Viruses.STRING: A Virus-Host Protein-Protein Interaction Database
Abstract
1. Introduction
2. Materials and Methods
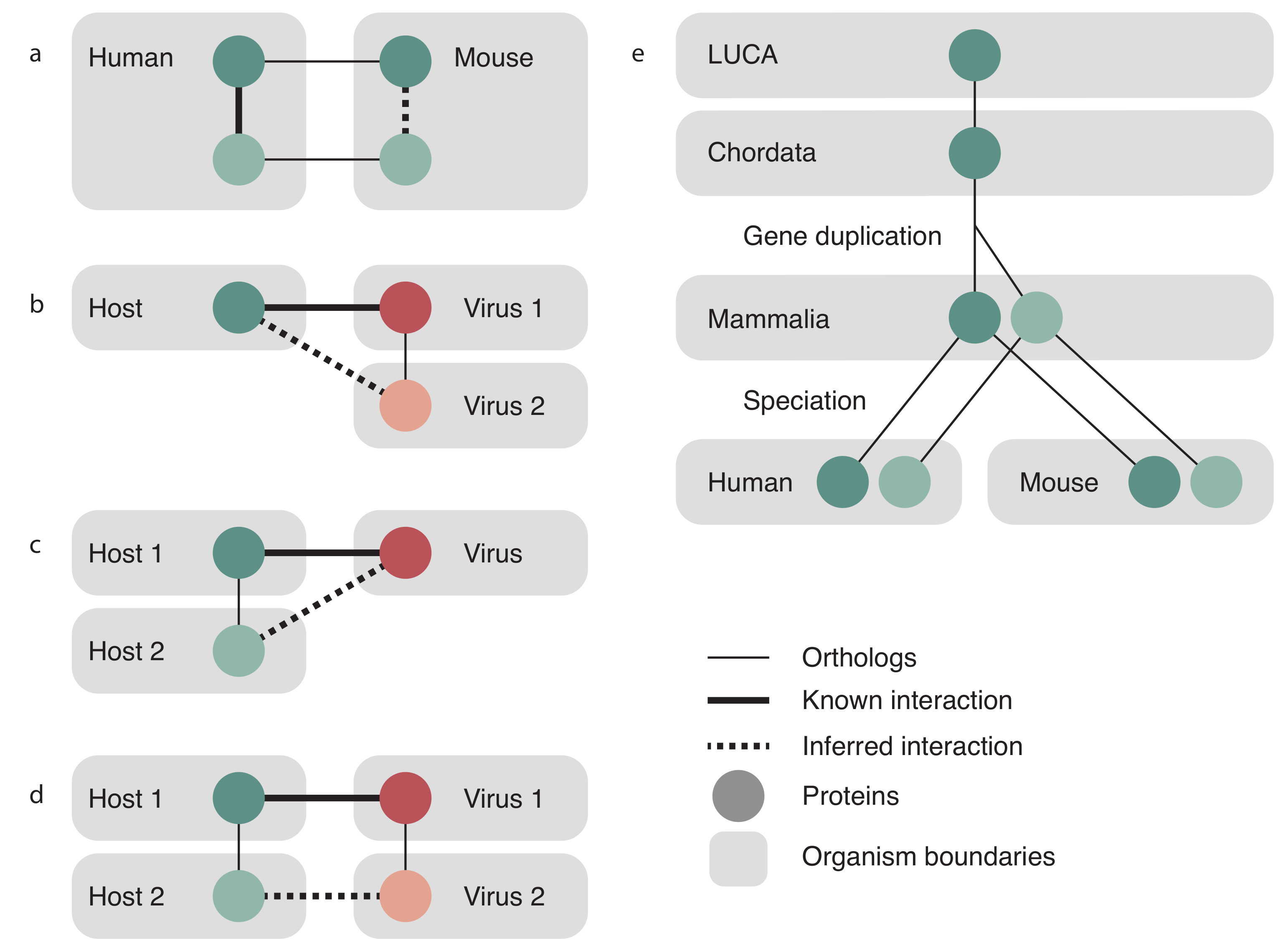
2.1. Text Mining Evidence
2.2. Experimental Evidence
2.3. Transfer Evidence
3. Results
3.1. Utility and Examples
3.1.1. Web Interface
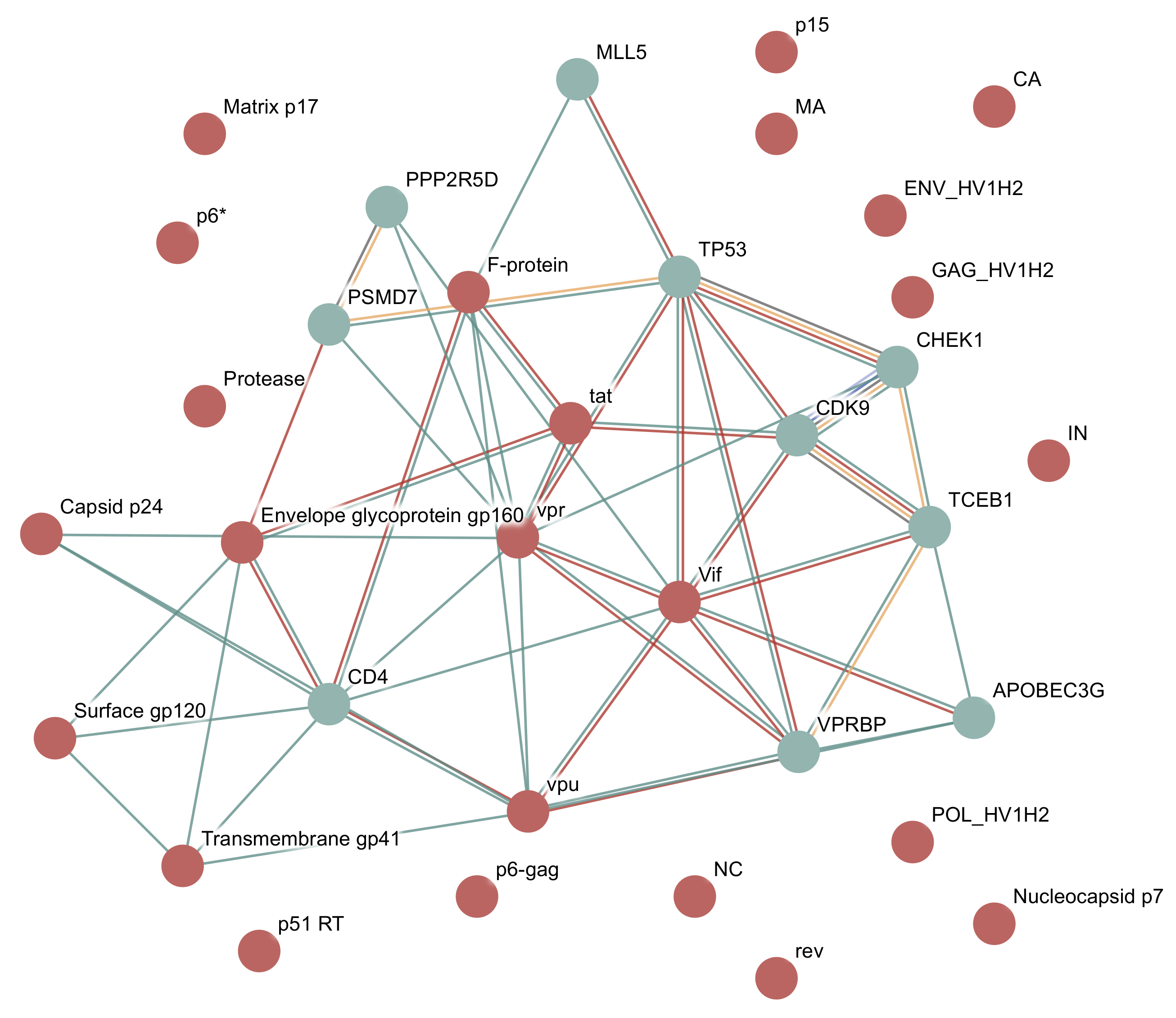
3.1.2. Example: HIV-1
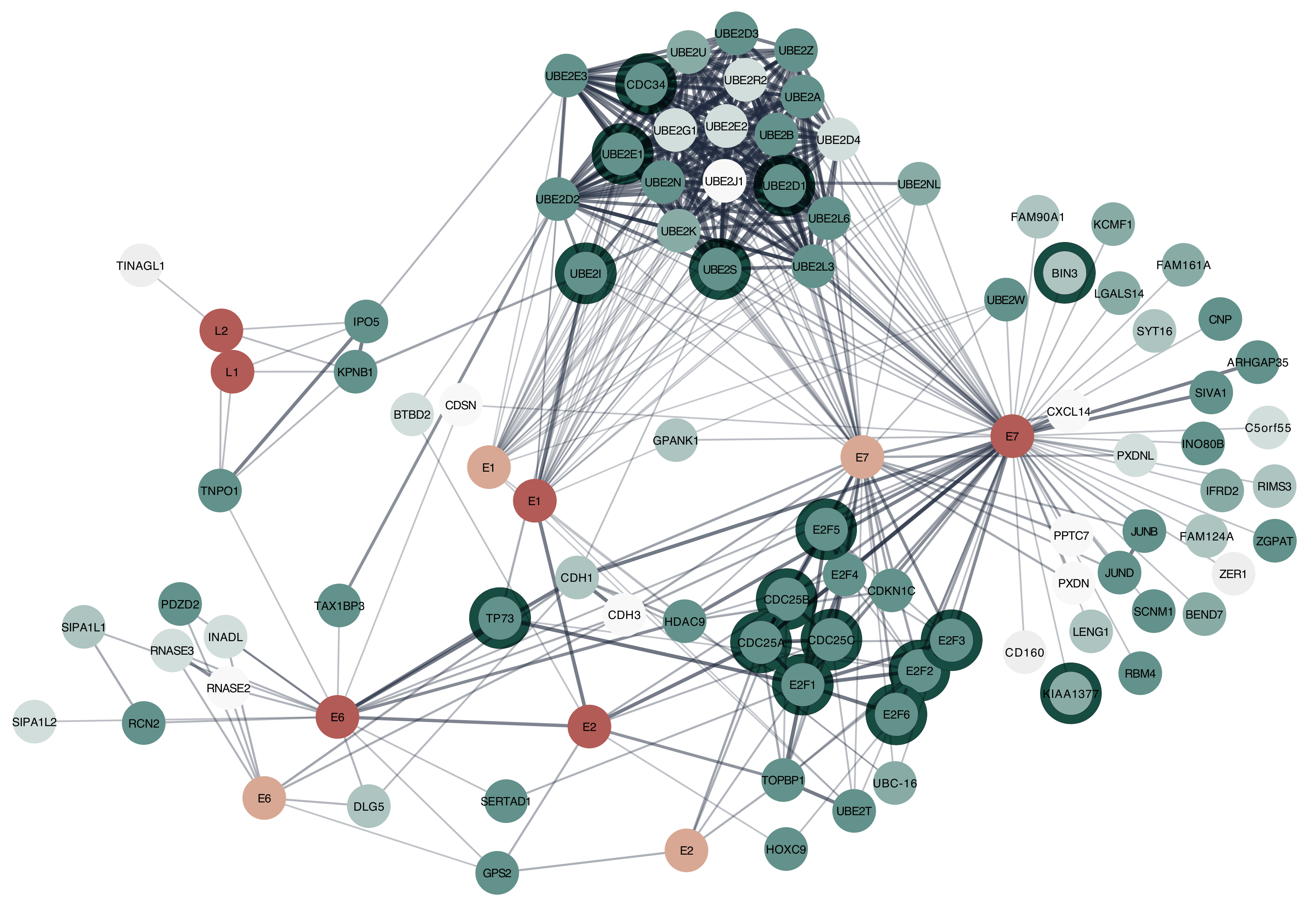
3.1.3. Cytoscape STRING App
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| HCMV | Human cytomegalovirus |
| HIV | Human immunodeficiency virus |
| MCMV | Murine cytomegalovirus |
| PPI | Protein–protein interaction |
References
- WHO. WHO Fact Sheets: Influenza, HCV, HPV; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Ceters for Disease Control and Prevention, Ebola (Ebola Virus Disease). Cost of the Ebola Epidemic. Available online: https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/cost-of-ebola.html (accessed on 7 July 2017).
- Molinari, N.A.M.; Ortega-Sanchez, I.R.; Messonnier, M.L.; Thompson, W.W.; Wortley, P.M.; Weintraub, E.; Bridges, C.B. The annual impact of seasonal influenza in the US: Measuring disease burden and costs. Vaccine 2007, 25, 5086–5096. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.N.; Gage, K.L.; Khan, A.S. Potential influence of climate change on vector-borne and zoonotic diseases: A review and proposed research plan. Environ. Health Perspect. 2010, 118, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Morens, D.M. Zika Virus in the Americas—Yet Another Arbovirus Threat. N. Engl. J. Med. 2016, 374, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Crameri, G. Emerging zoonotic viral diseases. Rev. Sci. Tech. Off. Int. Epizoot. 2014, 33, 569–581. [Google Scholar] [CrossRef]
- Maynard, N.D.; Gutschow, M.V.; Birch, E.W.; Covert, M.W. The virus as metabolic engineer. Biotechnol. J. 2010, 5, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Gulbahce, N.; Yan, H.; Dricot, A.; Padi, M.; Byrdsong, D.; Franchi, R.; Lee, D.S.; Rozenblatt-Rosen, O.; Mar, J.C.; Calderwood, M.A.; et al. Viral Perturbations of Host Networks Reflect Disease Etiology. PLoS Comput. Biol. 2012, 8, e1002531. [Google Scholar] [CrossRef] [PubMed]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef] [PubMed]
- Frentz, D.; Boucher, C.A.B.; Van De Vijver, D.A.M.C. Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS Rev. 2012, 14, 17–27. [Google Scholar] [PubMed]
- Razonable, R.R. Antiviral drugs for viruses other than human immunodeficiency virus. Mayo Clin. Proc. 2011, 86, 1009–1026. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.M. Hepatitis C Virus Resistance to Direct-Acting Antiviral Drugs in Interferon-Free Regimens. Gastroenterology 2016, 151, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Herpesvirus Resistance to Antiviral Drugs. In Antimicrobial Drug Resistance; Number 18; Academic Press: New York, NY, USA, 2009; pp. 171–181. [Google Scholar]
- Murali, T.M.; Dyer, M.D.; Badger, D.; Tyler, B.M.; Katze, M.G. Network-based prediction and analysis of HIV dependency factors. PLoS Comput. Biol. 2011, 7, e1002164. [Google Scholar] [CrossRef] [PubMed]
- Ehreth, J. The global value of vaccination. Vaccine 2003, 21, 596–600. [Google Scholar] [CrossRef]
- Soema, P.C.; Kompier, R.; Amorij, J.P.; Kersten, G.F.A. Current and next generation influenza vaccines: Formulation and production strategies. Eur. J. Pharm. Biopharm. 2015, 94, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Moyle, P.M.; Toth, I. Modern Subunit Vaccines: Development, Components, and Research Opportunities. ChemMedChem 2013, 8, 360–376. [Google Scholar] [CrossRef] [PubMed]
- Calderone, A.; Licata, L.; Cesareni, G. VirusMentha: A new resource for virus–host protein interactions. Nucleic Acids Res. 2014, 43, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ammari, M.G.; Gresham, C.R.; McCarthy, F.M.; Nanduri, B. HPIDB 2.0: A curated database for host-pathogen interactions. Database 2016, 2016, baw103. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z. PubMed and beyond: A survey of web tools for searching biomedical literature. Database 2011, 2011, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Attwood, T.; Agit, B.; Ellis, L. Longevity of Biological Databases. EMBnet J. 2015, 21, e803. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Pafilis, E.; Frankild, S.P.; Fanini, L.; Faulwetter, S.; Pavloudi, C.; Vasileiadou, A.; Arvanitidis, C.; Jensen, L.J. The SPECIES and ORGANISMS Resources for Fast and Accurate Identification of Taxonomic Names in Text. PLoS ONE 2013, 8, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009, 37, D5–D15. [Google Scholar] [CrossRef] [PubMed]
- Kibbe, W.A.; Arze, C.; Felix, V.; Mitraka, E.; Bolton, E.; Fu, G.; Mungall, C.J.; Binder, J.X.; Malone, J.; Vasant, D.; et al. Disease Ontology 2015 update: An expanded and updated database of Human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015, 43, D1071–D1078. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier Academic Press: New York, NY, USA, 2012. [Google Scholar]
- The UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212. [Google Scholar] [CrossRef]
- Cook, H.V.; Berzins, R.; Rodriguez, C.L.; Cejuela, J.M.; Jensen, L.J. Creation and evaluation of a dictionary-based tagger for virus species and proteins. In Proceedings of the BioNLP 2017 Workshop, Association for Computational Linguistics, Vancouver, BC, Canada, 4 August 2017; pp. 91–98. [Google Scholar]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D15. [Google Scholar] [CrossRef] [PubMed]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Oughtred, R.; Boucher, L.; Heinicke, S.; Chen, D.; Stark, C.; Breitkreutz, A.; Kolas, N.; O’Donnell, L.; et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015, 43, D470–D478. [Google Scholar] [CrossRef] [PubMed]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; Del-Toro, N.; et al. The MIntAct project - IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Xenarios, I.; Salwínski, L.; Duan, X.J.; Higney, P.; Kim, S.M.; Eisenberg, D. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002, 30, 303–305. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein–protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015, 44, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Epstein, J.H.; Murray, K.A.; Navarrete-Macias, I.; Zambrana-Torrelio, C.; Soloyvov, A.; Ojeda-Flores, R.; Arrigio, N.C.; Islam, A.; Kahn, S.A.; et al. A Strategy to Estimate Unknown Viral Diversity in Mammals. mBio 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Palasca, O.; Santos, A.; Stolte, C.; Gorodkin, J.; Jensen, L.J. TISSUES 2.0: An integrative web resource on mammalian tissue expression. Database 2018, 2018, bay003. [Google Scholar] [CrossRef] [PubMed]
- Reinson, T.; Henno, L.; Toots, M.; Ustav, M.; Ustav, M. The cell cycle timing of human papillomavirus DNA replication. PLoS ONE 2015, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- De Chassey, B.; Meyniel-Schicklin, L.; Vonderscher, J.; André, P.; Lotteau, V. Virus-host interactomics: New insights and opportunities for antiviral drug discovery. Genome Med. 2014, 6, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fu, B.; Li, W.; Patil, G.; Liu, L.; Dorf, M.E.; Li, S. Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat. Commun. 2017, 8, 13876. [Google Scholar] [CrossRef] [PubMed]
- Gaballa, A.; Newton, G.L.; Antelmann, H.; Parsonage, D.; Upton, H.; Rawat, M.; Claiborne, A.; Fahey, R.C.; Helmann, J.D. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc. Natl. Acad. Sci. USA 2010, 107, 6482–6486. [Google Scholar] [CrossRef] [PubMed]

| Number of Genes | FDR p-Value | GO Term |
|---|---|---|
| 549 | 2.28E-104 | positive regulation of macromolecule metabolic process |
| 718 | 5.96E-93 | positive regulation of cellular process |
| 452 | 3.02E-91 | cell surface receptor signaling pathway |
| 758 | 7.87E-91 | protein binding |
| 779 | 1.23E-90 | positive regulation of biological process |
| 539 | 1.61E-87 | positive regulation of cellular metabolic process |
| 459 | 3.86E-84 | multi-organism process |
| 277 | 4.56E-82 | innate immune response |
| 468 | 5.27E-82 | carbohydrate derivative binding |
| 595 | 1.99E-81 | response to stress |
| 240 | 2.07E-81 | multi-organism cellular process |
| 254 | 2.4E-81 | regulation of immune response |
| 239 | 2.55E-81 | viral process |
| 583 | 4.17E-81 | regulation of response to stimulus |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cook, H.V.; Doncheva, N.T.; Szklarczyk, D.; Von Mering, C.; Jensen, L.J. Viruses.STRING: A Virus-Host Protein-Protein Interaction Database. Viruses 2018, 10, 519. https://doi.org/10.3390/v10100519
Cook HV, Doncheva NT, Szklarczyk D, Von Mering C, Jensen LJ. Viruses.STRING: A Virus-Host Protein-Protein Interaction Database. Viruses. 2018; 10(10):519. https://doi.org/10.3390/v10100519
Chicago/Turabian StyleCook, Helen Victoria, Nadezhda Tsankova Doncheva, Damian Szklarczyk, Christian Von Mering, and Lars Juhl Jensen. 2018. "Viruses.STRING: A Virus-Host Protein-Protein Interaction Database" Viruses 10, no. 10: 519. https://doi.org/10.3390/v10100519
APA StyleCook, H. V., Doncheva, N. T., Szklarczyk, D., Von Mering, C., & Jensen, L. J. (2018). Viruses.STRING: A Virus-Host Protein-Protein Interaction Database. Viruses, 10(10), 519. https://doi.org/10.3390/v10100519





