Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Isolation
2.2. DNA Isolation and Sequencing
2.3. Assembly
2.4. Annotation
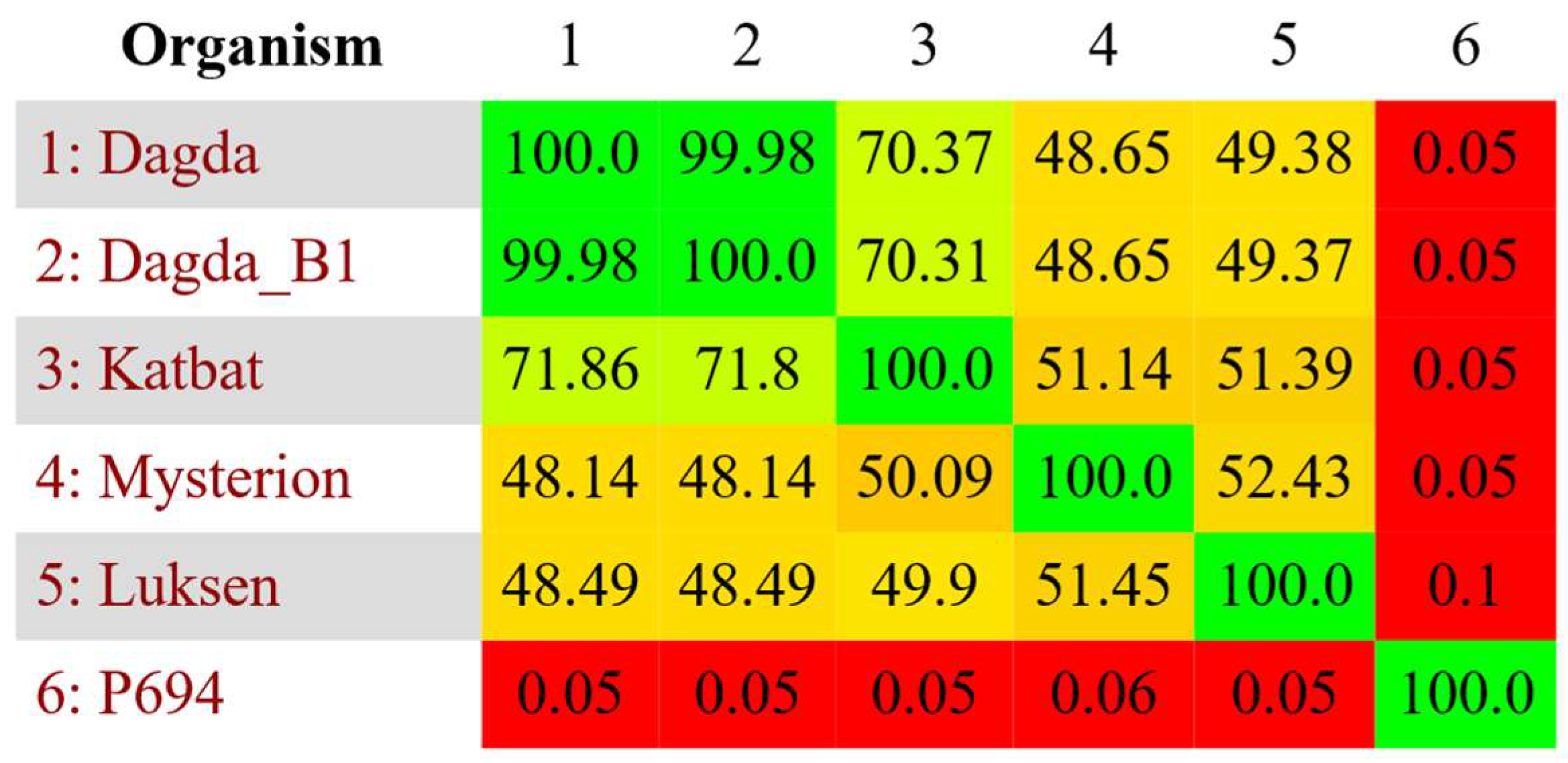
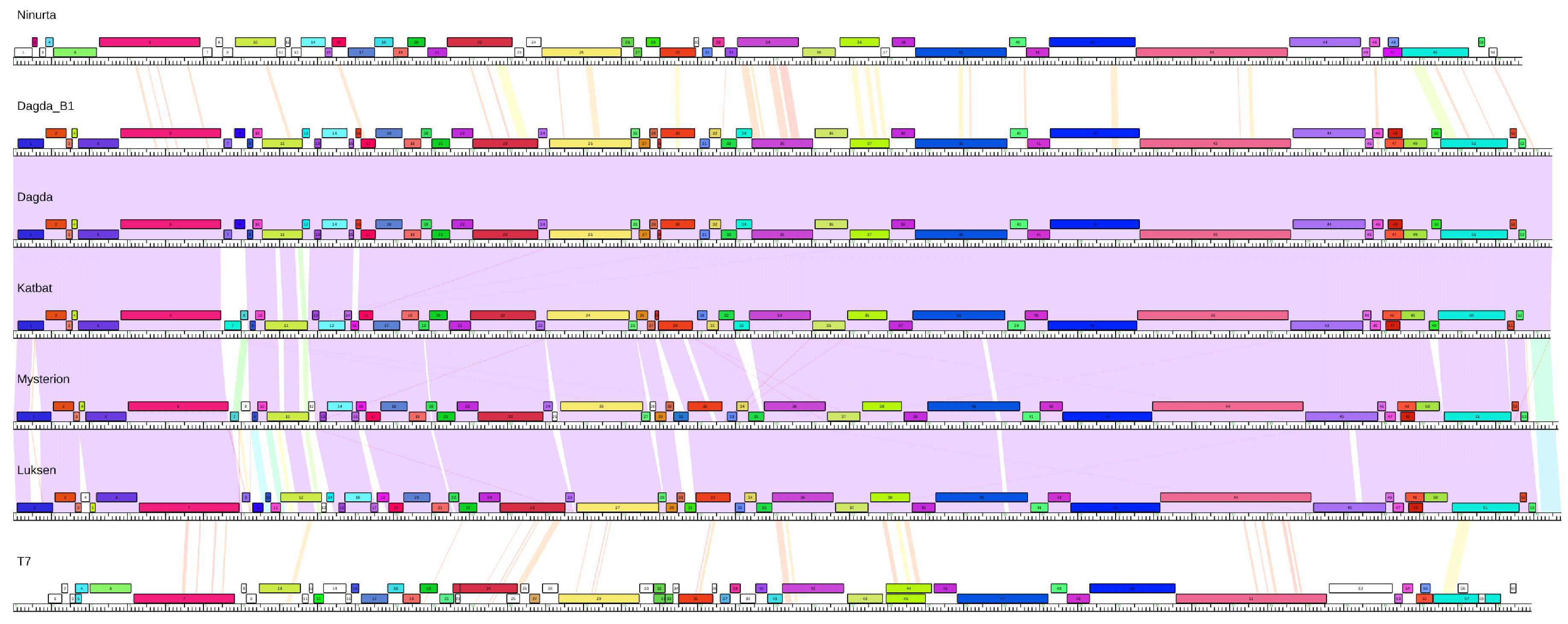
2.5. Comparative Genomics
2.6. Phage Stability
2.7. Potato Infection Model
3. Results and Discussion
3.1. Phage Characterization
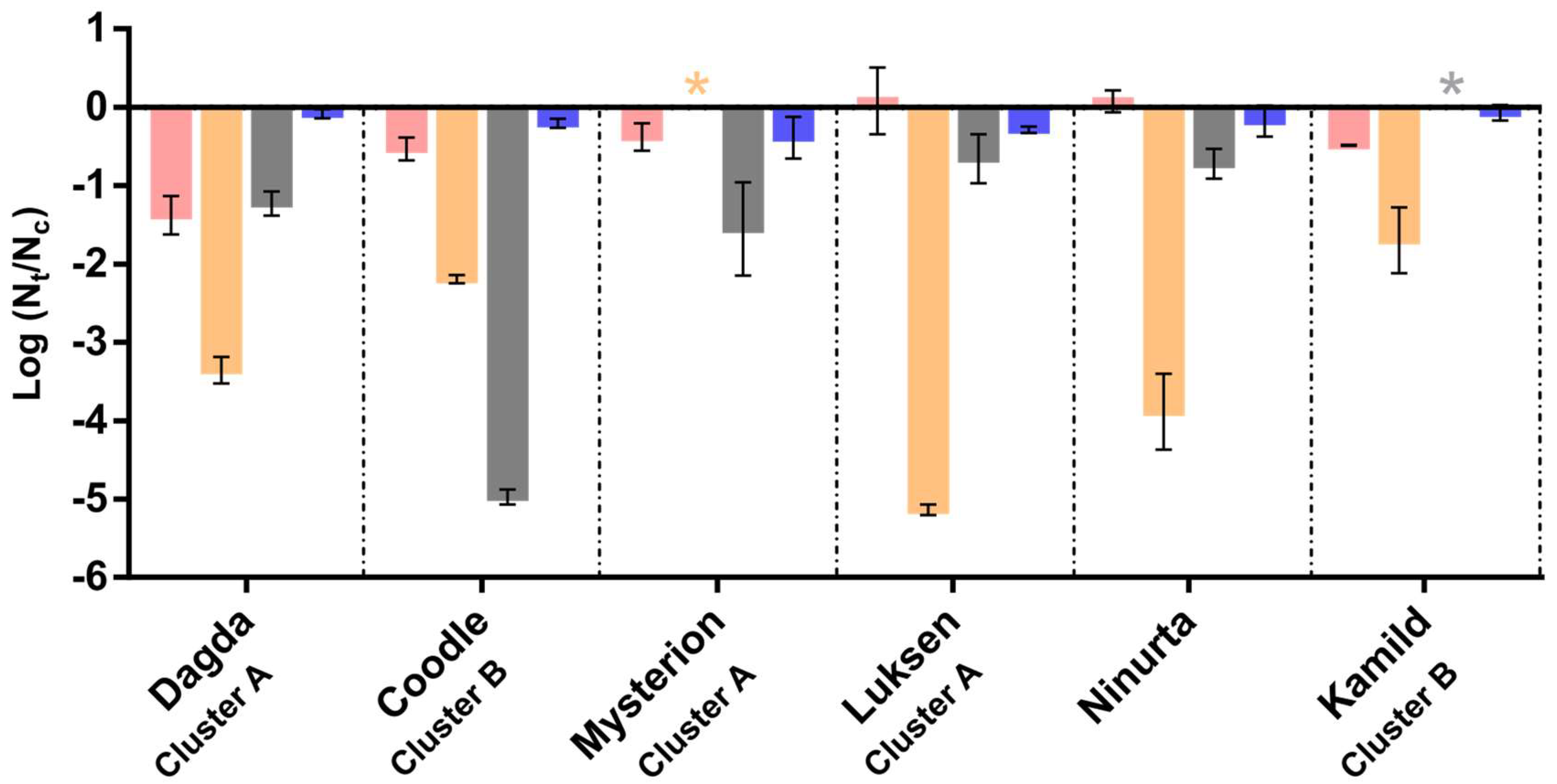
3.2. Formation of Phage Cocktail
3.3. Phage Stability
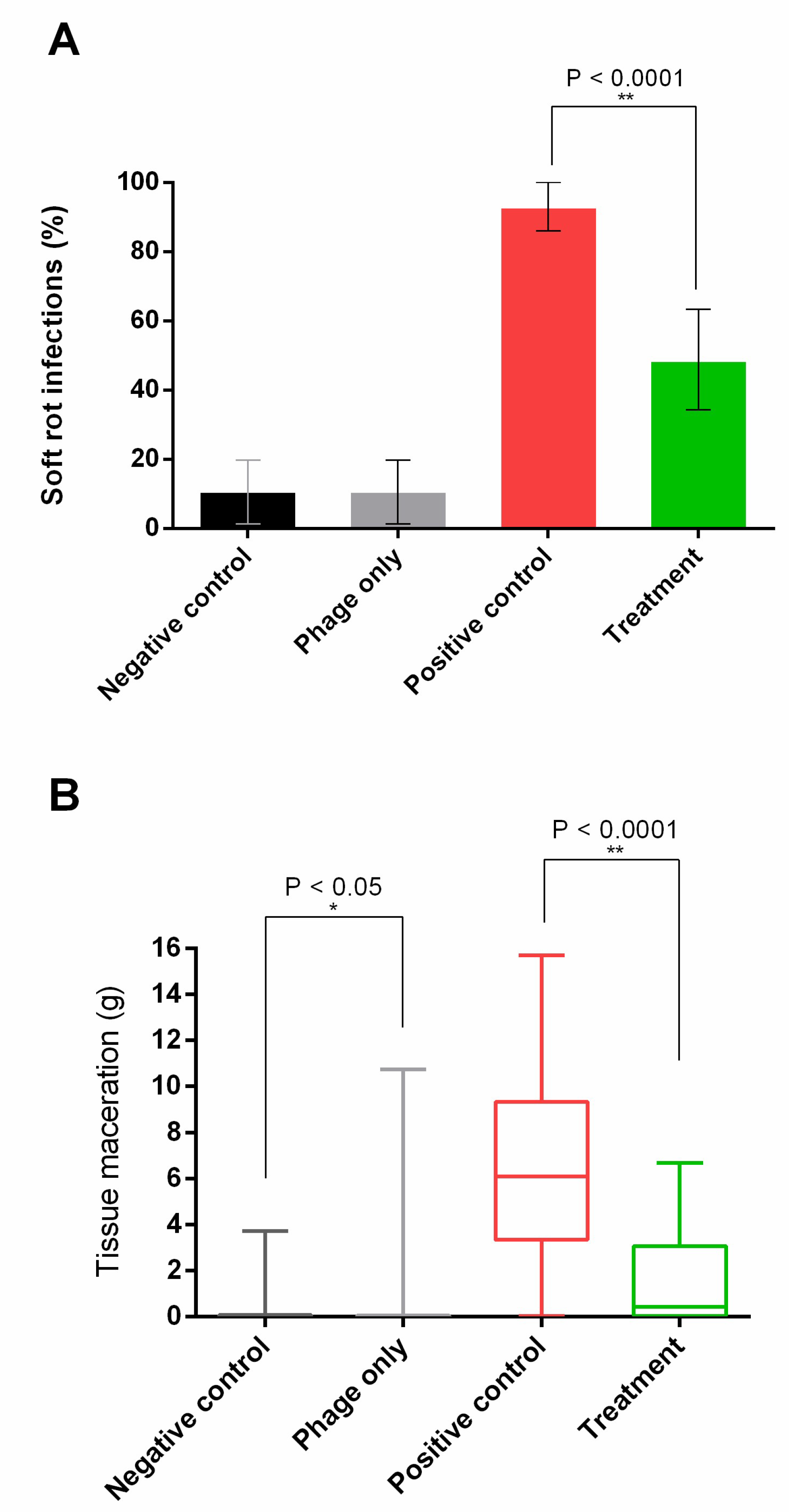
3.4. Potato Infection Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
References
- Ma, B.; Hibbing, M.E.; Kim, H.-S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera Pectobacterium and Dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier Academic Press: Cambridge, USA, 2005; ISBN 0120445654. [Google Scholar]
- Czajkowski, R.; Pérombelon, M.C.M.; van Veen, J.A.; van der Wolf, J.M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A. review. Plant Pathol. 2011, 60, 999–1013. [Google Scholar] [CrossRef]
- Toth, I.; Bell, K.; Holeva, M. Soft rot erwiniea: From genes to genomes. Mol. Plant Pathol. 2003, 4, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.; Miller, J.; Nolte, P. Diagnosis & Management of Potato Storage Diseases. Educ. Publ. Warehouse, Univ. Idaho 2006, CIS 1131, 1–8. [Google Scholar]
- Toth, I.K.; van der Wolf, J.M.; Saddler, G.; Lojkowska, E.; Hélias, V.; Pirhonen, M.; Tsror Lahkim, L.; Elphinstone, J.G. Dickeya species: an emerging problem for potato production in Europe. Plant Pathol. 2011, 60, 385–399. [Google Scholar] [CrossRef]
- Tsror, L.; Erlich, O.; Lebiush, S.; Hazanovsky, M.; Zig, U.; Slawiak, M.; Grabe, G.; van der Wolf, J.M.; van de Haar, J.J. Assessment of recent outbreaks of Dickeya sp. (syn. Erwinia chrysanthemi) slow wilt in potato crops in Israel. Eur. J. Plant Pathol. 2009, 123, 311–320. [Google Scholar] [CrossRef]
- Potrykus, M.; Golanowska, M.; Sledz, W.; Zoledowska, S.; Motyka, A.; Kolodziejska, A.; Butrymowicz, J.; Lojkowska, E. Biodiversity of Dickeya spp. isolated from potato plants and water sources in temperate climate. Plant Dis. 2016, 100, 408–417. [Google Scholar] [CrossRef]
- Czajkowski, R.; Ozymko, Z.; de Jager, V.; Siwinska, J.; Smolarska, A.; Ossowicki, A.; Narajczyk, M.; Lojkowska, E. Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages ΦPD10.3 and ΦPD23.1 infecting pectinolytic Pectobacterium spp. and Dickeya spp. PLoS ONE 2015, 10, e0119812. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; van Vaerenbergh, J.; Vandenheuvel, D.; Dunon, V.; Ceyssens, P.J.; de Proft, M.; Kropinski, A.M.; Noben, J.P.; Maes, M.; Lavigne, R. T4-related bacteriophage LIMEstone isolates for the control of soft rot on potato caused by “Dickeya solani”. PLoS ONE 2012, 7, e33227. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; Hendrix, H.; Lucid, A.; Neve, H.; Noben, J.-P.; Franz, C.; O’Mahony, J.; Lavigne, R.; Coffey, A. Novel N4-Like bacteriophages of Pectobacterium atrosepticum. Pharmaceuticals 2018, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Czajkowski, R.; Smolarska, A.; Ozymko, Z. The viability of lytic bacteriophage ΦD5 in potato-associated environments and its effect on Dickeya solani in potato (Solanum tuberosum L.) plants. PLoS ONE 2017, 12, e0183200. [Google Scholar] [CrossRef] [PubMed]
- Iriarte, F.B.; Balogh, B.; Momol, M.T.; Smith, L.M.; Wilson, M.; Jones, J.B. Factors affecting survival of bacteriophage on tomato leaf surfaces. Appl. Environ. Microbiol. 2007, 73, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Jackson, L.E.; Balogh, B.; Obradovic, A.; Iriarte, F.B.; Momol, M.T. Bacteriophages for plant disease control. Annu. Rev. Phytopathol. 2007, 45, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular cloning: A laboratory manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1990; ISBN 0-87969-309-6. [Google Scholar]
- Nielsen, T.K.; Carstens, A.B.; Browne, P.; Lametsch, R.; Neve, H.; Kot, W.; Hansen, L.H. The first characterized phage against a member of the ecologically important sphingomonads reveals high dissimilarity against all other known phages. Sci. Rep. 2017, 7, 13566. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- DNA Master. Available online: http://cobamide2.bio.pitt.edu (accessed on 7 November 2018).
- Borodovsky, M.; McIninch, J. GENMARK: Parallel gene recognition for both DNA strands. Comput. Chem. 1993, 17, 123–133. [Google Scholar] [CrossRef]
- Salzberg, S.L.; Delcher, A.L.; Kasif, S.; White, O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998, 26, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Agren, J.; Sundström, A.; Håfström, T.; Segerman, B. Gegenees: fragmented alignment of multiple genomes for determining phylogenomic distances and genetic signatures unique for specified target groups. PLoS ONE 2012, 7, e39107. [Google Scholar] [CrossRef] [PubMed]
- Cresawn, S.G.; Bogel, M.; Day, N.; Jacobs-Sera, D.; Hendrix, R.W.; Hatfull, G.F. Phamerator: A bioinformatic tool for comparative bacteriophage genomics. BMC Bioinform. 2011, 12, 395. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Goeker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. bioRxiv 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Dees, M.W.; Lebecka, R.; Perminow, J.I.S.; Czajkowski, R.; Grupa, A.; Motyka, A.; Zoledowska, S.; Śliwka, J.; Lojkowska, E.; Brurberg, M.B. Characterization of Dickeya and Pectobacterium strains obtained from diseased potato plants in different climatic conditions of Norway and Poland. Eur. J. Plant Pathol. 2017, 148, 839–851. [Google Scholar] [CrossRef]
- Rocha, E.P.C.; Danchin, A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002, 18, 291–294. [Google Scholar] [CrossRef]
- Chen, M.; Xu, J.; Yao, H.; Lu, C.; Zhang, W. Isolation, genome sequencing and functional analysis of two T7-like coliphages of avian pathogenic Escherichia coli. Gene 2016, 582, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.; Summer, E.J.; Struck, D.K.; Young, R. The final step in the phage infection cycle: The Rz and Rz1 lysis proteins link the inner and outer membranes. Mol. Microbiol. 2008, 70, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.-W.; Kropinski, A.M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Edgell, D.R.; Gibb, E.A.; Belfort, M. Mobile DNA elements in T4 and related phages. Virol. J. 2010. [Google Scholar] [CrossRef] [PubMed]
- Goodrich-Blair, H.; Shub, D.A. Beyond homing: Competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell 1996, 84, 211–221. [Google Scholar] [CrossRef]
- Brok-Volchanskaya, V.S.; Kadyrov, F.A.; Sivogrivov, D.E.; Kolosov, P.M.; Sokolov, A.S.; Shlyapnikov, M.G.; Kryukov, V.M.; Granovsky, I.E. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 2008, 36, 2094–2105. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and bacterial plant diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Casey, E.; van Sinderen, D.; Mahony, J. In Vitro characteristics of phages to guide “real life” phage therapy suitability. Viruses 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.; Brister, J.R. How to name and classify your phage: an informal guide. Viruses 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Pérombelon, M.C.M.; Hyman, L.J. Serological methods to quantify potato seed contamination by Erwinia carotovora subsp. atroseptica. EPPO Bull. 1995, 25, 195–202. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carstens, A.B.; Djurhuus, A.M.; Kot, W.; Jacobs-Sera, D.; Hatfull, G.F.; Hansen, L.H. Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses 2018, 10, 621. https://doi.org/10.3390/v10110621
Carstens AB, Djurhuus AM, Kot W, Jacobs-Sera D, Hatfull GF, Hansen LH. Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses. 2018; 10(11):621. https://doi.org/10.3390/v10110621
Chicago/Turabian StyleCarstens, Alexander B., Amaru M. Djurhuus, Witold Kot, Deborah Jacobs-Sera, Graham F. Hatfull, and Lars H. Hansen. 2018. "Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani" Viruses 10, no. 11: 621. https://doi.org/10.3390/v10110621
APA StyleCarstens, A. B., Djurhuus, A. M., Kot, W., Jacobs-Sera, D., Hatfull, G. F., & Hansen, L. H. (2018). Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses, 10(11), 621. https://doi.org/10.3390/v10110621




