HIV-1 Protease: Structural Perspectives on Drug Resistance
Abstract
1. Introduction
2. HIV protease and structure guided design of PIs
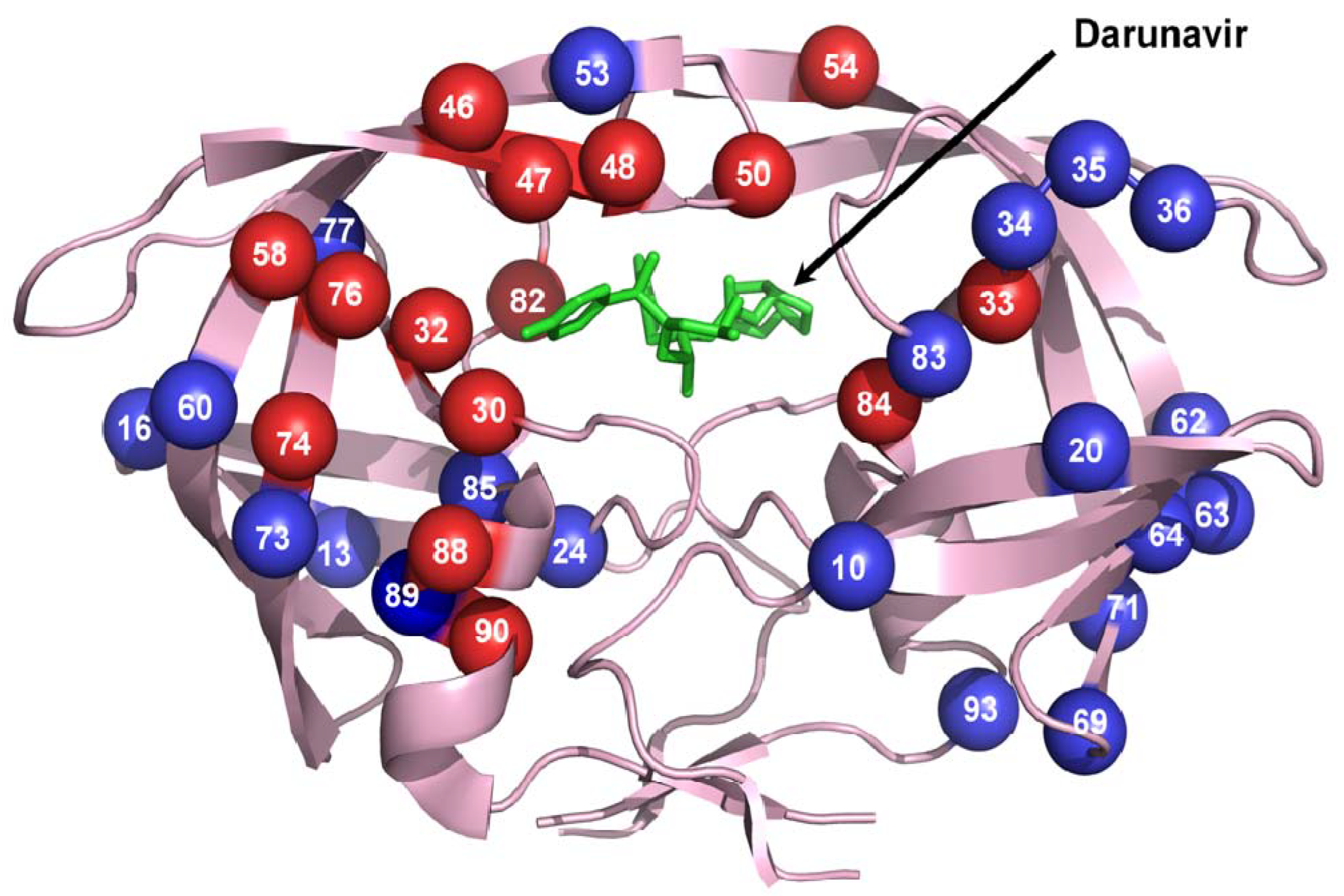
3. Evolution of Drug resistance to PIs
4. Molecular mechanisms of drug resistance
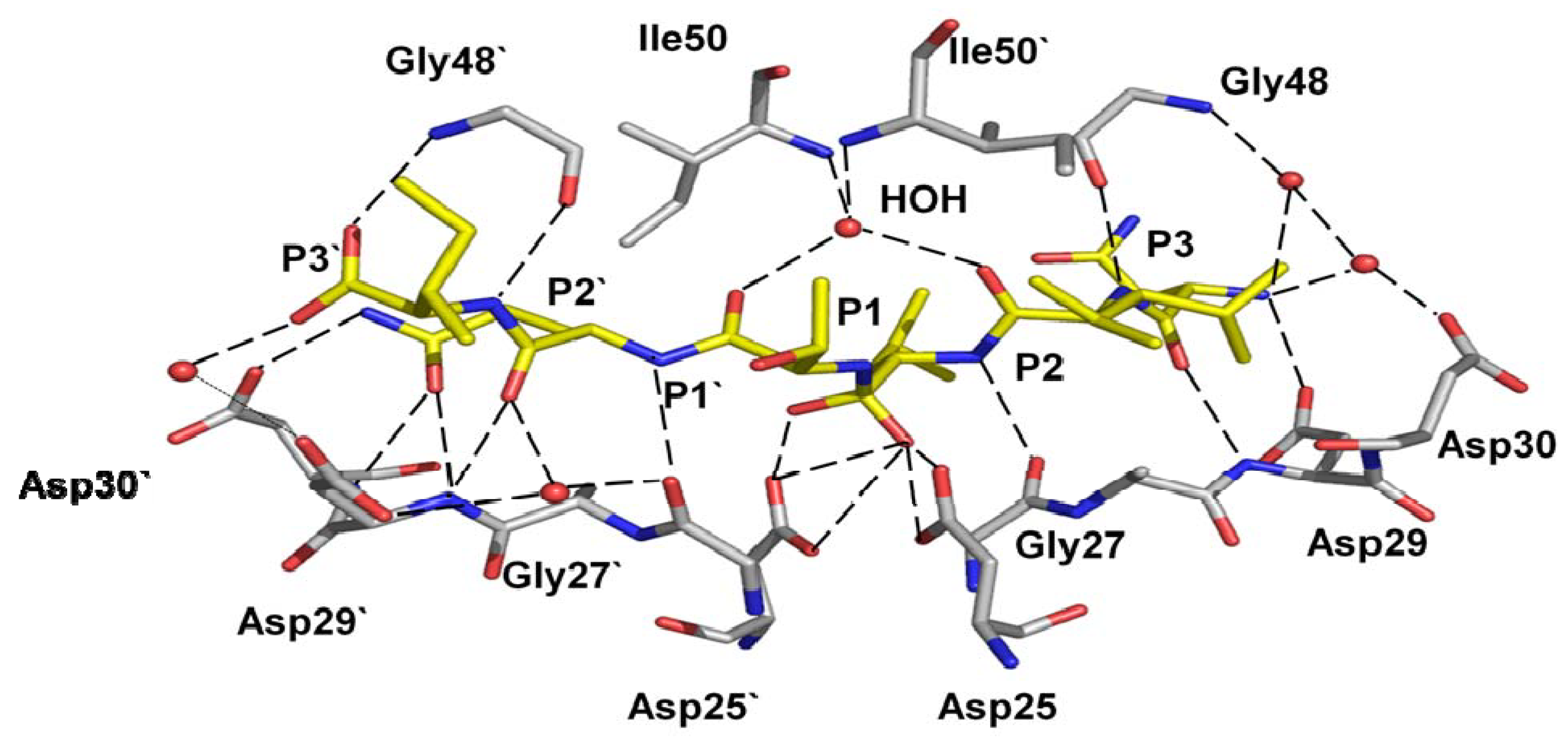
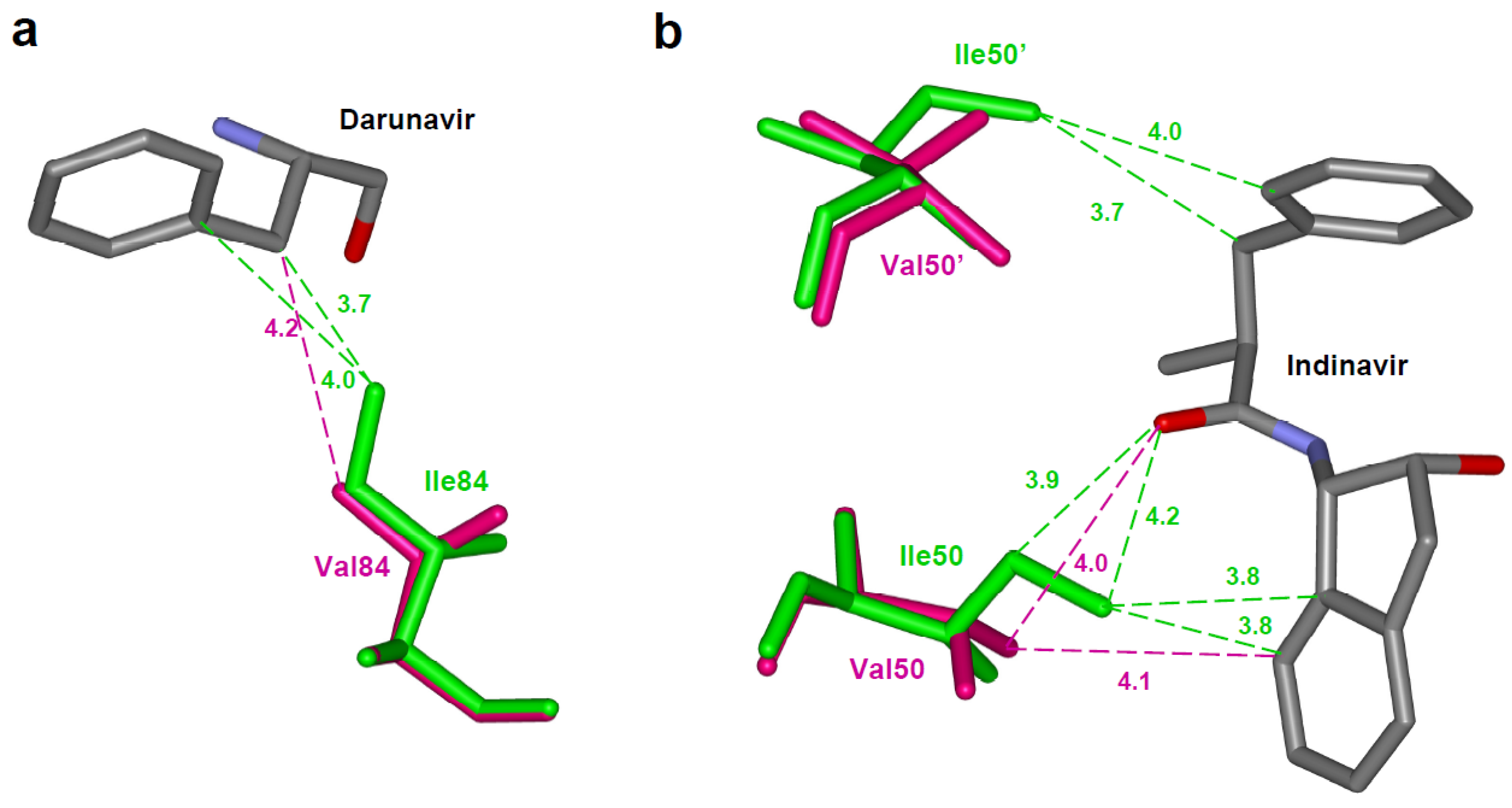
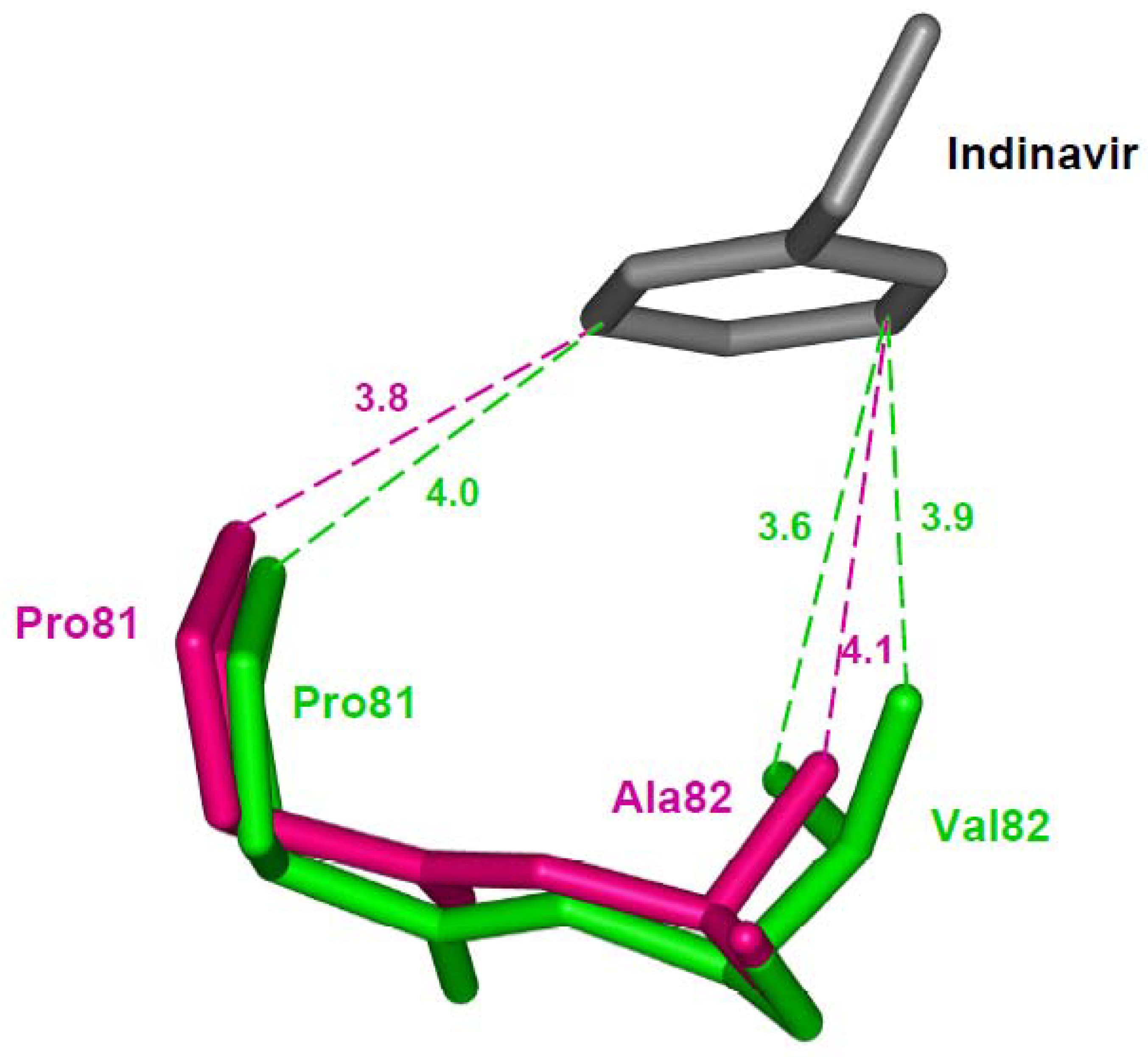
4.1. Reduced interactions with inhibitor
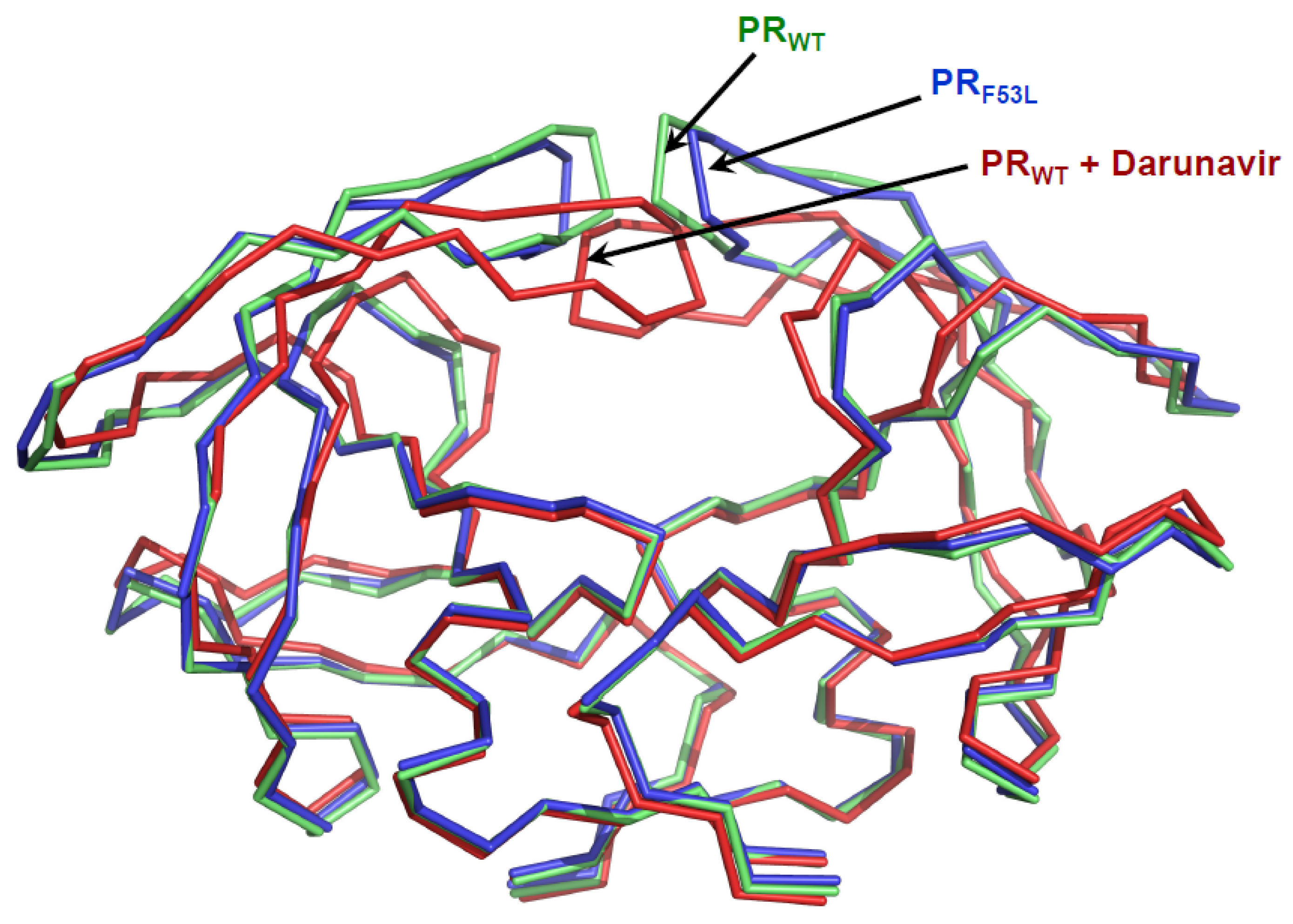
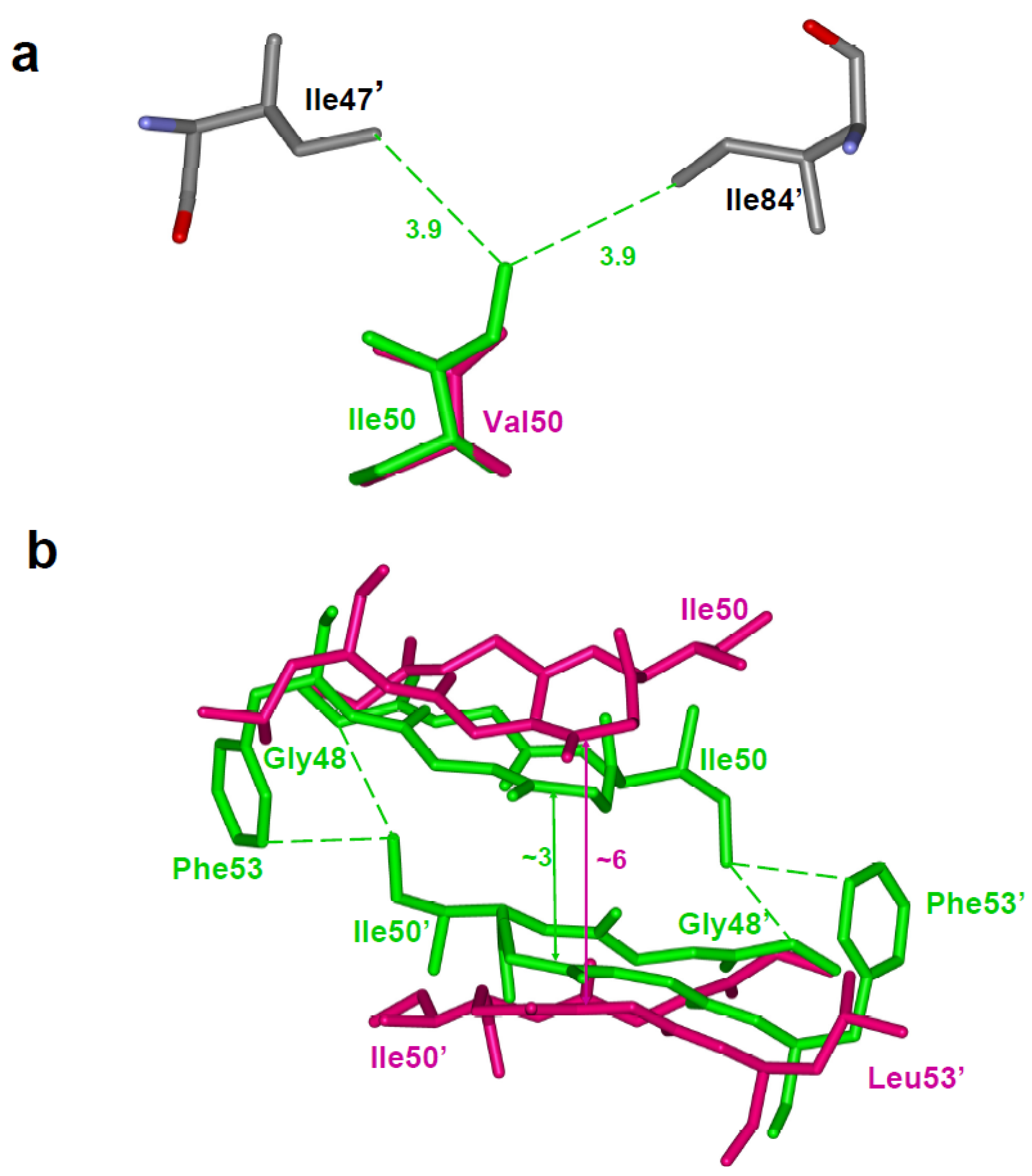
4.2. Main chain shift due to mutation
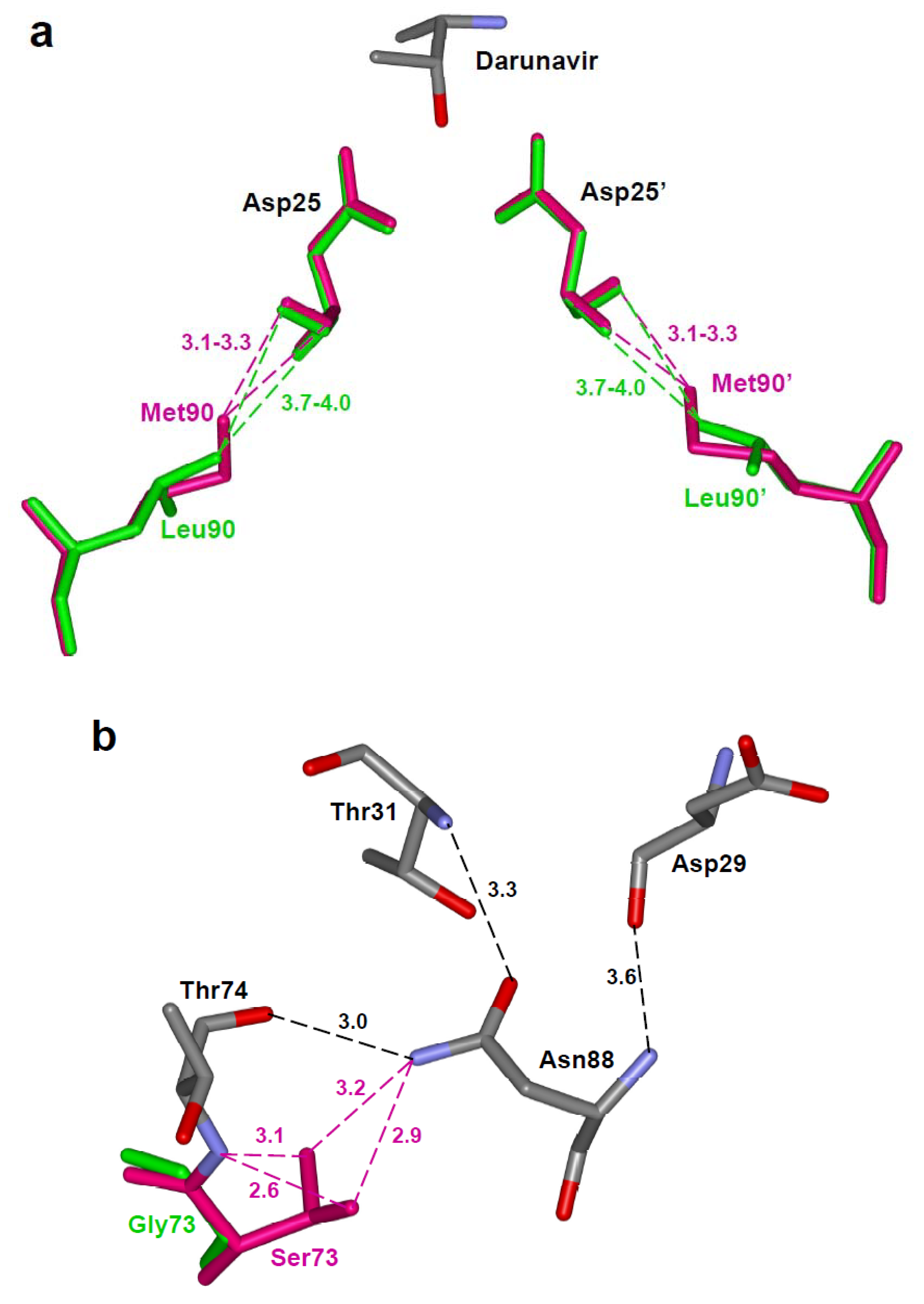
4.3. Mutations that alter the dimer interface
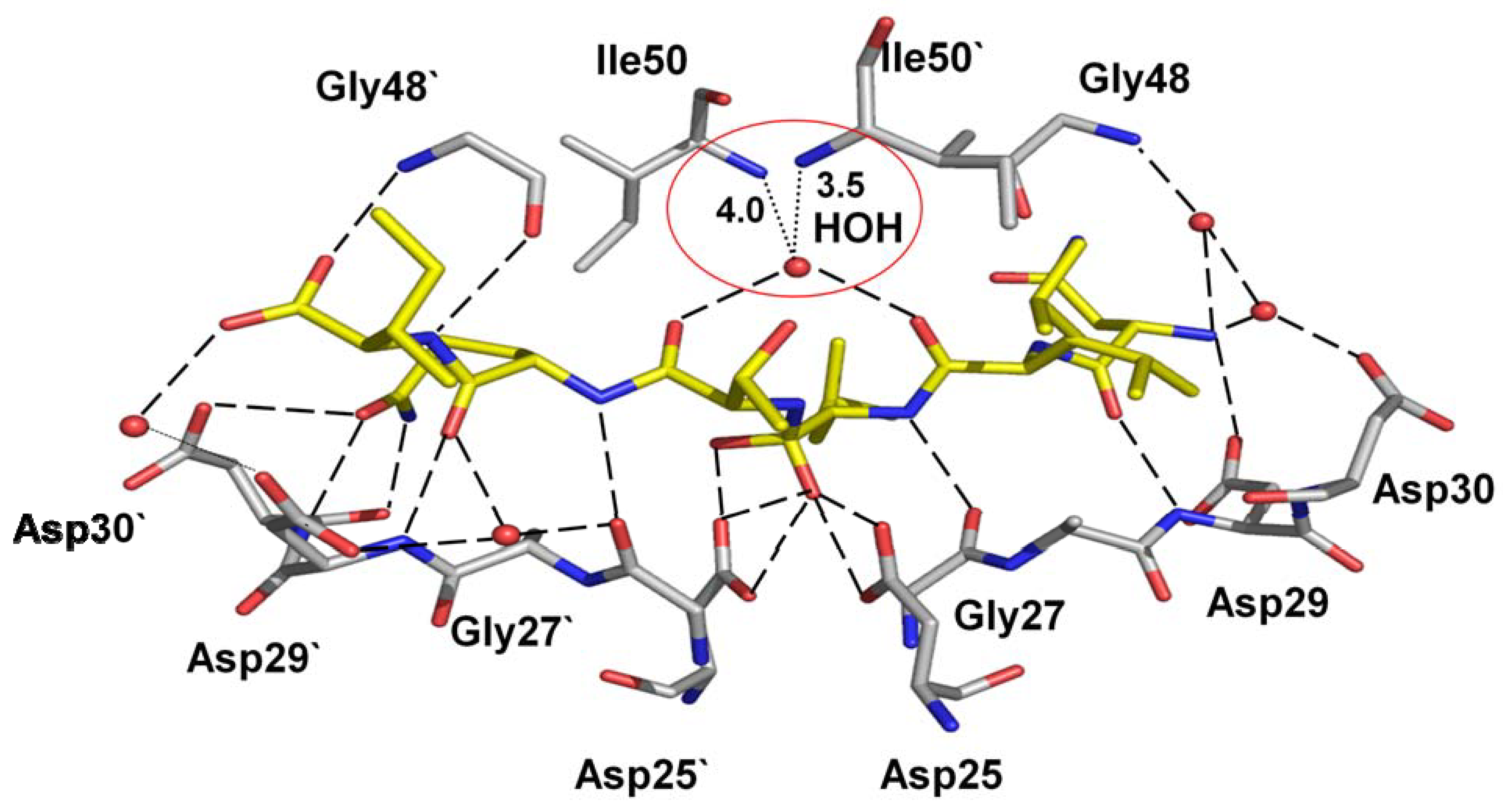
4.4. Distal mutations that transmit changes to the active site cavity
4.5. Reduced interaction with intermediate
5. Studies with clinical isolates containing multiple mutations
6. Structure-guided design of PIs for resistant HIV protease
7. Conclusions
Acknowledgments
References
- Weber, I.T.; Zhang, Y.; Tozser, J. HIV-1 Protease and AIDS Therapy. In Viral Proteases And Antiviral Protease Inhibitor Therapy; Lendeckel, U., Hooper, N., Eds.; in press.[Green Version]
- Palella, F.J., Jr.; Delaney, K.M.; Moorman, A.C.; Loveless, M.O.; Fuhrer, J.; Satten, G.A.; Aschman, D.J.; Holmberg, S.D. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N. Engl. J. Med. 1998, 338, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.M.; Squires, K.E.; Hughes, M.D.; Grimes, J.M.; Demeter, L.M.; Currier, J.S.; Eron, J.J., Jr.; Feinberg, J.E.; Balfour, H.H., Jr.; Deyton, L.R.; Chodakewitz, J.A.; Fischl, M.A. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 1997, 337, 725–733. [Google Scholar] [CrossRef]
- Sepkowitz, K.A. AIDS--the first 20 years. N. Eng. J. Med. 2001, 344, 1764–1772. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Chapsal, B.D.; Weber, I.T.; Mitsuya, H. Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance. Acc. Chem. Res. 2008, 41, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Leshchenko-Yashchuk, S.; Anderson, D.D.; Baldridge, A.; Noetzel, M.; Miller, H.B.; Tie, Y.; Wang, Y.F.; Koh, Y.; Weber, I.T.; Mitsuya, H. Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1’-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2009, 52, 3902–3914. [Google Scholar] [CrossRef] [PubMed]
- Mocroft, A.; Ruiz, L.; Reiss, P.; Ledergerber, B.; Katlama, C.; Lazzarin, A.; Goebel, F.D.; Phillips, A.N.; Clotet, B.; Lundgren, J.D. Virological rebound after suppression on highly active antiretroviral therapy. AIDS 2003, 17, 1741–1751. [Google Scholar] [CrossRef]
- Leigh Brown, A.J.; Frost, S.D.; Mathews, W.C.; Dawson, K.; Hellmann, N.S.; Daar, E.S.; Richman, D.D.; Little, S. J. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J. Infect. Dis. 2003, 187, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Richman, D.D.; Morton, S.C.; Wrin, T.; Hellmann, N.; Berry, S.; Shapiro, M.F.; Bozzette, S.A. The prevalence of antiretroviral drug resistance in the United States. AIDS 2004, 18, 1393–1401. [Google Scholar] [CrossRef]
- Hurt, C.B.; McCoy, S.I.; Kuruc, J.; Nelson, J.A.; Kerkau, M.; Fiscus, S.; McGee, K.; Sebastian, J.; Leone, P.; Pilcher, C.; Hicks, C.; Eron, J. Transmitted antiretroviral drug resistance among acute and recent HIV infections in North Carolina from 1998 to 2007. Antivir. Ther. 2009, 14, 673–678. [Google Scholar] [CrossRef]
- Grossman, Z.; Lorber, M.; Maayan, S.; Bar-Yacov, N.; Levy, I.; Averbuch, D.; Istomin, V.; Chowers, M.; Sthoeger, Z.; Ram, D.; Rudich, H.; Mileguir, F.; Pavel, R.; Almaliach, R.; Schlaeffer, F.; Kra-Oz, Z.; Mendelson, E.; Schapiro, J. M.; Riesenberg, K. Drug-resistant HIV infection among drug-naive patients in Israel. Clin. Infect. Dis. 2005, 40, 294–302. [Google Scholar] [CrossRef]
- Grant, R.M.; Hecht, F.M.; Warmerdam, M.; Liu, L.; Liegler, T.; Petropoulos, C.J.; Hellmann, N.S.; Chesney, M.; Busch, M.P.; Kahn, J.O. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 2002, 288, 181–188. [Google Scholar] [CrossRef]
- Miller, V. International perspectives on antiretroviral resistance. Resistance to protease inhibitors. J. Acquir. Immune. Defic. Syndr. 2001, 26 Suppl 1, S34–S50. [Google Scholar] [CrossRef]
- Shafer, R.W.; Schapiro, J.M. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008, 10, 67–84. [Google Scholar]
- Hammer, S.M.; Saag, M.S.; Schechter, M.; Montaner, J.S.; Schooley, R.T.; Jacobsen, D.M.; Thompson, M.A.; Carpenter, C.C.; Fischl, M.A.; Gazzard, B.G.; Gatell, J.M.; Hirsch, M.S.; Katzenstein, D.A.; Richman, D.D.; Vella, S.; Yeni, P.G.; Volberding, P.A. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society--USA panel. Top. HIV. Med. 2006, 14, 827–843. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Zack, J.A.; Knigge, M.; Paul, D.A.; Kempf, D.J.; Norbeck, D.W.; Swanstrom, R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 1993, 67, 4050–4055. [Google Scholar] [CrossRef]
- Rose, J.R.; Babe, L. M.; Craik, C.S. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 1995, 69, 2751–2758. [Google Scholar] [CrossRef]
- Kohl, N.E.; Emini, E.A.; Schleif, W.A.; Davis, L.J.; Heimbach, J.C.; Dixon, R.A.; Scolnick, E.M.; Sigal, I.S. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. U.S.A 1988, 85, 4686–4690. [Google Scholar] [CrossRef]
- Peng, C.; Ho, B.K.; Chang, T.W.; Chang, N.T. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J. Virol. 1989, 63, 2550–2556. [Google Scholar] [CrossRef] [PubMed]
- Wlodawer, A.; Vondrasek, J. Inhibitors of HIV-1 protease: a major success of structure-assisted drug design. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 249–284. [Google Scholar] [CrossRef]
- Noble, M.E.; Endicott, J.A.; Johnson, L.N. Protein kinase inhibitors: insights into drug design from structure. Science 2004, 303, 1800–1805. [Google Scholar] [CrossRef] [PubMed]
- Alymova, I.V.; Taylor, G.; Portner, A. Neuraminidase inhibitors as antiviral agents. Curr. Drug Targets. Infect. Disord. 2005, 5, 401–409. [Google Scholar] [CrossRef]
- Mittl, P.R.; Grutter, M.G. Opportunities for structure-based design of protease-directed drugs. Curr. Opin. Struct. Biol. 2006, 16, 769–775. [Google Scholar] [CrossRef]
- Ghosh, A.K. Harnessing nature’s insight: design of aspartyl protease inhibitors from treatment of drug-resistant HIV to Alzheimer’s disease. J. Med. Chem. 2009, 52, 2163–2176. [Google Scholar] [CrossRef]
- Rose, R.B.; Craik, C.S.; Stroud, R.M. Domain flexibility in retroviral proteases: structural implications for drug resistant mutations. Biochemistry 1998, 37, 2607–2621. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.R.; Schiffer, C.A. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure 2000, 8, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Perryman, A.L.; Lin, J.H.; McCammon, J. A. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004, 13, 1108–1123. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Kovalevsky, A.Y.; Louis, J.M.; Boross, P.I.; Wang, Y.F.; Harrison, R.W.; Weber, I.T. Mechanism of drug resistance revealed by the crystal structure of the unliganded HIV-1 protease with F53L mutation. J. Mol. Biol. 2006, 358, 1191–1199. [Google Scholar] [CrossRef]
- Spinelli, S.; Liu, Q.Z.; Alzari, P.M.; Hirel, P.H.; Poljak, R.J. The three-dimensional structure of the aspartyl protease from the HIV-1 isolate BRU. Biochimie 1991, 73, 1391–1396. [Google Scholar] [CrossRef]
- Tie, Y.; Boross, P.I.; Wang, Y.F.; Gaddis, L.; Hussain, A.K.; Leshchenko, S.; Ghosh, A.K.; Louis, J.M.; Harrison, R.W.; Weber, I.T. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol.Biol. 2004, 338, 341–352. [Google Scholar] [CrossRef]
- Gustchina, A.; Sansom, C.; Prevost, M.; Richelle, J.; Wodak, S.Y.; Wlodawer, A.; Weber, I.T. Energy calculations and analysis of HIV-1 protease-inhibitor crystal structures. Protein Eng 1994, 7, 309–317. [Google Scholar] [CrossRef]
- Kovalevsky, A. Y.; Chumanevich, A. A.; Liu, F.; Louis, J.M.; Weber, I.T. Caught in the Act: the 1.5 A resolution crystal structures of the HIV-1 protease and the I54V mutant reveal a tetrahedral reaction intermediate. Biochemistry 2007, 46, 14854–14864. [Google Scholar] [CrossRef]
- Kumar, M.; Prashar, V.; Mahale, S.; Hosur, M.V. Observation of a tetrahedral reaction intermediate in the HIV-1 protease-substrate complex. Biochem. J. 2005, 389, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Tie, Y.; Boross, P.I.; Wang, Y.F.; Gaddis, L.; Liu, F.; Chen, X.; Tozser, J.; Harrison, R.W.; Weber, I.T. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005, 272, 5265–5277. [Google Scholar] [CrossRef]
- Beck, Z.Q.; Morris, G.M.; Elder, J.H. Defining HIV-1 Protease Substrate Selectivity. Curr. Drug Targets. Infect. Disord. 2002, 2, 37–50. [Google Scholar] [CrossRef]
- Tozser, J. Comparative Studies on retroviral Proteases: Substrate Specificity. Viruses submitted for publication. 2009. [Google Scholar][Green Version]
- Roberts, N.A.; Martin, J.A.; Kinchington, D.; Broadhurst, A.V.; Craig, J.C.; Duncan, I.B.; Galpin, S.A.; Handa, B.K.; Kay, J.; Krohn, A. Rational design of peptide-based HIV proteinase inhibitors. Science 1990, 248, 358–361. [Google Scholar] [CrossRef]
- Mehandru, S.; Markowitz, M. Tipranavir: a novel non-peptidic protease inhibitor for the treatment of HIV infection. Expert. Opin. Investig. Drugs 2003, 12, 1821–1828. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.; Nakata, H.; Maeda, K.; Ogata, H.; Bilcer, G.; Devasamudram, T.; Kincaid, J.F.; Boross, P.; Wang, Y.F.; Tie, Y.; Volarath, P.; Gaddis, L.; Harrison, R.W.; Weber, I.T.; Ghosh, A.K.; Mitsuya, H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2003, 47, 3123–3129. [Google Scholar] [CrossRef]
- Jacobsen, H.; Yasargil, K.; Winslow, D.L.; Craig, J.C.; Krohn, A.; Duncan, I.B.; Mous, J. Characterization of human immunodeficiency virus type 1 mutants with decreased sensitivity to proteinase inhibitor Ro 31-8959. Virology 1995, 206, 527–534. [Google Scholar] [CrossRef]
- Harrigan, P.R.; Hogg, R.S.; Dong, W.W.; Yip, B.; Wynhoven, B.; Woodward, J.; Brumme, C.J.; Brumme, Z.L.; Mo, T.; Alexander, C.S.; Montaner, J.S. Predictors of HIV drug-resistance mutations in a large antiretroviral-naive cohort initiating triple antiretroviral therapy. J. Infect. Dis. 2005, 191, 339–347. [Google Scholar] [CrossRef]
- Weiser, S.D.; Guzman, D.; Riley, E.D.; Clark, R.; Bangsberg, D.R. Higher rates of viral suppression with nonnucleoside reverse transcriptase inhibitors compared to single protease inhibitors are not explained by better adherence. HIV. Clin. Trials 2004, 5, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, J. M.; Miller, C.; Currier, J.S.; Forthal, D.N.; Kemper, C.A.; Beall, G.N.; Tilles, J.G.; Capparelli, E.V.; McCutchan, J.A.; Haubrich, R.H. The correlation between plasma concentrations of protease inhibitors, medication adherence and virological outcome in HIV-infected patients. Antivir. Ther. 2004, 9, 753–761. [Google Scholar] [PubMed]
- Paterson, D.L.; Swindells, S.; Mohr, J.; Brester, M.; Vergis, E.N.; Squier, C.; Wagener, M.M.; Singh, N. Adherence to protease inhibitor therapy and outcomes in patients with HIV infection. Ann. Intern. Med. 2000, 133, 21–30. [Google Scholar] [CrossRef]
- Nolan, D.; Reiss, P.; Mallal, S. Adverse effects of antiretroviral therapy for HIV infection: a review of selected topics. Expert. Opin. Drug Saf. 2005, 4, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Wensing, A.M.; Boucher, C.A. Worldwide transmission of drug-resistant HIV. AIDS Rev. 2003, 5, 140–155. [Google Scholar]
- Kuritzkes, D.R. Resistance to protease inhibitors. J. HIV. Ther. 2002, 7, 87–91. [Google Scholar]
- Shafer, R.W. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 2002, 15, 247–277. [Google Scholar] [CrossRef]
- Vergne, L.; Peeters, M.; Mpoudi-Ngole, E.; Bourgeois, A.; Liegeois, F.; Toure-Kane, C.; Mboup, S.; Mulanga-Kabeya, C.; Saman, E.; Jourdan, J.; Reynes, J.; Delaporte, E. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J. Clin. Microbiol. 2000, 38, 3919–3925. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cajas, J.L.; Wainberg, M.A. Protease inhibitor resistance in HIV-infected patients: molecular and clinical perspectives. Antiviral Res. 2007, 76, 203–221. [Google Scholar] [CrossRef]
- Nijhuis, M.; van Maarseveen, N.M.; Lastere, S.; Schipper, P.; Coakley, E.; Glass, B.; Rovenska, M.; de Jong, D.; Chappey, C.; Goedegebuure, I.W.; Heilek-Snyder, G.; Dulude, D.; Cammack, N.; Brakier-Gingras, L.; Konvalinka, J.; Parkin, N.; Krausslich, H.G.; Brun-Vezinet, F.; Boucher, C.A. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS. Med. 2007, 4, e36. [Google Scholar] [CrossRef]
- Pereira-Vaz, J.; Duque, V.; Trindade, L.; Saraiva-da-Cunha, J.; Melico-Silvestre, A. Detection of the protease codon 35 amino acid insertion in sequences from treatment-naive HIV-1 subtype C infected individuals in the Central Region of Portugal. J. Clin. Virol. 2009, 46, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Kozisek, M.; Saskova, K.G.; Rezacova, P.; Brynda, J.; van Maarseveen, N.M.; de Jong, D.; Boucher, C.A.; Kagan, R.M.; Nijhuis, M.; Konvalinka, J. Ninety-nine is not enough: molecular characterization of inhibitor-resistant human immunodeficiency virus type 1 protease mutants with insertions in the flap region. J. Virol. 2008, 82, 5869–5878. [Google Scholar] [CrossRef] [PubMed]
- Tramuto, F.; Bonura, F.; Mancuso, S.; Romano, N.; Vitale, F. Detection of a new 3-base pair insertion mutation in the protease gene of human immunodeficiency virus type 1 during highly active antiretroviral therapy (HAART). AIDS Res. Hum. Retroviruses 2005, 21, 420–423. [Google Scholar] [CrossRef]
- Friend, J.; Parkin, N.; Liegler, T.; Martin, J.N.; Deeks, S.G. Isolated lopinavir resistance after virological rebound of a ritonavir/lopinavir-based regimen. AIDS 2004, 18, 1965–1966. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Campoy, A.; Muzammil, S.; Ohtaka, H.; Schon, A.; Vega, S.; Freire, E. Structural and thermodynamic basis of resistance to HIV-1 protease inhibition: implications for inhibitor design. Curr. Drug Targets. Infect. Disord. 2003, 3, 311–328. [Google Scholar] [CrossRef] [PubMed]
- Kovalevsky, A.Y.; Liu, F.; Leshchenko, S.; Ghosh, A.K.; Louis, J.M.; Harrison, R.W.; Weber, I.T. Ultra-high resolution crystal structure of HIV-1 protease mutant reveals two binding sites for clinical inhibitor TMC114. J. Mol. Biol. 2006, 363, 161–173. [Google Scholar] [CrossRef]
- Weber, I.T.; Kovalevsky, A.Y.; Harrison, R.W. Structures Of HIV Protease Guide Inhibitor Design To Overcome Drug Resistance. In Frontiers in Drug Design and Discovery: Structure-Based Drug Design in the 21st Century, 3rd ed.; Cladwell, G.W., Atta-ur-Rahman Player, M.R., Choudhry, M.I., Eds.; Bentham Science Publishers Ltd.; pp. 45–62.
- Tie, Y.; Kovalevsky, A.Y.; Boross, P.; Wang, Y.F.; Ghosh, A.K.; Tozser, J.; Harrison, R.W.; Weber, I.T. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins 2007, 67, 232–242. [Google Scholar] [CrossRef]
- Mahalingam, B.; Louis, J.M.; Reed, C.C.; Adomat, J.M.; Krouse, J.; Wang, Y.F.; Harrison, R.W.; Weber, I.T. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 1999, 263, 238–245. [Google Scholar] [CrossRef]
- Liu, F.; Boross, P.I.; Wang, Y.F.; Tozser, J.; Louis, J.M.; Harrison, R.W.; Weber, I.T. Kinetic, stability, and structural changes in high-resolution crystal structures of HIV-1 protease with drug-resistant mutations L24I, I50V, and G73S. J. Mol. Biol. 2005, 354, 789–800. [Google Scholar] [CrossRef]
- Hong, L.; Zhang, X.C.; Hartsuck, J.A.; Tang, J. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Sci. 2000, 9, 1898–1904. [Google Scholar] [CrossRef]
- Kozisek, M.; Bray, J.; Rezacova, P.; Saskova, K.; Brynda, J.; Pokorna, J.; Mammano, F.; Rulisek, L.; Konvalinka, J. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J. Mol. Biol. 2007, 374, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Mahalingam, B.; Wang, Y.F.; Boross, P.I.; Tozser, J.; Louis, J.M.; Harrison, R.W.; Weber, I.T. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur. J. Biochem. 2004, 271, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Prabu-Jeyabalan, M.; Nalivaika, E.A.; King, N.M.; Schiffer, C.A. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy. J. Virol. 2003, 77, 1306–1315. [Google Scholar] [CrossRef]
- Sayer, J. M.; Liu, F.; Ishima, R.; Weber, I.T.; Louis, J.M. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J. Biol. Chem. 2008, 283, 13459–13470. [Google Scholar] [CrossRef]
- Liu, F.; Kovalevsky, A.Y.; Tie, Y.; Ghosh, A.K.; Harrison, R.W.; Weber, I.T. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J. Mol. Biol. 2008, 381, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Kovalevsky, A.Y.; Tie, Y.; Liu, F.; Boross, P.I.; Wang, Y.F.; Leshchenko, S.; Ghosh, A.K.; Harrison, R.W.; Weber, I.T. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J. Med. Chem. 2006, 49, 1379–1387. [Google Scholar] [CrossRef]
- Mahalingam, B.; Louis, J.M.; Hung, J.; Harrison, R.W.; Weber, I.T. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 2001, 43, 455–464. [Google Scholar] [CrossRef]
- Ode, H.; Ota, M.; Neya, S.; Hata, M.; Sugiura, W.; Hoshino, T. Resistant mechanism against nelfinavir of human immunodeficiency virus type 1 proteases. J. Phys. Chem. B 2005, 109, 565–574. [Google Scholar] [CrossRef]
- Clemente, J.C.; Moose, R.E.; Hemrajani, R.; Whitford, L.R.; Govindasamy, L.; Reutzel, R.; McKenna, R.; Agbandje-McKenna, M.; Goodenow, M.M.; Dunn, B.M. Comparing the accumulation of active- and nonactive-site mutations in the HIV-1 protease. Biochemistry 2004, 43, 12141–12151. [Google Scholar] [CrossRef]
- Saskova, K.G.; Kozisek, M.; Rezacova, P.; Brynda, J.; Yashina, T.; Kagan, R.M.; Konvalinka, J. Molecular characterization of clinical isolates of human immunodeficiency virus resistant to the protease inhibitor darunavir. J. Virol. 2009, 83, 8810–8818. [Google Scholar] [CrossRef]
- Logsdon, B.C.; Vickrey, J.F.; Martin, P.; Proteasa, G.; Koepke, J.I.; Terlecky, S.R.; Wawrzak, Z.; Winters, M.A.; Merigan, T.C.; Kovari, L.C. Crystal structures of a multidrug-resistant human immunodeficiency virus type 1 protease reveal an expanded active-site cavity. J. Virol. 2004, 78, 3123–3132. [Google Scholar] [CrossRef]
- Martin, P.; Vickrey, J.F.; Proteasa, G.; Jimenez, Y.L.; Wawrzak, Z.; Winters, M.A.; Merigan, T.C.; Kovari, L. C. “Wide-open” 1.3 A structure of a multidrug-resistant HIV-1 protease as a drug target. Structure 2005, 13, 1887–1895. [Google Scholar] [CrossRef] [PubMed]
- Prabu-Jeyabalan, M.; King, N.M.; Nalivaika, E.A.; Heilek-Snyder, G.; Cammack, N.; Schiffer, C.A. Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother. 2006, 50, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, S.; Kairys, V.; Fernandes, M.X.; Schiffer, C.; Gilson, M.K. Evaluation of the substrate envelope hypothesis for inhibitors of HIV-1 protease. Proteins 2007, 68, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Altman, M.D.; Ali, A.; Reddy, G.S.; Nalam, M.N.; Anjum, S.G.; Cao, H.; Chellappan, S.; Kairys, V.; Fernandes, M.X.; Gilson, M.K.; Schiffer, C.A.; Rana, T.M.; Tidor, B. HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J. Am. Chem. Soc. 2008, 130, 6099–6113. [Google Scholar] [CrossRef]
- Bottcher, J.; Blum, A.; Dorr, S.; Heine, A.; Diederich, W. E.; Klebe, G. Targeting the open-flap conformation of HIV-1 protease with pyrrolidine-based inhibitors. ChemMedChem 2008, 3, 1337–1344. [Google Scholar] [CrossRef]
- Kozisek, M.; Cigler, P.; Lepsik, M.; Fanfrlik, J.; Rezacova, P.; Brynda, J.; Pokorna, J.; Plesek, J.; Gruner, B.; Grantz, S. K.; Vaclavikova, J.; Kral, V.; Konvalinka, J. Inorganic polyhedral metallacarborane inhibitors of HIV protease: a new approach to overcoming antiviral resistance. J. Med. Chem. 2008, 51, 4839–4843. [Google Scholar] [CrossRef]
- King, N.M.; Prabu-Jeyabalan, M.; Nalivaika, E.A.; Wigerinck, P.; de Bethune, M.P.; Schiffer, C.A. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 2004, 78, 12012–12021. [Google Scholar] [CrossRef]
- McKeage, K.; Perry, C.M.; Keam, S.J. Darunavir: a review of its use in the management of HIV infection in adults. Drugs 2009, 69, 477–503. [Google Scholar] [CrossRef]
- Katlama, C.; Esposito, R.; Gatell, J.M.; Goffard, J.C.; Grinsztejn, B.; Pozniak, A.; Rockstroh, J.; Stoehr, A.; Vetter, N.; Yeni, P.; Parys, W.; Vangeneugden, T. Efficacy and safety of TMC114/ritonavir in treatment-experienced HIV patients: 24-week results of POWER 1. AIDS 2007, 21, 395–402. [Google Scholar] [CrossRef]
- Haubrich, R.; Berger, D.; Chiliade, P.; Colson, A.; Conant, M.; Gallant, J.; Wilkin, T.; Nadler, J.; Pierone, G.; Saag, M.; van Baelen, B.; Lefebvre, E. Week 24 efficacy and safety of TMC114/ritonavir in treatment-experienced HIV patients. AIDS 2007, 21, F11–F18. [Google Scholar] [CrossRef]
- Kovalevsky, A.Y.; Ghosh, A.K.; Weber, I.T. Solution kinetics measurements suggest HIV-1 protease has two binding sites for darunavir and amprenavir. J. Med. Chem. 2008, 51, 6599–6603. [Google Scholar] [CrossRef] [PubMed]
- Kovalevsky, A.Y.; Louis, J.M.; Aniana, A.; Ghosh, A.K.; Weber, I. T. Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease. J. Mol. Biol. 2008, 384, 178–192. [Google Scholar] [CrossRef]
- Ghosh, A.K.; Gemma, S.; Baldridge, A.; Wang, Y.F.; Kovalevsky, A.Y.; Koh, Y.; Weber, I.T.; Mitsuya, H. Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2008, 51, 6021–6033. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Gemma, S.; Takayama, J.; Baldridge, A.; Leshchenko-Yashchuk, S.; Miller, H.B.; Wang, Y.F.; Kovalevsky, A.Y.; Koh, Y.; Weber, I.T.; Mitsuya, H. Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies. Org. Biomol. Chem. 2008, 6, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Koh, Y.; Das, D.; Li, J.; Leschenko, S.; Wang, Y.F.; Boross, P.I.; Weber, I.T.; Ghosh, A.K.; Mitsuya, H. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2007, 51, 2143–2155. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Kulkarni, S.; Anderson, D.D.; Hong, L.; Baldridge, A.; Wang, Y.F.; Chumanevich, A.A.; Kovalevsky, A.Y.; Tojo, Y.; Masayuki, A.; Koh, Y.; Tang, J.; Weber, I.T.; Mitsuya, H. Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance. J. Med. Chem. Epub 11 September 2009. [CrossRef]

| Drugs | Major mutations | Minor mutations |
|
L V I G K L D V L E E M K M I G I F I Q D I L I H A G T L V V N I I N L L I 10 11 13 16 20 24 30 32 33 34 35 36 43 46 47 48 50 53 54 58 60 62 63 64 69 71 73 74 76 77 82 83 84 85 88 89 90 93 | ||
Saquinavir/r | G L 48 90 V M |
L L I I A G V V I 10 24 54 62 71 73 77 82 84 I I V V V S I A V R L T F V T S |
Indinavir/r | M V I 46 82 84 I A V L F T |
L K L V M I A G L V L 10 20 24 32 36 54 71 73 76 77 90 I M I I I V V S V I M R R T A V |
Nelfinavir/r |
D L 30 90 N M |
L M M A V V I N 10 36 46 71 77 82 84 88 F I I V I A V D I L T F S T S |
Fosamprenavir/r |
I I 50 84 V V |
L V M I I G L V L 10 32 46 47 54 73 76 82 90 F I I V L S V A M I L V F R M T V S |
Lopinavir/r | V I V 32 47 82 I V A A F T S |
L K L L M I I F I L A G L I L 10 20 24 33 46 47 50 53 54 63 71 73 76 84 90 F M I F I V V L V P V S V V M I R L A L T R A V M T S |
Atazanavir/r |
I I N 50 84 88 L V S |
L G K L V L E M M G F I D I I A G V I I N L I 10 16 20 24 32 33 34 36 46 48 53 54 60 62 64 71 73 82 84 85 88 90 93 F E M I I I Q I I V L L E V L V C A V V S M L I R F L L Y V M I S F M V V V M V T T T C T L A S A |
Tipranavir/r |
L I Q T V I 33 47 58 74 82 84 F V E P L V T | L I K E M K M I H N L 10 13 20 35 36 43 46 54 69 83 90 V V M G I T L A K D M R M V |
Darunavir/r |
I I L I 50 54 76 84 V M V V L |
V V L I T L L 11 32 33 47 74 89 90 I I F V P V M |
| Mutant | Inhibitor | Ki (nM) | Ki /Ki(WT) | Reference |
| L24I | Indinavir | 1.4 | 2.6 | [61] |
| D30N | Darunavir | 6.6 | 30.0 | [68] |
| D30N | Indinavir | 7.0 | 32.0 | [68] |
| G48V | Darunavir | 17.0 | 29.0 | [67] |
| G48V | Saquinavir | 36.0 | 86.0 | [67] |
| I50V | Darunavir | 18.0 | 82.0 | [68] |
| I50V | Saquinavir | 36.0 | 164.0 | [68] |
| I50V | Indinavir | 27.0 | 50.0 | [61] |
| I54V | Darunavir | 5.0 | 8.6 | [67] |
| I54V | Saquinavir | 6.0 | 14.3 | [67] |
| I54M | Darunavir | 1.6 | 2.8 | [67] |
| I54M | Saquinavir | 2.2 | 5.2 | [67] |
| F53L | Indinavir | 11.1 | 20.6 | [28] |
| G73S | Indinavir | 0.55 | 1.0 | [61] |
| V82A | Darunavir | 1.3 | 1.3 | [30] |
| V82A | Saquinavir | 4.3 | 1.1 | [59] |
| V82A | Indinavir | 1.8 | 3.4 | [64] |
| I84V | Darunavir | 3.2 | 3.2 | [30] |
| I84V | Saquinavir | 4.3 | 1.1 | [59] |
| D30N/N88D | Nelfinavir | 7.0 | 100.0 | [63] |
| N88D | Nelfinavir | 0.2 | 2.9 | [63] |
| L90M | Darunavir | 0.03 | 0.14 | [68] |
| L90M | Indinavir | 0.09 | 0.17 | [64] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2009 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weber, I.T.; Agniswamy, J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses 2009, 1, 1110-1136. https://doi.org/10.3390/v1031110
Weber IT, Agniswamy J. HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses. 2009; 1(3):1110-1136. https://doi.org/10.3390/v1031110
Chicago/Turabian StyleWeber, Irene T., and Johnson Agniswamy. 2009. "HIV-1 Protease: Structural Perspectives on Drug Resistance" Viruses 1, no. 3: 1110-1136. https://doi.org/10.3390/v1031110
APA StyleWeber, I. T., & Agniswamy, J. (2009). HIV-1 Protease: Structural Perspectives on Drug Resistance. Viruses, 1(3), 1110-1136. https://doi.org/10.3390/v1031110




