An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Infection
2.2. Plasmids and Bacteria Strains
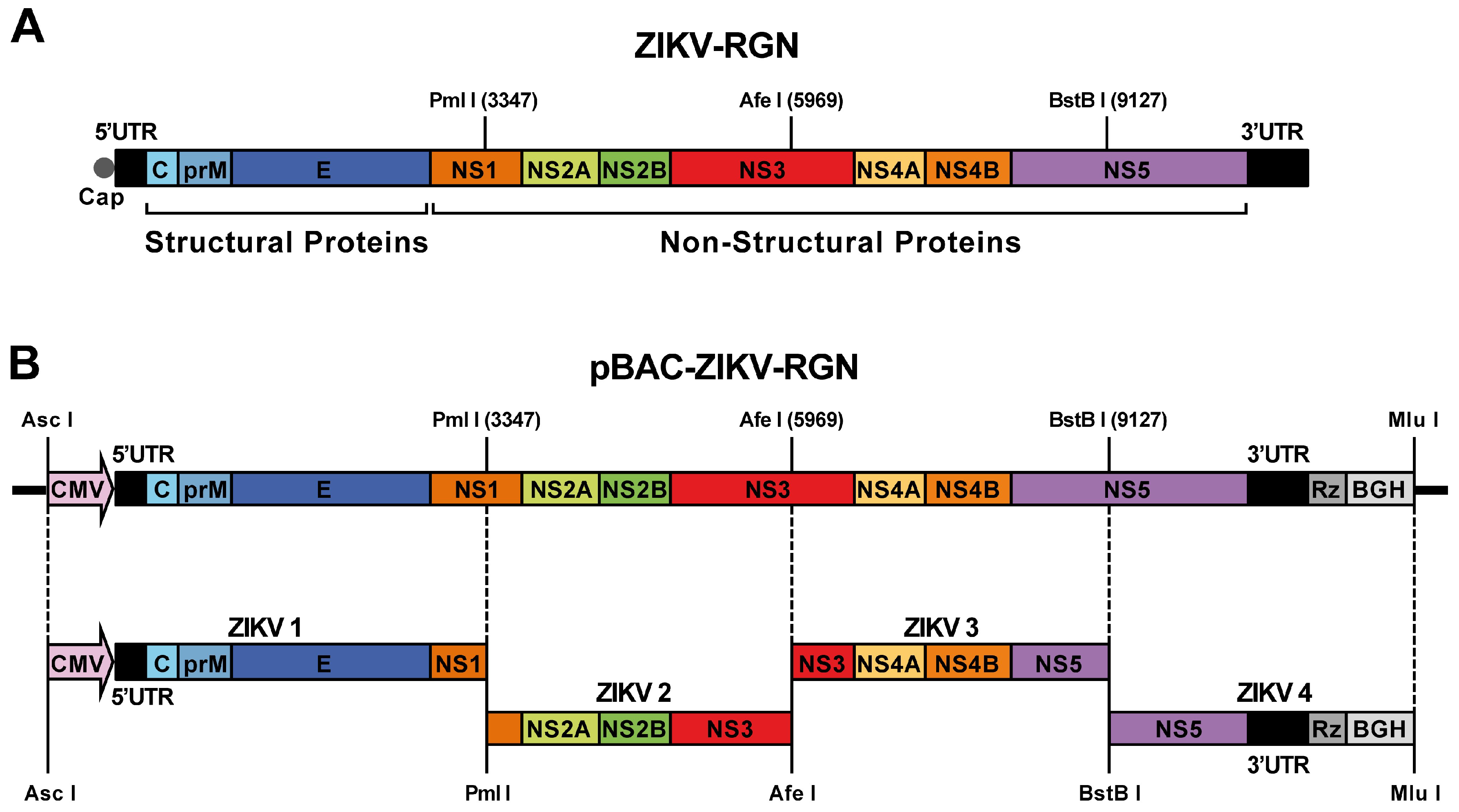
2.3. Construction of ZIKV-RGN Infectious cDNA Clone
2.4. Recovery of Infectious Virus from the BAC cDNA Clones
2.5. Sequencing of Viral RNA
2.6. Virus Titrations
2.7. Virus Growth Kinetics
2.8. Analysis of Viral RNA Synthesis
2.9. Indirect Immunofluorescence Assay
2.10. Enzyme-Linked Immunosorbent Assay
2.11. Mice Experiments
2.12. Statistical Analysis
2.13. Ethics Statement
3. Results
3.1. Development of a ZIKV-RGN Reverse Genetic System Using a BAC
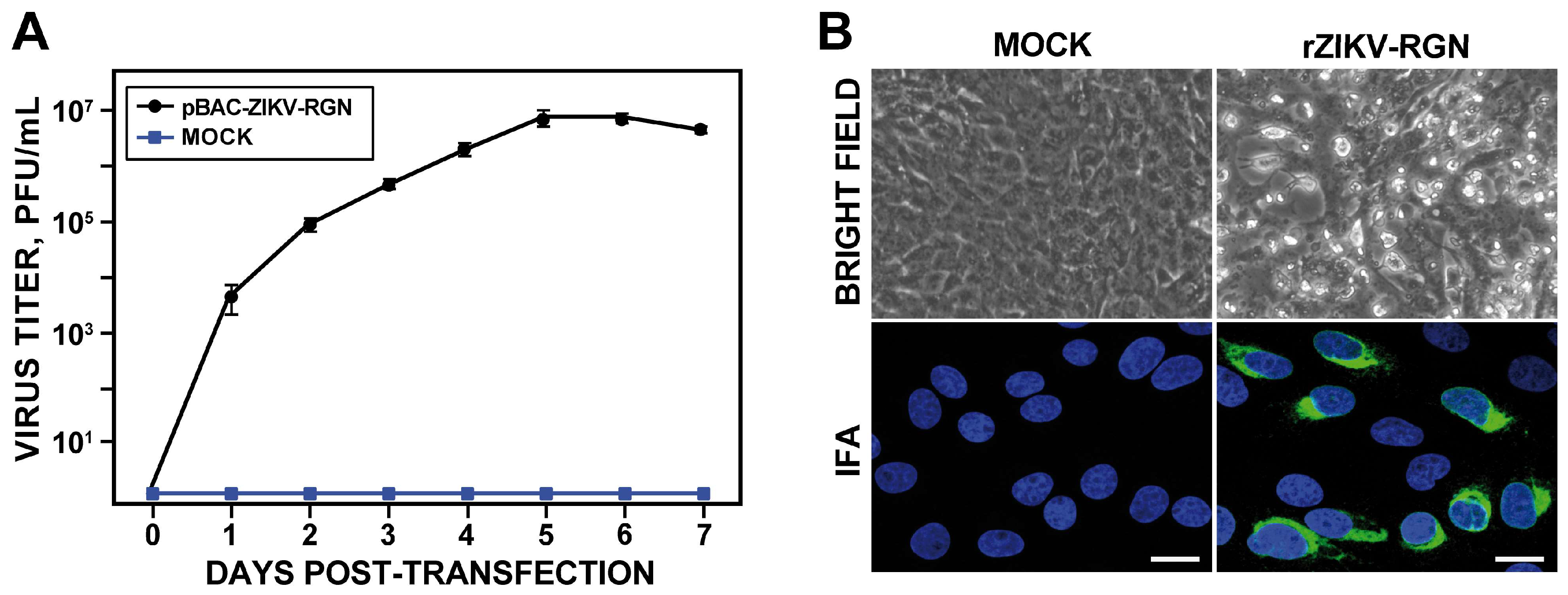
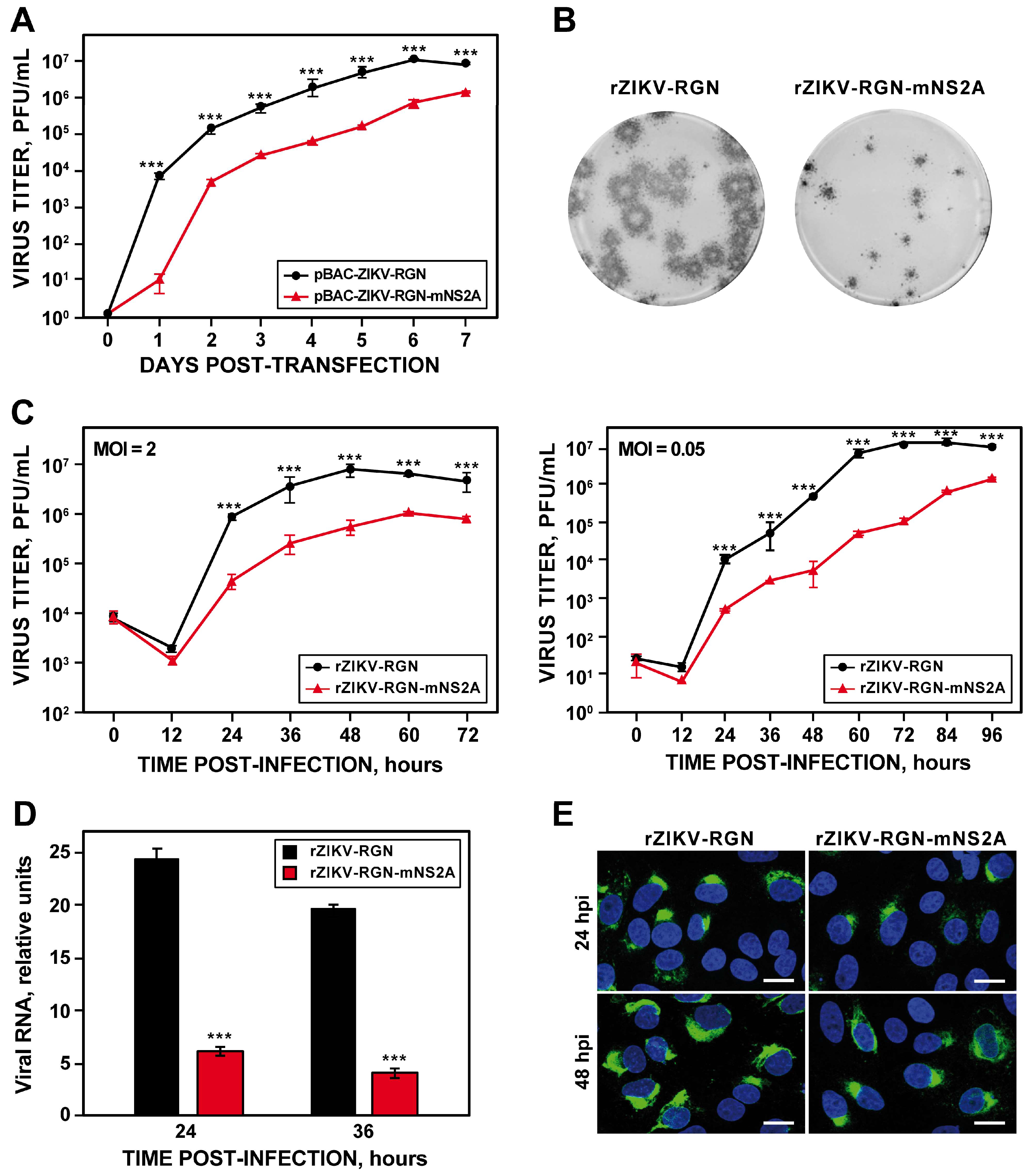
3.2. Rescue and In Vitro Characterization of rZIKV-RGN
3.3. Pathogenesis of rZIKV-RGN in Mice
3.4. Rescue and In Vitro Charazterization of a rZIKV-RGN Harboring a Point Mutation in the NS2A Protein
3.5. Pathogenesis of rZIKV-RGN-mNS2A in Mice
3.6. Analysis of the Protection Efficacy in Mice of rZIKV-RGN-mNS2A
3.7. Genetic Stability of rZIKV-RGN-mNS2A In Vero Cells
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Baud, D.; Gubler, D.J.; Schaub, B.; Lanteri, M.C.; Musso, D. An update on zika virus infection. Lancet 2017, 390, 2099–2109. [Google Scholar] [CrossRef]
- Friedrich, M.J. Who calls off global zika emergency. J. Am. Med. Assoc. 2017, 317, 246. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 å resolution cryo-em structure of zika virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Bos, S.; Li, G.; Wang, S.; Gadea, G.; Despres, P.; Zhao, R.Y. Probing molecular insights into zika virus(-)host interactions. Viruses 2018, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Murray, C.J.; Thiel, H.J.; Rice, C.M. Flaviviridae. In Fields virology, 6th ed.; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer, Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 712–748. [Google Scholar]
- Macnamara, F.N. Zika virus: A report on three cases of human infection during an epidemic of jaundice in nigeria. Trans. R. Soc. Trop. Med. Hyg. 1954, 48, 139–145. [Google Scholar] [CrossRef]
- Moulin, E.; Selby, K.; Cherpillod, P.; Kaiser, L.; Boillat-Blanco, N. Simultaneous outbreaks of dengue, chikungunya and zika virus infections: Diagnosis challenge in a returning traveller with nonspecific febrile illness. New Microbes New Infect. 2016, 11, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Boeuf, P.; Drummer, H.E.; Richards, J.S.; Scoullar, M.J.; Beeson, J.G. The global threat of zika virus to pregnancy: Epidemiology, clinical perspectives, mechanisms, and impact. BMC Med. 2016, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika virus outbreak on yap island, federated states of micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.L.; Mallet, H.P.; Sall, A.A.; Musso, D. Zika virus, french polynesia, south pacific, 2013. Emerg. Infect. Dis. 2014, 20, 1085–1086. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Azevedo Rdo, S.; Kraemer, M.U.; Souza, R.; Cunha, M.S.; Hill, S.C.; Theze, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika virus in the americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M. Four in florida are infected with zika from local mosquitoes. Br. Med. J. 2016, 354, i4235. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, M. Zika virus was transmitted by sexual contact in texas, health officials report. Br. Med. J. 2016, 352, i720. [Google Scholar] [CrossRef] [PubMed]
- Costello, A.; Dua, T.; Duran, P.; Gulmezoglu, M.; Oladapo, O.T.; Perea, W.; Pires, J.; Ramon-Pardo, P.; Rollins, N.; Saxena, S. Defining the syndrome associated with congenital zika virus infection. Bull. World Health Organ. 2016, 94, 406–406a. [Google Scholar] [CrossRef] [PubMed]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.; Guimaraes, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The brazilian zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Do Rosario, M.S.; de Jesus, P.A.; Vasilakis, N.; Farias, D.S.; Novaes, M.A.; Rodrigues, S.G.; Martins, L.C.; Vasconcelos, P.F.; Ko, A.I.; Alcantara, L.C.; et al. Guillain-barre syndrome after zika virus infection in brazil. Am. J. Trop. Med. Hyg. 2016, 95, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell. Stem Cell 2016, 19, 672. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.J.; Cao, B.; Govero, J.; Smith, A.M.; Fernandez, E.; Cabrera, O.H.; Garber, C.; Noll, M.; Klein, R.S.; Noguchi, K.K.; et al. Zika virus infection during pregnancy in mice causes placental damage and fetal demise. Cell 2016, 165, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popovic, M.; Poljsak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodusek, V.; et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Diagne, C.T.; Diallo, D.; Faye, O.; Ba, Y.; Faye, O.; Gaye, A.; Dia, I.; Faye, O.; Weaver, S.C.; Sall, A.A.; et al. Potential of selected senegalese aedes spp. Mosquitoes (diptera: Culicidae) to transmit zika virus. BMC Infect. Dis. 2015, 15, 492. [Google Scholar] [CrossRef] [PubMed]
- Tham, H.W.; Balasubramaniam, V.; Ooi, M.K.; Chew, M.F. Viral determinants and vector competence of zika virus transmission. Front. Microbiol. 2018, 9, 1040. [Google Scholar] [CrossRef] [PubMed]
- Colt, S.; Garcia-Casal, M.N.; Pena-Rosas, J.P.; Finkelstein, J.L.; Rayco-Solon, P.; Weise Prinzo, Z.C.; Mehta, S. Transmission of zika virus through breast milk and other breastfeeding-related bodily-fluids: A systematic review. PLoS Negl. Trop. Dis. 2017, 11, e0005528. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Morales, A.J.; Bandeira, A.C.; Franco-Paredes, C. The expanding spectrum of modes of transmission of zika virus: A global concern. Ann. Clin. Microbiol. Antimicrob. 2016, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Aubry, F.; Nougairede, A.; Gould, E.A.; de Lamballerie, X. Flavivirus reverse genetic systems, construction techniques and applications: A historical perspective. Antivir. Res. 2015, 114, 67–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pu, S.Y.; Wu, R.H.; Yang, C.C.; Jao, T.M.; Tsai, M.H.; Wang, J.C.; Lin, H.M.; Chao, Y.S.; Yueh, A. Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. J. Virol. 2011, 85, 2927–2941. [Google Scholar] [CrossRef] [PubMed]
- Ruggli, N.; Rice, C.M. Functional cDNA clones of the flaviviridae: Strategies and applications. Adv. Virus Res. 1999, 53, 183–207. [Google Scholar] [PubMed]
- Deng, C.-L.; Zhang, Q.-Y.; Chen, D.-D.; Liu, S.-Q.; Qin, C.-F.; Zhang, B.; Ye, H.-Q. Recovery of the zika virus through an in vitro ligation approach. J. Gen. Virol. 2017, 98, 1739–1743. [Google Scholar] [CrossRef] [PubMed]
- Widman, D.G.; Young, E.; Yount, B.L.; Plante, K.S.; Gallichotte, E.N.; Carbaugh, D.L.; Peck, K.M.; Plante, J.; Swanstrom, J.; Heise, M.T.; et al. A reverse genetics platform that spans the zika virus family tree. mBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, A.S.; Pattnaik, A.; Sahoo, B.R.; Muthukrishnan, E.; Natarajan, S.K.; Steffen, D.; Vu, H.L.X.; Delhon, G.; Osorio, F.A.; Petro, T.M.; et al. Zika virus encoding nonglycosylated envelope protein is attenuated and defective in neuroinvasion. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Xie, X.; Muruato, A.E.; Rossi, S.L.; Roundy, C.M.; Azar, S.R.; Yang, Y.; Tesh, R.B.; Bourne, N.; Barrett, A.D.; et al. An infectious cDNA clone of zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell. Host Microbe. 2016, 19, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-Y.; Yu, J.-Y.; Huang, X.-Y.; Fan, H.; Li, X.-F.; Deng, Y.-Q.; Ji, X.; Cheng, M.-L.; Ye, Q.; Zhao, H.; et al. Characterization of cis-acting rna elements of zika virus by using a self-splicing ribozyme-dependent infectious clone. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, M.C.; Sourisseau, M.; Espino, M.M.; Gray, E.S.; Chambers, M.T.; Tortorella, D.; Evans, M.J. Rescue of the 1947 zika virus prototype strain with a cytomegalovirus promoter-driven cDNA clone. mSphere 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Kenney, H.; Chen, R.; Liu, G.; Manukyan, H.; Whitehead, S.S.; Laassri, M.; Chumakov, K.; Pletnev, A.G. A full-length infectious cDNA clone of zika virus from the 2015 epidemic in brazil as a genetic platform for studies of virus-host interactions and vaccine development. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Weger-Lucarelli, J.; Duggal, N.K.; Bullard-Feibelman, K.; Veselinovic, M.; Romo, H.; Nguyen, C.; Ruckert, C.; Brault, A.C.; Bowen, R.A.; Stenglein, M.; et al. Development and characterization of recombinant virus generated from a new world zika virus infectious clone. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Atieh, T.; Baronti, C.; de Lamballerie, X.; Nougairede, A. Simple reverse genetics systems for asian and african zika viruses. Sci. Rep. 2016, 6, 39384. [Google Scholar] [CrossRef] [PubMed]
- Gadea, G.; Bos, S.; Krejbich-Trotot, P.; Clain, E.; Viranaicken, W.; El-Kalamouni, C.; Mavingui, P.; Desprès, P. A robust method for the rapid generation of recombinant zika virus expressing the gfp reporter gene. Virology 2016, 497, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Munster, M.; Plaszczyca, A.; Cortese, M.; Neufeldt, C.J.; Goellner, S.; Long, G.; Bartenschlager, R. A reverse genetics system for zika virus based on a simple molecular cloning strategy. Viruses 2018, 10, 368. [Google Scholar] [CrossRef] [PubMed]
- Setoh, Y.X.; Prow, N.A.; Peng, N.; Hugo, L.E.; Devine, G.; Hazlewood, J.E.; Suhrbier, A.; Khromykh, A.A. De novo generation and characterization of new zika virus isolate using sequence data from a microcephaly case. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Usme-Ciro, J.A.; Lopera, J.A.; Enjuanes, L.; Almazan, F.; Gallego-Gomez, J.C. Development of a novel DNA-launched dengue virus type 2 infectious clone assembled in a bacterial artificial chromosome. Virus Res. 2014, 180, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Boysen, C.; Shizuya, H.; Simon, M.I.; Hood, L. Complete nucleotide sequence of two generations of a bacterial artificial chromosome cloning vector. BioTechniques 1997, 23, 992–994. [Google Scholar] [CrossRef] [PubMed]
- Shizuya, H.; Birren, B.; Kim, U.J.; Mancino, V.; Slepak, T.; Tachiiri, Y.; Simon, M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an f-factor-based vector. Proc. Natl. Acad. Sci. USA 1992, 89, 8794–8797. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The miqe guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Huang, K.; Chauche, C.; DeDiego, M.L.; Murcia, P.R.; Parrish, C.R.; Martinez-Sobrido, L. Canine influenza viruses with modified ns1 proteins for the development of live-attenuated vaccines. Virology 2017, 500, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dowall, S.D.; Graham, V.A.; Rayner, E.; Atkinson, B.; Hall, G.; Watson, R.J.; Bosworth, A.; Bonney, L.C.; Kitchen, S.; Hewson, R. A susceptible mouse model for zika virus infection. PLoS Negl. Trop. Dis. 2016, 10, e0004658. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Govero, J.; Smith, A.M.; Platt, D.J.; Fernandez, E.; Miner, J.J.; Diamond, M.S. A mouse model of zika virus pathogenesis. Cell. Host Microbe 2016, 19, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.L.; Tesh, R.B.; Azar, S.R.; Muruato, A.E.; Hanley, K.A.; Auguste, A.J.; Langsjoen, R.M.; Paessler, S.; Vasilakis, N.; Weaver, S.C. Characterization of a novel murine model to study zika virus. Am. J. Trop. Med. Hyg. 2016, 94, 1362–1369. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (U.S.). Committee for the Update of the Guide for the Care and Use of Laboratory Animals. In Guide for the Care and Use of Laboratory Animals, 8th ed.; Institute for Laboratory Animal Research (U.S.) & National Academies Press (U.S.): Washington, DC, USA, 2011. [Google Scholar]
- Dubensky, T.W., Jr.; Driver, D.A.; Polo, J.M.; Belli, B.A.; Latham, E.M.; Ibanez, C.E.; Chada, S.; Brumm, D.; Banks, T.A.; Mento, S.J.; et al. Sindbis virus DNA-based expression vectors: Utility for in vitro and in vivo gene transfer. J. Virol. 1996, 70, 508–519. [Google Scholar] [PubMed]
- Virus Pathogen Resource (ViPR), Faviviridae. Available online: https://www.viprbrc.org/brc/home.spg?decorator=flavi (accessed on 24 May 2018).
- Almazan, F.; Gonzalez, J.M.; Penzes, Z.; Izeta, A.; Calvo, E.; Plana-Duran, J.; Enjuanes, L. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 2000, 97, 5516–5521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jamsai, D.; Orford, M.; Nefedov, M.; Fucharoen, S.; Williamson, R.; Ioannou, P.A. Targeted modification of a human β-globin locus bac clone using get recombination and an i-scei counterselection cassette. Genomics 2003, 82, 68–77. [Google Scholar] [CrossRef]
- Lee, E.C.; Yu, D.; Martinez de Velasco, J.; Tessarollo, L.; Swing, D.A.; Court, D.L.; Jenkins, N.A.; Copeland, N.G. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of bac DNA. Genomics 2001, 73, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 2006, 40, 191–197. [Google Scholar] [PubMed]
- Zhang, Y.; Buchholz, F.; Muyrers, J.P.; Stewart, A.F. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 1998, 20, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Almazan, F.; Dediego, M.L.; Galan, C.; Escors, D.; Alvarez, E.; Ortego, J.; Sola, I.; Zuniga, S.; Alonso, S.; Moreno, J.L.; et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 2006, 80, 10900–10906. [Google Scholar] [CrossRef] [PubMed]
- Almazan, F.; DeDiego, M.L.; Sola, I.; Zuniga, S.; Nieto-Torres, J.L.; Marquez-Jurado, S.; Andres, G.; Enjuanes, L. Engineering a replication-competent, propagation-defective middle east respiratory syndrome coronavirus as a vaccine candidate. mBio 2013, 4, e00650-13. [Google Scholar] [CrossRef] [PubMed]
- Balint, A.; Farsang, A.; Zadori, Z.; Hornyak, A.; Dencso, L.; Almazan, F.; Enjuanes, L.; Belak, S. Molecular characterization of feline infectious peritonitis virus strain df-2 and studies of the role of orf3abc in viral cell tropism. J. Virol. 2012, 86, 6258–6267. [Google Scholar] [CrossRef] [PubMed]
- St-Jean, J.R.; Desforges, M.; Almazan, F.; Jacomy, H.; Enjuanes, L.; Talbot, P.J. Recovery of a neurovirulent human coronavirus oc43 from an infectious cDNA clone. J. Virol. 2006, 80, 3670–3674. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus kunjin nonstructural proteins ns2a and ns4a. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.L.; Fayzulin, R.; Dewsbury, N.; Bourne, N.; Mason, P.W. Mutations in west nile virus nonstructural proteins that facilitate replicon persistence in vitro attenuate virus replication in vitro and in vivo. Virology 2007, 364, 184–195. [Google Scholar] [CrossRef] [PubMed]
- Kummerer, B.M.; Rice, C.M. Mutations in the yellow fever virus nonstructural protein ns2a selectively block production of infectious particles. J. Virol. 2002, 76, 4773–4784. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.Y.; Pijlman, G.P.; Kondratieva, N.; Hyde, J.; Mackenzie, J.M.; Khromykh, A.A. Role of nonstructural protein ns2a in flavivirus assembly. J. Virol. 2008, 82, 4731–4741. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Chen, H.B.; Wang, X.J.; Huang, H.; Khromykh, A.A. Analysis of adaptive mutations in kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein ns2a in inhibition of β interferon promoter-driven transcription. J. Virol. 2004, 78, 12225–12235. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Wang, X.J.; Clark, D.C.; Lobigs, M.; Hall, R.A.; Khromykh, A.A. A single amino acid substitution in the west nile virus nonstructural protein ns2a disables its ability to inhibit α/β interferon induction and attenuates virus virulence in mice. J. Virol. 2006, 80, 2396–2404. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Jordan, J.L. Subversion of interferon by dengue virus. Curr. Top. Microbiol. Immunol. 2010, 338, 35–44. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Márquez-Jurado, S.; Nogales, A.; Ávila-Pérez, G.; Iborra, F.J.; Martínez-Sobrido, L.; Almazán, F. An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo. Viruses 2018, 10, 547. https://doi.org/10.3390/v10100547
Márquez-Jurado S, Nogales A, Ávila-Pérez G, Iborra FJ, Martínez-Sobrido L, Almazán F. An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo. Viruses. 2018; 10(10):547. https://doi.org/10.3390/v10100547
Chicago/Turabian StyleMárquez-Jurado, Silvia, Aitor Nogales, Ginés Ávila-Pérez, Francisco J. Iborra, Luis Martínez-Sobrido, and Fernando Almazán. 2018. "An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo" Viruses 10, no. 10: 547. https://doi.org/10.3390/v10100547
APA StyleMárquez-Jurado, S., Nogales, A., Ávila-Pérez, G., Iborra, F. J., Martínez-Sobrido, L., & Almazán, F. (2018). An Alanine-to-Valine Substitution in the Residue 175 of Zika Virus NS2A Protein Affects Viral RNA Synthesis and Attenuates the Virus In Vivo. Viruses, 10(10), 547. https://doi.org/10.3390/v10100547






