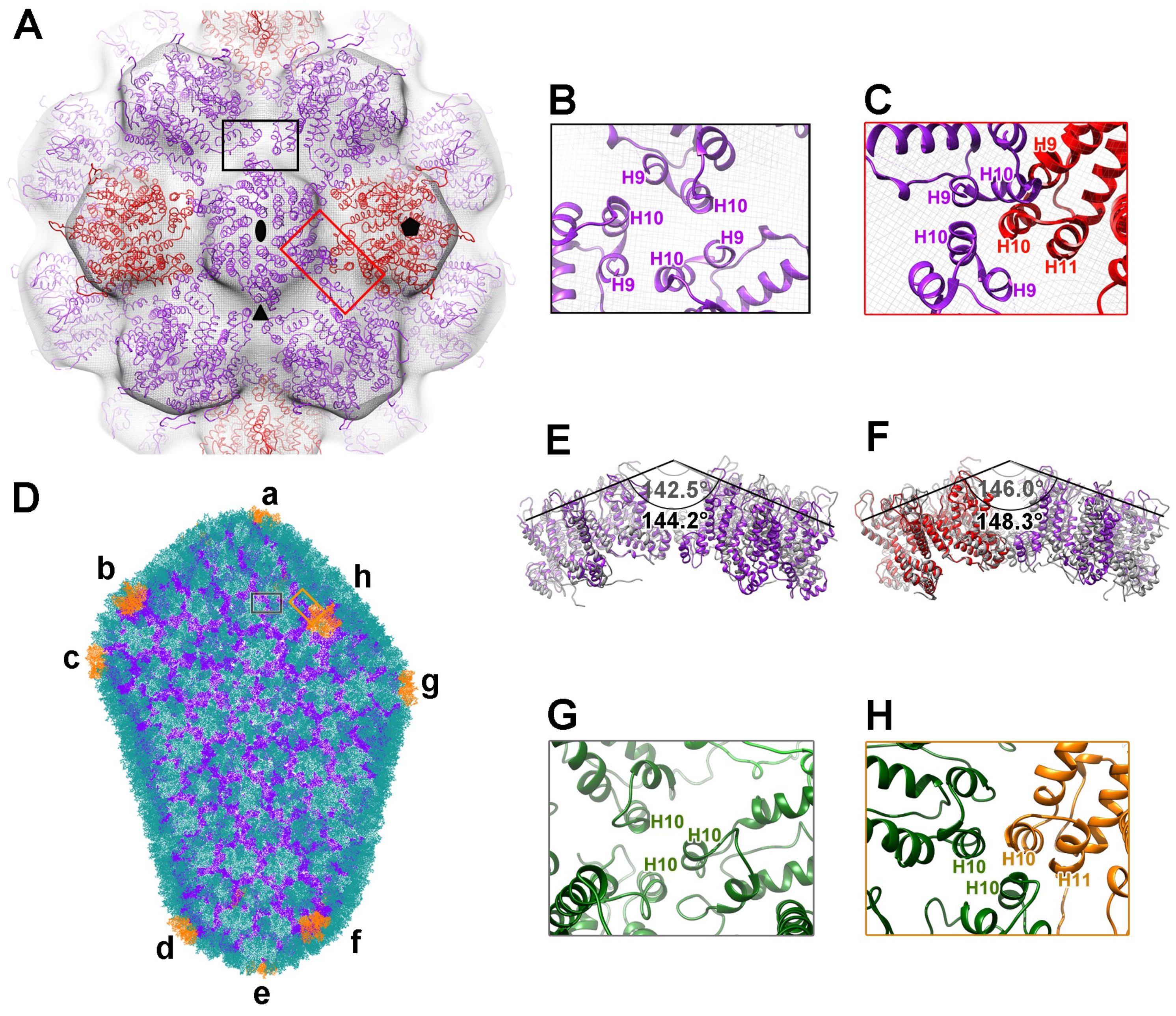
3.3. Inter-Subunit Interactions in Particles
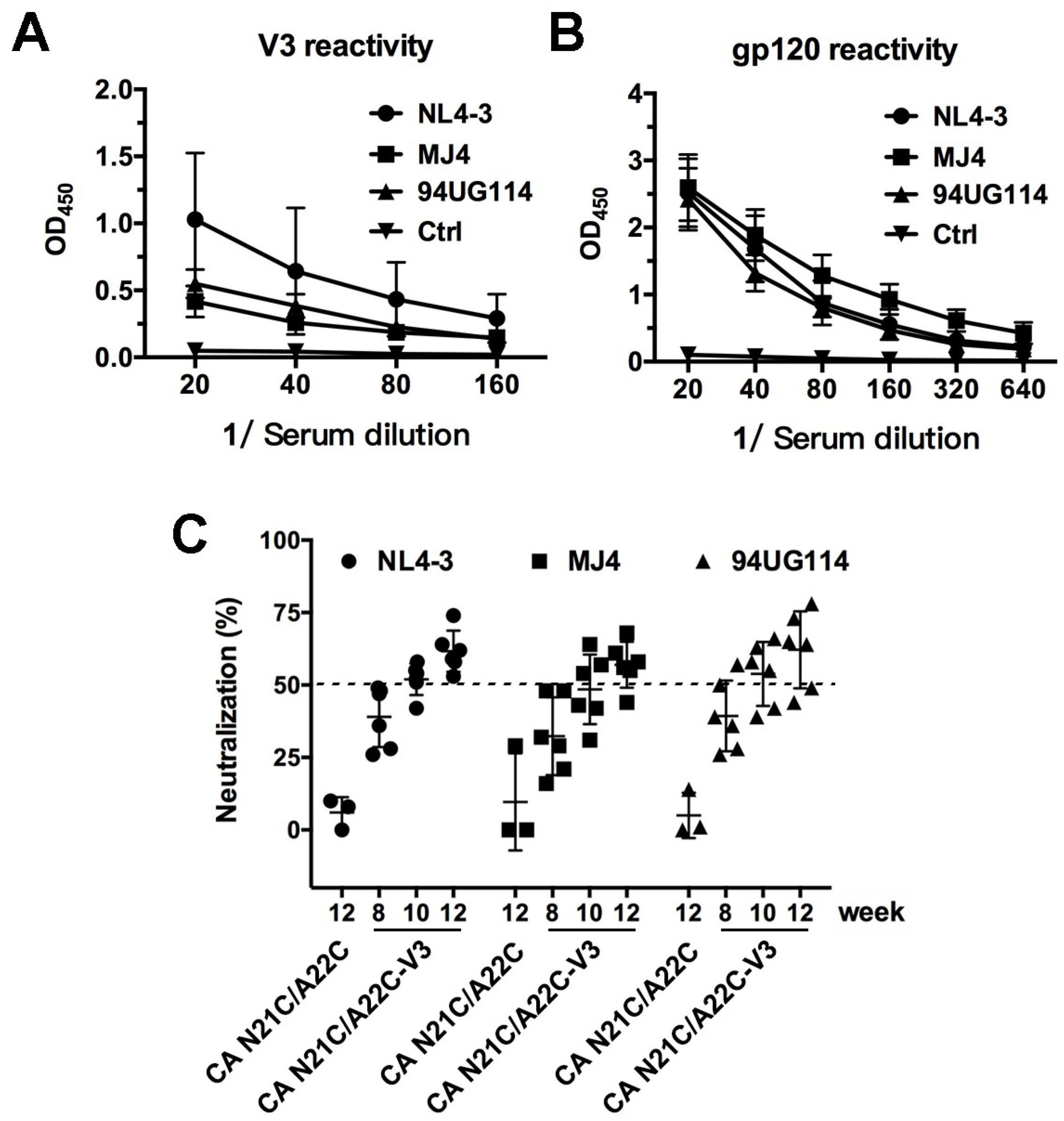
3.4. V3 Loop Grafted to CA Particles Induces Neutralizing Antibodies
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Bell, N.M.; Lever, A.M. HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 2013, 21, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Yeager, M.; Sundquist, W.I. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 2008, 18, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Cheng, A.; Yeager, M. Structure of full-length HIV-1 CA: A model for the mature capsid lattice. Cell 2007, 131, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Mortuza, G.B.; Haire, L.F.; Stevens, A.; Smerdon, S.J.; Stoye, J.P.; Taylor, I.A. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature 2004, 431, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Gamble, T.R.; Yoo, S.; Vajdos, F.F.; von Schwedler, U.K.; Worthylake, D.K.; Wang, H.; McCutcheon, J.P.; Sundquist, W.I.; Hill, C.P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 1997, 278, 849–853. [Google Scholar] [CrossRef] [PubMed]
- Worthylake, D.K.; Wang, H.; Yoo, S.; Sundquist, W.I.; Hill, C.P. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D Biol. Crystallogr. 1999, 55, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Byeon, I.J.; Meng, X.; Jung, J.; Zhao, G.; Yang, R.; Ahn, J.; Shi, J.; Concel, J.; Aiken, C.; Zhang, P.; et al. Structural convergence between Cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 2009, 139, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; von Schwedler, U.K.; Stray, K.M.; Aiken, C.; Sundquist, W.I. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 2004, 78, 2545–2552. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Banumathi, S.; Hua, Y.; Yeager, M. Disulfide bond stabilization of the hexameric capsomer of human immunodeficiency virus. J. Mol. Biol. 2010, 401, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Pornillos, O.; Ganser-Pornillos, B.K.; Yeager, M. Atomic-level modelling of the HIV capsid. Nature 2011, 469, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Yeager, M. Design of in vitro symmetric complexes and analysis by hybrid methods reveal mechanisms of HIV capsid assembly. J. Mol. Biol. 2011, 410, 534–552. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hill, C.P.; Sundquist, W.I.; Finch, J.T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 2000, 407, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Grunewald, K.; Glass, B.; Forster, F.; Krausslich, H.G.; Fuller, S.D. The mechanism of HIV-1 core assembly: Insights from three-dimensional reconstructions of authentic virions. Structure 2006, 14, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Glass, B.; Hagen, W.J.; Krausslich, H.G.; Briggs, J.A. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef] [PubMed]
- Campbell, E.M.; Hope, T.J. HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 2015, 13, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Tedbury, P.R.; Freed, E.O. HIV-1 Gag: An Emerging Target for Antiretroviral Therapy. Curr. Top. Microbiol. Immunol. 2015. [Google Scholar] [CrossRef]
- Blair, W.S.; Pickford, C.; Irving, S.L.; Brown, D.G.; Anderson, M.; Bazin, R.; Cao, J.; Ciaramella, G.; Isaacson, J.; Jackson, L.; et al. HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog. 2010, 6, e1001220. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.A.; Freed, E.O. HIV type 1 Gag as a target for antiviral therapy. AIDS Res. Hum. Retroviruses 2012, 28, 54–75. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Xu, K. Structural Features of Broadly Neutralizing Antibodies and Rational Design of Vaccine. Adv. Exp. Med. Biol. 2018, 1075, 73–95. [Google Scholar] [PubMed]
- Kwong, P.D.; Mascola, J.R. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 2018, 48, 855–871. [Google Scholar] [CrossRef] [PubMed]
- De Taeye, S.W.; Moore, J.P.; Sanders, R.W. HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol. 2016, 37, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Borducchi, E.N.; Liu, J.; Nkolola, J.P.; Cadena, A.M.; Yu, W.H.; Fischinger, S.; Broge, T.; Abbink, P.; Mercado, N.B.; Chandrashekar, A.; et al. Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 2018. [Google Scholar] [CrossRef] [PubMed]
- Schoofs, T.; Klein, F.; Braunschweig, M.; Kreider, E.F.; Feldmann, A.; Nogueira, L.; Oliveira, T.; Lorenzi, J.C.; Parrish, E.H.; Learn, G.H.; et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 2016, 352, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Lynch, R.M.; Boritz, E.; Coates, E.E.; DeZure, A.; Madden, P.; Costner, P.; Enama, M.E.; Plummer, S.; Holman, L.; Hendel, C.S.; et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci. Transl. Med. 2015, 7, 319ra206. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Acharya, P.; Kong, R.; Cheng, C.; Chuang, G.Y.; Liu, K.; Louder, M.K.; O’Dell, S.; Rawi, R.; Sastry, M.; et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 2018, 24, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y. Recent update in HIV vaccine development. Clin. Exp. Vaccine Res. 2016, 5, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, K.E.; D’Couto, H.T.; Barouch, D.H. New concepts in HIV-1 vaccine development. Curr. Opin. Immunol. 2016, 41, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Williamson, A.L.; Rybicki, E.P. Justification for the inclusion of Gag in HIV vaccine candidates. Expert Rev. Vaccines 2016, 15, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Nyombayire, J.; Anzala, O.; Gazzard, B.; Karita, E.; Bergin, P.; Hayes, P.; Kopycinski, J.; Omosa-Manyonyi, G.; Jackson, A.; Bizimana, J.; et al. First-in-Human Evaluation of the Safety and Immunogenicity of an Intranasally Administered Replication-Competent Sendai Virus-Vectored HIV Type 1 Gag Vaccine: Induction of Potent T-Cell or Antibody Responses in Prime-Boost Regimens. J. Infect. Dis. 2017, 215, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Hraber, P.; Wagh, K.; Hahn, B.H. Polyvalent vaccine approaches to combat HIV-1 diversity. Immunol. Rev. 2017, 275, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Cao, F.; Wang, L.; Hou, W.; Zhang, J.; Hew, C.L.; Li, S.; Yuan, Y.A.; Xia, N. Structure of a novel shoulder-to-shoulder p24 dimer in complex with the broad-spectrum antibody A10F9 and its implication in capsid assembly. PLoS ONE 2013, 8, e61314. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.Q.; Palovcak, E.; Armache, J.P.; Verba, K.A.; Cheng, Y.; Agard, D.A. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 2017, 14, 331–332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Sinkovits, R.S.; Baker, T.S. AUTO3DEM—An automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol. 2007, 157, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012, 180, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL User’s Manual; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Zhao, G.; Perilla, J.R.; Yufenyuy, E.L.; Meng, X.; Chen, B.; Ning, J.; Ahn, J.; Gronenborn, A.M.; Schulten, K.; Aiken, C.; et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 2013, 497, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Decker, J.M.; Liu, H.; Zhang, Z.; Arani, R.B.; Kilby, J.M.; Saag, M.S.; Wu, X.; Shaw, G.M.; Kappes, J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002, 46, 1896–1905. [Google Scholar] [CrossRef] [PubMed]
- Sarzotti-Kelsoe, M.; Bailer, R.T.; Turk, E.; Lin, C.L.; Bilska, M.; Greene, K.M.; Gao, H.; Todd, C.A.; Ozaki, D.A.; Seaman, M.S.; et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 2014, 409, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Bocanegra, R.; Rodríguez-Huete, A.; Fuertes, M.Á.; del Álamo, M.; Mateu, M.G. Molecular recognition in the human immunodeficiency virus capsid and antiviral design. Virus Res. 2012, 169, 388–410. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.; Obr, M.; Hagen, W.J.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Krausslich, H.G.; Briggs, J.A. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016. [Google Scholar] [CrossRef] [PubMed]
- Cardone, G.; Purdy, J.G.; Cheng, N.; Craven, R.C.; Steven, A.C. Visualization of a missing link in retrovirus capsid assembly. Nature 2009, 457, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, S.; Gu, Y.; Xia, N. Antiviral Therapy by HIV-1 Broadly Neutralizing and Inhibitory Antibodies. Int. J. Mol. Sci. 2016, 17, E1901. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Jin, G.; Wang, J.Y.; Li, H.N.; Liu, H.; Chang, X.Y.; Wang, F.X.; Liu, S.L. Live attenuated Salmonella displaying HIV-1 10E8 epitope on fimbriae: Systemic and mucosal immune responses in BALB/c mice by mucosal administration. Sci. Rep. 2016, 6, 29556. [Google Scholar] [CrossRef] [PubMed]
- Krebs, S.J.; McBurney, S.P.; Kovarik, D.N.; Waddell, C.D.; Jaworski, J.P.; Sutton, W.F.; Gomes, M.M.; Trovato, M.; Waagmeester, G.; Barnett, S.J.; et al. Multimeric scaffolds displaying the HIV-1 envelope MPER induce MPER-specific antibodies and cross-neutralizing antibodies when co-immunized with gp160 DNA. PLoS ONE 2014, 9, e113463. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.; Tu, X.; Bharaj, P.; Guo, H.; Zhang, J.; Shankar, P.; Manjunath, N. Human Rhinovirus Presenting 4E10 Epitope of HIV-1 MPER Elicits Neutralizing Antibodies in Human ICAM-1 Transgenic Mice. Mol. Ther. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hartley, O.; Klasse, P.J.; Sattentau, Q.J.; Moore, J.P. V3: HIV’s switch-hitter. AIDS Res. Hum. Retroviruses 2005, 21, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Moseri, A.; Tantry, S.; Sagi, Y.; Arshava, B.; Naider, F.; Anglister, J. An optimally constrained V3 peptide is a better immunogen than its linear homolog or HIV-1 gp120. Virology 2010, 401, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Durani, V.; Miranda, E.R.; Citron, M.; Liang, X.; Schleif, W.; Joyce, J.G.; Varadarajan, R. Design of immunogens that present the crown of the HIV-1 V3 loop in a conformation competent to generate 447-52D-like antibodies. Biochem. J. 2006, 399, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Totrov, M.; Jiang, X.; Kong, X.P.; Cohen, S.; Krachmarov, C.; Salomon, A.; Williams, C.; Seaman, M.S.; Abagyan, R.; Cardozo, T.; et al. Structure-guided design and immunological characterization of immunogens presenting the HIV-1 gp120 V3 loop on a CTB scaffold. Virology 2010, 405, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Moseri, A.; Sinha, E.; Zommer, H.; Arshava, B.; Naider, F.; Anglister, J. Immunofocusing using conformationally constrained V3 peptide immunogens improves HIV-1 neutralization. Vaccine 2017, 35, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Zhang, R.; Orwenyo, J.; Giddens, J.; Yang, Q.; LaBranche, C.C.; Montefiori, D.C.; Wang, L.X. Multivalent Antigen Presentation Enhances the Immunogenicity of a Synthetic Three-Component HIV-1 V3 Glycopeptide Vaccine. ACS Cent. Sci. 2018, 4, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Tagliamonte, M.; Marasco, D.; Ruggiero, A.; De Stradis, A.; Tornesello, M.L.; Totrov, M.; Buonaguro, F.M.; Buonaguro, L. HIV p24 as scaffold for presenting conformational HIV Env antigens. PLoS ONE 2012, 7, e43318. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; He, M.; Bai, S.; Zhang, F.; Jiang, J.; Zheng, Q.; Gao, S.; Yan, X.; Li, S.; Gu, Y.; et al. T = 4 Icosahedral HIV-1 Capsid As an Immunogenic Vector for HIV-1 V3 Loop Epitope Display. Viruses 2018, 10, 667. https://doi.org/10.3390/v10120667
Zhang Z, He M, Bai S, Zhang F, Jiang J, Zheng Q, Gao S, Yan X, Li S, Gu Y, et al. T = 4 Icosahedral HIV-1 Capsid As an Immunogenic Vector for HIV-1 V3 Loop Epitope Display. Viruses. 2018; 10(12):667. https://doi.org/10.3390/v10120667
Chicago/Turabian StyleZhang, Zhiqing, Maozhou He, Shimeng Bai, Feng Zhang, Jie Jiang, Qingbing Zheng, Shuangquan Gao, Xiaodong Yan, Shaowei Li, Ying Gu, and et al. 2018. "T = 4 Icosahedral HIV-1 Capsid As an Immunogenic Vector for HIV-1 V3 Loop Epitope Display" Viruses 10, no. 12: 667. https://doi.org/10.3390/v10120667
APA StyleZhang, Z., He, M., Bai, S., Zhang, F., Jiang, J., Zheng, Q., Gao, S., Yan, X., Li, S., Gu, Y., & Xia, N. (2018). T = 4 Icosahedral HIV-1 Capsid As an Immunogenic Vector for HIV-1 V3 Loop Epitope Display. Viruses, 10(12), 667. https://doi.org/10.3390/v10120667




