Expanding the Research Frontiers of Pinus Species in Wood Biology
Abstract
1. Introduction
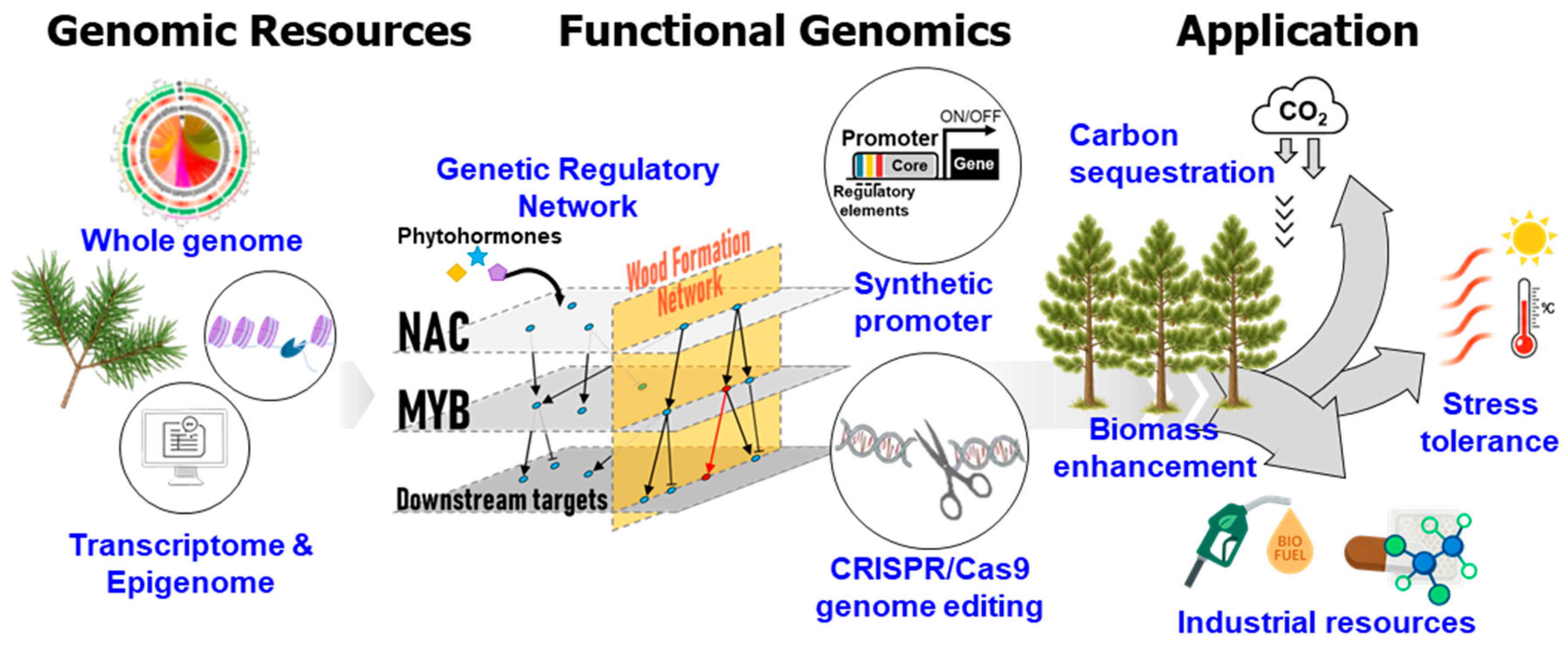
2. Integrative Genomics in Pinus: From Structural Assembly to Functional Characterization
2.1. Advances in Genome Sequencing and Assembly
2.2. Comparative Genomics and Evolutionary Insight
2.3. Transcriptomic, Small RNA, and Epigenomic Landscapes
2.4. Overcoming Barriers in Functional Genomics
2.5. Genome Editing and Synthetic Biology Horizons
3. Molecular Mechanisms of Wood Formation
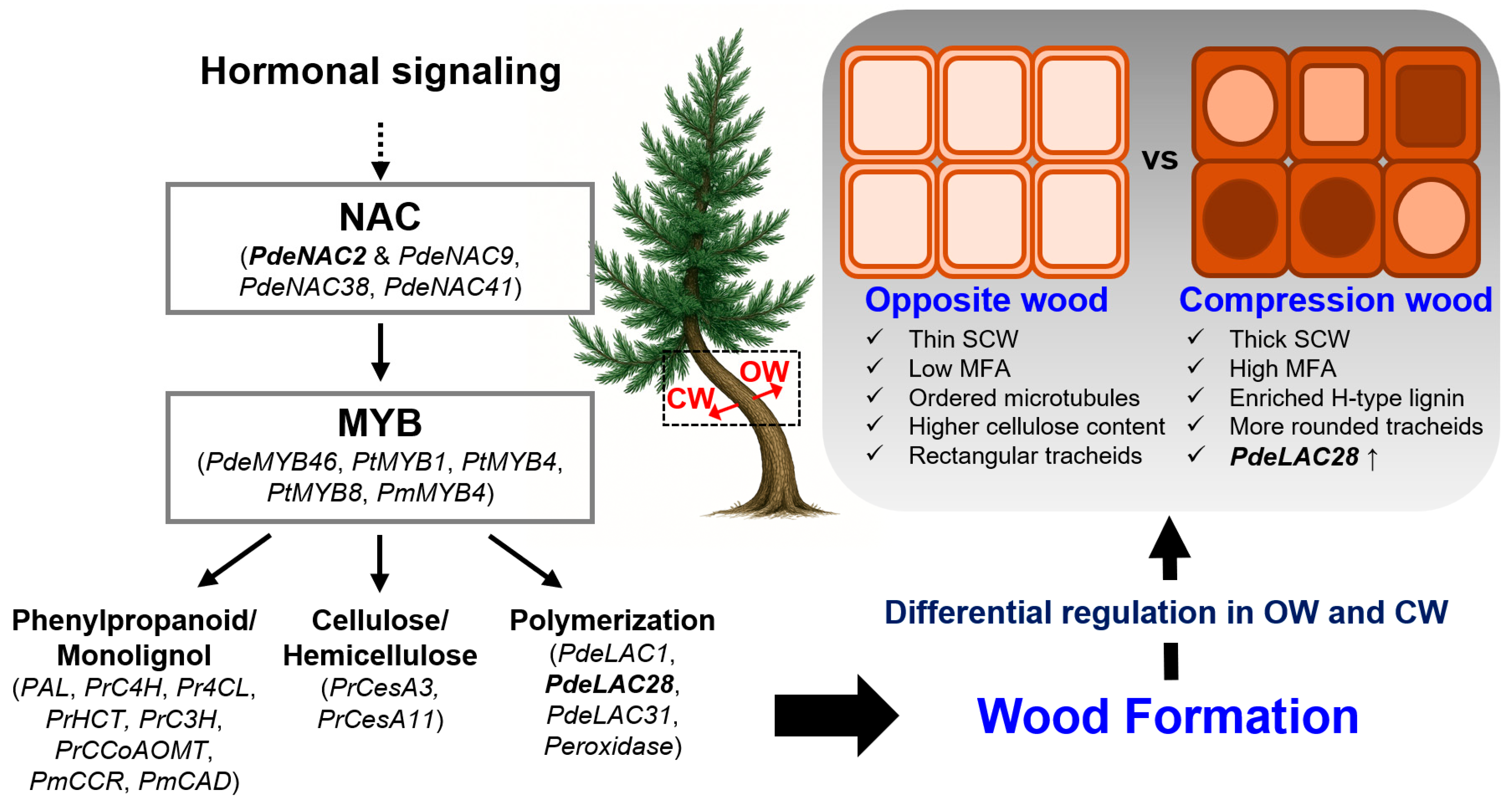
3.1. Regulatory Networks: NAC–MYB and Downstream Cascades
3.2. Lignin Biosynthesis
3.3. Hormonal Regulation of Secondary Growth
3.4. Compression Wood Formation and Adaptive Responses
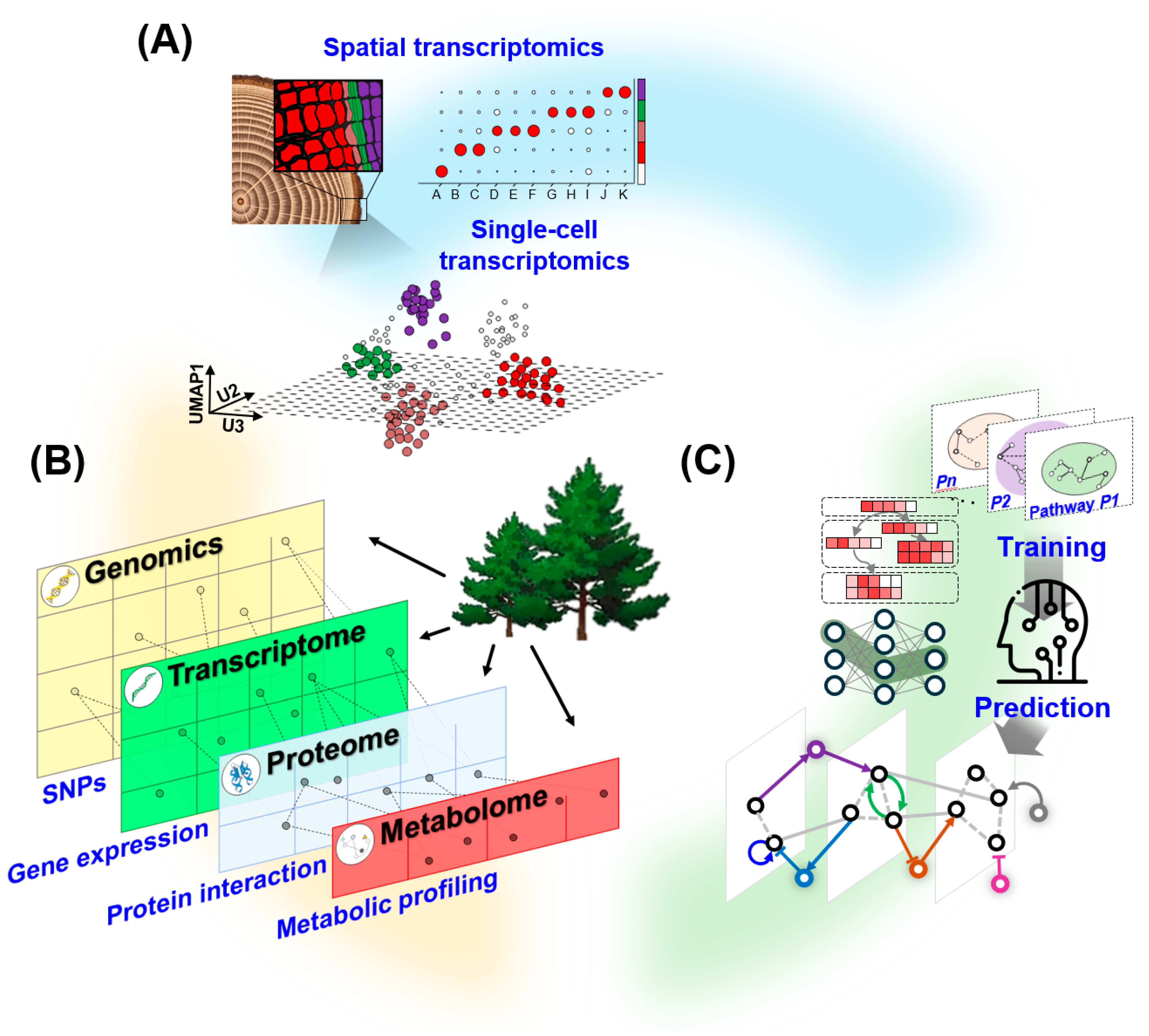
4. Emerging Tools and Integrative Approaches
4.1. Single-Cell Genomics and Spatial Transcriptomics
4.2. Multi-Omics Integration
4.3. Computational Biology and Network Prediction
5. Applications and Translational Perspectives
5.1. Genetic Engineering for Biomass and Lignin Modification
5.2. Stress Resilience, Adaptation, and Carbon Sequestration
5.3. Genome-Wide Association Studies and Genomic Selection in Molecular Breeding of Pinus
5.4. Industrial and Bioenergy Applications
6. Challenges and Future Directions
- (1)
- Expand genomic foundations: Improve reference-quality genome assemblies across diverse Pinus species. Haplotype-resolved genomes and pan-genomes will capture allelic diversity and structural variation critical for breeding climate-resilient varieties and will enable more comprehensive functional annotation [13,14].
- (2)
- Develop functional toolkits: Establish efficient protoplast systems, stable transformation pipelines, and conifer-optimized promoters. Optimize CRISPR/Cas systems and explore novel Cas variants, viral delivery platforms, and DNA-free ribonucleoprotein delivery into meristematic tissues to enable rapid genome editing assays while bypassing stable transformation constraints.
- (3)
- Apply single-cell and multi-omics: Use single-cell transcriptomics, proteomics, and metabolomics to map wood formation at high resolution. Integrate RNA-seq data with high-resolution wood anatomy images using deep-learning approaches, together with machine learning and GRN modeling, to directly link cell-expansion gene expression to tracheid lumen size and predict key regulators of growth and stress responses.
- (1)
- Advance synthetic biology: Prioritize development of modular circuits, inducible promoters, and multigene stacking strategies. Integrate synthetic regulation with genome editing for precise control of lignin content, cellulose composition, and tracheid architecture.
- (2)
- Validate in the field: Conduct long-term trials to test growth, resilience, and genetic stability under varied environments. Combine molecular outcomes with ecological impact assessments to ensure sustainability and safety.
- (3)
- Toward predictive forestry: Integrate genomic prediction, climate modeling, and carbon accounting into forestry management. This will allow selection and deployment of Pinus genotypes optimized for both productivity and climate resilience.
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christenhusz, M.J.M.; Reveal, J.L.; Farjon, A.; Gardner, M.F.; Mill, R.R.; Chase, M.W. A New Classification and Linear Sequence of Extant Gymnosperms. Phytotaxa 2011, 70, 55–70. [Google Scholar] [CrossRef]
- Gernandt, D.S.; Geada López, G.; Ortiz García, S.; Liston, A. Phylogeny and Classification of Pinus. Taxon 2005, 54, 29–42. [Google Scholar] [CrossRef]
- Dziedziński, M.; Kobus-Cisowska, J.; Stachowiak, B. Pinus Species as Prospective Reserves of Bioactive Compounds with Potential Use in Functional Food-Current State of Knowledge. Plants 2021, 10, 1306. [Google Scholar] [CrossRef]
- Brown, C.; Ball, J. World View of Plantation Grown Wood. In FAO Forestry Working Paper; Food and Agriculture Organization of the United Nations: Rome, Italy, 1999; 12p. [Google Scholar]
- Pan, Y.; Birdsey, R.A.; Phillips, O.L.; Jackson, R.B. The Structure, Distribution, and Biomass of the World’s Forests. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 593–622. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Bae, E.K.; Tran, T.N.A.; Lee, H.; Ko, J.H. Exploring the Seasonal Dynamics and Molecular Mechanism of Wood Formation in Gymnosperm Trees. Int. J. Mol. Sci. 2023, 24, 8624. [Google Scholar] [CrossRef]
- Borthakur, D.; Busov, V.; Cao, X.H.; Du, Q.; Gailing, O.; Isik, F.; Ko, J.H.; Li, C.; Li, Q.; Niu, S.; et al. Current Status and Trends in Forest Genomics. For. Res. 2022, 2, 11. [Google Scholar] [CrossRef]
- Farjon, A. The Kew Review Conifers of the World. Kew Bull. 2018, 73, 8. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Beckham, G.T.; Biddy, M.J.; Chandra, R.; Chen, F.; Davis, M.F.; Davison, B.H.; Dixon, R.A.; Gilna, P.; Keller, M.; et al. Lignin Valorization: Improving Lignin Processing in the Biorefinery. Science 2014, 344, 1246843. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Kim, M.H.; Park, E.J.; Lee, H.; Ko, J.H. Seasonal Developing Xylem Transcriptome Analysis of Pinus densiflora Unveils Novel Insights for Compression Wood Formation. Genes 2023, 14, 1698. [Google Scholar] [CrossRef]
- Kovach, A.; Wegrzyn, J.L.; Parra, G.; Holt, C.; Bruening, G.E.; Loopstra, C.A.; Hartigan, J.; Yandell, M.; Langley, C.H.; Korf, I.; et al. The Pinus taeda Genome Is Characterized by Diverse and Highly Diverged Repetitive Sequences. BMC Genom. 2010, 11, 420. [Google Scholar] [CrossRef]
- Kim, M.H.; Cho, J.S.; Tran, T.N.A.; Nguyen, T.T.T.; Park, E.J.; Im, J.H.; Han, K.H.; Lee, H.; Ko, J.H. Comparative functional analysis of PdeNAC2 and AtVND6 in the tracheary element formation. Tree Physiol. 2023, 43, 1201–1217. [Google Scholar] [CrossRef]
- Richardson, D.M. (Ed.) Ecology and Biogeography of Pinus; Cambridge University Press: Cambridge, UK, 1998; pp. 1–527. [Google Scholar]
- Zimin, A.; Stevens, K.A.; Crepeau, M.W.; Holtz-Morris, A.; Koriabine, M.; Marçais, G.; Puiu, D.; Roberts, M.; Wegrzyn, J.L.; de Jong, P.J.; et al. Sequencing and Assembly of the 22-Gb Loblolly Pine Genome. Genetics 2014, 196, 875–890. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, J.L.; Liechty, J.D.; Stevens, K.A.; Wu, L.S.; Loopstra, C.A.; Vasquez-Gross, H.A.; Dougherty, W.M.; Lin, B.Y.; Zieve, J.J.; Martínez-García, P.J.; et al. Unique Features of the Loblolly Pine (Pinus taeda L.) Megagenome Revealed through Sequence Annotation. Genetics 2014, 196, 891–909. [Google Scholar] [CrossRef] [PubMed]
- Ohri, D. Karyotype evolution in conifers. Feddes Repert. 2021, 132, 232–262. [Google Scholar] [CrossRef]
- Jang, M.J.; Cho, H.J.; Park, Y.S.; Lee, H.Y.; Bae, E.K.; Jung, S.; Jin, H.; Woo, J.; Park, E.; Kim, S.J.; et al. Haplotype-resolved genome assembly and resequencing analysis provide insights into genome evolution and allelic imbalance in Pinus densiflora. Nat. Genet. 2024, 56, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Neale, D.B.; Wegrzyn, J.L.; Stevens, K.A.; Zimin, A.V.; Puiu, D.; Crepeau, M.W.; Cardeno, C.; Koriabine, M.; Holtz-Morris, A.E.; Liechty, J.D.; et al. Decoding the Massive Genome of Loblolly Pine Using Haploid DNA and Novel Assembly Strategies. Genome Biol. 2014, 15, R59. [Google Scholar] [CrossRef]
- Stevens, K.A.; Wegrzyn, J.L.; Zimin, A.; Puiu, D.; Crepeau, M.; Cardeno, C.; Paul, R.; Gonzalez-Ibeas, D.; Koriabine, M.; Holtz-Morris, A.E.; et al. Sequence of the Sugar Pine Megagenome. Genetics 2016, 204, 1613–1626. [Google Scholar] [CrossRef]
- Sperry, J.S. Evolution of water transport and xylem structure. Int. J. Plant Sci. 2003, 164, 115–127. [Google Scholar] [CrossRef]
- Nguyen, T.T.T.; Kim, M.H.; Pyo, S.W.; Jang, H.A.; Kim, H.J.; Kim, D.G.; Ko, J.H. A Comprehensive Analysis of the Laccase Gene Family of Pinus densiflora Reveals a Functional Role of PdeLAC28 in Lignin Biosynthesis for Compression Wood Formation. Forests 2024, 15, 2220. [Google Scholar] [CrossRef]
- Chen, B.; Xu, H.; Guo, Y.; Grünhofer, P.; Schreiber, L.; Lin, J.; Li, R. Transcriptomic and epigenomic remodeling occurs during vascular cambium periodicity in Populus tomentosa. Hortic. Res. 2021, 8, 102. [Google Scholar] [CrossRef]
- Nagle, M.; Déjardin, A.; Pilate, G.; Strauss, S.H. Opportunities for Innovation in Genetic Transformation of Forest Trees. Front. Plant Sci. 2018, 9, 1443. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.T.; Choi, N.Y.; Pyo, S.W.; Choi, Y.I.; Ko, J.H. Optimized and Reliable Protoplast Isolation for Transient Gene Expression Studies in the Gymnosperm Tree Species Pinus densiflora. Forests 2025, 16, 1373. [Google Scholar] [CrossRef]
- Ralph, S.G.; Chun, H.J.E.; Kolosova, N.; Cooper, D.; Oddy, C.; Ritland, C.E.; Kirkpatrick, R.; Moore, R.; Barber, S.; Holt, R.A.; et al. A Conifer Genomics Resource of 200,000 Spruce (Picea spp.) ESTs and 6,464 High-Quality, Sequence-Finished Full-Length cDNAs for Sitka Spruce (Picea sitchensis). BMC Genom. 2008, 9, 484. [Google Scholar] [CrossRef]
- Caballero, M.; Lauer, E.; Bennett, J.; Zaman, S.; McEvoy, S.; Acosta, J.; Jackson, C.; Townsend, L.; Eckert, A.; Whetten, R.W.; et al. Toward genomic selection in Pinus taeda: Integrating resources to support array design in a complex conifer genome. Appl. Plant Sci. 2021, 9, e11439. [Google Scholar] [CrossRef]
- Moran, E.V.; DeSilva, R.; Canning, C.; Wright, J.W. Testing source elevation versus genotype as predictors of sugar pine performance in a post-fire restoration planting. Ecosphere 2024, 15, e70010. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.J.; Glassman, S.I.; Bruns, T.D.; Aronson, E.L.; Hart, S.C. Soil microbial communities associated with giant sequoia: How does the world’s largest tree affect some of the world’s smallest organisms? Ecol. Evol. 2020, 10, 6593–6609. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Jiao, L.; Wang, J.; Ma, L.; Lu, Y.; Zhang, Y.; Guo, J.; Yin, Y. Analyses of high spatial resolution datasets identify genes associated with multi-layered secondary cell wall thickening in Pinus bungeana. Ann. Bot. 2024, 133, 953–968. [Google Scholar] [CrossRef]
- Han, F.; Wang, P.; Chen, X.; Zhao, H.; Zhu, Q.; Song, Y.; Nie, Y.; Li, Y.; Guo, M.; Niu, S. An ethylene-induced NAC transcription factor acts as a multiple abiotic stress responsor in conifer. Hortic. Res. 2023, 10, uhad130. [Google Scholar] [CrossRef]
- Xing, B.; Li, S.; Qi, J.; Yang, L.; Yin, D.; Sun, S. Integrated transcriptomic and metabolic analyses reveal the early response mechanism of Pinus tabulaeformis to pine wood nematodes. BMC Genom. 2024, 25, 865. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, T.; Liu, J.; Zhang, R.; Yu, Y.; Zhou, G.; Liu, J.; Gao, B. The Key Role of Plant Hormone Signaling Transduction and Flavonoid Biosynthesis Pathways in the Response of Chinese Pine (Pinus tabuliformis) to Feeding Stimulation by Pine Caterpillar (Dendrolimus tabulaeformis). Int. J. Mol. Sci. 2024, 25, 6354. [Google Scholar] [CrossRef] [PubMed]
- Sturrock, S.; Frickey, T.; Freeman, J.; Butler, J.; Fritsche, S.; Gea, P.; Graham, N.; Macdonald, L.; Mercier, C.; Paget, M.; et al. Pinus radiata genome reveals a downward demographic trajectory and opportunities for genomics-assisted breeding. G3 Genes Genomes Genet. 2025, 15, jkaf125. [Google Scholar] [CrossRef]
- Addison, S.L.; Mendoza, L.L.; Rúa, M.A.; Clinton, P.W.; Wakelin, S.A. A genotype-by-sequencing dataset and identity-by-state matrix of genetic variation in 821 Pinus radiata trees from 16 counties. Data Brief 2025, 63, 112123. [Google Scholar] [CrossRef]
- Jokipii-Lukkari, S.; Delhomme, N.; Schiffthaler, B.; Mannapperuma, C.; Prestele, J.; Nilsson, O.; Street, R.; Tuominen, H. Transcriptional Roadmap to Seasonal Variation in Wood Formation of Norway Spruce. Plant Physiol. 2018, 176, 2851–2870. [Google Scholar] [CrossRef]
- Hall, D.; Zhao, W.; Wennström, U.; Andersson Gull, B.; Wang, X.R. Parentage and relatedness reconstruction in Pinus sylvestris using genotyping-by-sequencing. Heredity 2020, 124, 633–646. [Google Scholar] [CrossRef]
- Mishcherikova, V.; Lynikienė, J.; Marčiulynas, A.; Gedminas, A.; Prylutskyi, O.; Marčiulynienė, D.; Menkis, A. Biogeography of Fungal Communities Associated with Pinus sylvestris L. and Picea abies (L.) H. Karst. along the Latitudinal Gradient in Europe. J. Fungi 2023, 9, 829. [Google Scholar] [CrossRef]
- Dolgosheina, E.V.; Mardis, E.R.; Unrau, P.J. Conifers have a unique small RNA silencing signature. RNA 2008, 14, 1490–1506. [Google Scholar] [CrossRef]
- Hori, C.; Gaskell, J.; Cullen, D.; Sabat, G.; Stewart, P.E.; Lail, K.; Peng, Y.; Barry, K.; Grigoriev, I.V.; Kohler, A.; et al. Multi-omic Analyses of Extensively Decayed Pinus contorta Reveal Expression of a Diverse Array of Lignocellulose-Degrading Enzymes. Appl. Environ. Microbiol. 2018, 84, e01133-18. [Google Scholar] [CrossRef] [PubMed]
- Parchman, T.L.; Geist, K.S.; Grahnen, J.A.; Benkman, C.W.; Buerkle, C.A. Transcriptome sequencing in an ecologically important tree species: Assembly, annotation, and marker discovery. BMC Genom. 2010, 11, 180. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Pionneau, C.; Brach, J.; Costa, P.; Baillères, H. Compression Wood-Responsive Proteins in Developing Xylem of Maritime Pine (Pinus pinaster Ait.). Plant Physiol. 2000, 123, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Escribano, L.; Morales Clemente, M.T.; Fariña-Flores, D.; Raposo, R. A delayed response in phytohormone signaling and production contributes to pine susceptibility to Fusarium circinatum. BMC Plant Biol. 2024, 24, 727. [Google Scholar] [CrossRef]
- Montero-Pau, J.; Pérez-Oliver, M.A.; Rodríguez-Cuesta, Á.; Arrillaga, I.; Sales, E. Temperature-induced variation in the transcriptome of maritime pine (Pinus pinaster Ait.) embryogenic masses modulates the phenotype of the derived plants. BMC Genom. 2025, 26, 467. [Google Scholar] [CrossRef]
- Liu, J.J.; Sniezko, R.A.; Sturrock, R.N.; Chen, H. Western white pine SNP discovery and high-throughput genotyping for breeding and conservation applications. BMC Plant Biol. 2014, 14, 380. [Google Scholar] [CrossRef]
- Liu, J.J.; Sturrock, R.N.; Sniezko, R.A.; Williams, H.; Benton, R.; Zamany, A. Transcriptome analysis of the white pine blister rust pathogen Cronartium ribicola: De novo assembly, expression profiling, and identification of candidate effectors. BMC Genom. 2015, 16, 678. [Google Scholar] [CrossRef]
- Liu, J.J.; Fernandes, H.; Zamany, A.; Sikorski, M.; Jaskolski, M.; Sniezko, R.A. In-vitro anti-fungal assay and association analysis reveal a role for the Pinus monticola PR10 gene (PmPR10-3.1) in quantitative disease resistance to white pine blister rust. Genome 2021, 64, 693–704. [Google Scholar] [CrossRef]
- Yao, S.; Chen, P.; Yu, Y.; Zhang, M.; Wang, D.; Liu, J.; Hao, Q.; Ji, K. PmMYB4, a Transcriptional Activator from Pinus massoniana, Regulates Secondary Cell Wall Formation and Lignin Biosynthesis. Forests 2021, 12, 1618. [Google Scholar] [CrossRef]
- Hu, X.; Wang, S.; Wang, Z.; Ju, S.; Liu, X.; Li, G.; Zhang, Y.; Zhang, F.; Li, M. Transcriptomic and metabolomic insights into pine wood nematode resistance mechanisms in Pinus massoniana. Tree Physiol. 2025, 45, 10. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Tu, J.; Wang, H.; Xu, Y.; Wu, F. Transcriptomic and targeted metabolomic unravelling the molecular mechanism of sugar metabolism regulating heteroblastic changes in Pinus massoniana seedlings. Plant Physiol. Biochem. 2023, 203, 108029. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, Z.; Fan, F.; Qin, H.; Ding, G. Effects of exogenous GA3 on stem secondary growth of Pinus massoniana seedlings. Plant Physiol. Biochem. 2024, 206, 108254. [Google Scholar] [CrossRef]
- Chen, H.; Qin, X.; Chen, Y.; Zhang, H.; Feng, Y.; Tan, J.; Chen, X.; Hu, L.; Xie, J.; Xie, J.; et al. Chromosome-level genome assembly of Pinus massoniana provides insights into conifer adaptive evolution. GigaScience 2025, 14, giaf056. [Google Scholar] [CrossRef]
- Bullington, L.S.; Lekberg, Y.; Sniezko, R.; Larkin, B. The influence of genetics, defensive chemistry and the fungal microbiome on disease outcome in whitebark pine trees. Mol. Plant Pathol. 2018, 19, 1847–1858. [Google Scholar] [CrossRef]
- Liu, J.J.; Sniezko, R.; Murray, M.; Wang, N.; Chen, H.; Zamany, A.; Sturrock, R.N.; Savin, D.; Kegley, A. Genetic Diversity and Population Structure of Whitebark Pine (Pinus albicaulis Engelm.) in Western North America. PLoS ONE 2016, 11, e0167986. [Google Scholar] [CrossRef]
- Gan, P.; Li, P.; Zhang, X.; Li, H.; Ma, S.; Zong, D.; He, C. Comparative Transcriptomic and Metabolomic Analyses of Differences in Trunk Spiral Grain in Pinus yunnanensis. Int. J. Mol. Sci. 2023, 24, 14658. [Google Scholar] [CrossRef]
- He, H.; Xu, J.; Cai, N.; Xu, Y. Analysis of the molecular mechanism endogenous hormone regulating axillary bud development in Pinus yunnanensis. BMC Plant Biol. 2024, 24, 1219. [Google Scholar] [CrossRef]
- Lu, Z.; Pan, Z.; Chen, L.; Chen, S.; Tang, J.; Cai, N.; Wang, X.; Xu, Y. Integrating analysis of nutrient elements, endogenous phytohormones, and transcriptomics reveals factors influencing variation of growth in height in Pinus yunnanensis Franch. Plant Physiol. Biochem. 2025, 223, 109866. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, M.; Cai, K.; Liu, L.; Han, R.; Pei, X.; Zhang, L.; Zhao, X. Phytohormone biosynthesis and transcriptional analyses provide insight into the main growth stage of male and female cones Pinus koraiensis. Front. Plant Sci. 2023, 14, 1273409. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.; Zhang, M.; Xiong, C. Spatial specificity of metabolism regulation of abscisic acid-imposed seed germination inhibition in Korean pine (Pinus koraiensis sieb et zucc). Front. Plant Sci. 2024, 15, 1417632. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, J.; Ye, L.; Liu, N.; Wang, F. Methyl jasmonate induced tolerance effect of Pinus koraiensis to Bursaphelenchus xylophilus. Pest Manag. Sci. 2025, 81, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Pinosio, S.; González-Martínez, S.C.; Bagnoli, F.; Cattonaro, F.; Grivet, D.; Marroni, F.; Lorenzo, Z.; Pausas, J.G.; Verdú, M.; Vendramin, G.G. First insights into the transcriptome and development of new genomic tools of a widespread circum-Mediterranean tree species, Pinus halepensis Mill. Mol. Ecol. Resour. 2014, 14, 846–856. [Google Scholar] [CrossRef]
- Fox, H.; Doron-Faigenboim, A.; Kelly, G.; Bourstein, R.; Attia, Z.; Zhou, J.; Moshe, Y.; Moshelion, M.; David-Schwartz, R. Transcriptome analysis of Pinus halepensis under drought stress and during recovery. Tree Physiol. 2018, 38, 423–441. [Google Scholar] [CrossRef]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway Spruce Genome Sequence and Conifer Genome Evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef]
- Zimin, A.V.; Stevens, K.A.; Crepeau, M.W.; Puiu, D.; Wegrzyn, J.L.; Yorke, J.A.; Langley, C.H.; Neale, D.B.; Salzberg, S.L. An Improved Assembly of the Loblolly Pine Mega-Genome Using Long-Read Single-Molecule Sequencing. Gigascience 2017, 6, giw016. [Google Scholar] [CrossRef]
- Niu, S.; Li, J.; Bo, W.; Yang, W.; Zuccolo, A.; Giacomello, S.; Chen, X.; Han, F.; Yang, J.; Song, Y.; et al. The Chinese Pine Genome and Methylome Unveil Key Features of Conifer Evolution. Cell 2022, 185, 204–217.e14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Ran, J.H. Evolution and Biogeography of Gymnosperms. Mol. Phylogenet. Evol. 2014, 75, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Tan, T.; Wu, D.; Li, C.; Jing, H.; Wu, J. Seasonal Variations in Carbon, Nitrogen, and Phosphorus of Pinus Yunnanenis at Different Stand Ages. Front. Plant Sci. 2023, 14, 1107961. [Google Scholar] [CrossRef]
- Keeling, C.I.; Bohlmann, J. Genes, Enzymes and Chemicals of Terpenoid Diversity in the Constitutive and Induced Defence of Conifers against Insects and Pathogens. New Phytol. 2006, 170, 657–675. [Google Scholar] [CrossRef]
- Jokipii-Lukkari, S.; Sundell, D.; Nilsson, O.; Hvidsten, T.R.; Street, N.R.; Tuominen, H. NorWood: A gene expression resource for evo-devo studies of conifer wood development. New Phytol. 2017, 216, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, W.W.; Ayyampalayam, S.; Bordeaux, J.M.; Howe, G.T.; Jermstad, K.D.; Neale, D.B.; Rogers, D.L.; Dean, J.F.D. Conifer DBMagic: A database housing multiple de novo transcriptome assemblies for 12 diverse conifer species. Tree Genet. Genomes 2012, 8, 1477–1485. [Google Scholar] [CrossRef]
- Kim, M.H.; Tran, T.N.A.; Cho, J.S.; Park, E.J.; Lee, H.; Kim, D.G.; Hwang, S.; Ko, J.H. Wood Transcriptome Analysis of Pinus densiflora Identifies Genes Critical for Secondary Cell Wall Formation and NAC Transcription Factors Involved in Tracheid Formation. Tree Physiol. 2021, 41, 1289–1305. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.H.; Amerson, H.; Chiang, V.L. MicroRNAs in Loblolly Pine (Pinus taeda L.) and Their Association with Fusiform Rust Gall Development. Plant J. 2007, 51, 1077–1098. [Google Scholar] [CrossRef]
- Qiu, D.; Pan, X.; Wilson, I.W.; Li, F.; Liu, M.; Teng, W.; Zhang, B. High Throughput Sequencing Technology Reveals That the Taxoid Elicitor Methyl Jasmonate Regulates MicroRNA Expression in Chinese Yew (Taxus chinensis). Gene 2009, 436, 37–44. [Google Scholar] [CrossRef]
- Bedon, F.; Grima-Pettenati, J.; Mackay, J. Conifer R2R3-MYB transcription factors: Sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol. 2007, 7, 17. [Google Scholar] [CrossRef]
- Ausin, I.; Feng, S.; Yu, C.; Liu, W.; Kuo, H.Y.; Jacobsen, E.L.; Zhai, J.; Gallego-Bartolome, J.; Wang, L.; Egertsdotter, U.; et al. DNA methylome of the 20-gigabase Norwayspruce genome. Proc. Natl. Acad. Sci. USA 2016, 113, E8106–E8113. [Google Scholar] [CrossRef]
- Shuai, P.; Hsieh, J.-W.A.; Kao, C.-T.; Hu, C.-W.; Wang, R.; Kuo, S.-C.; Yen, M.-R.; Liou, P.-C.; Ho, Y.-C.; Chu, C.-C.; et al. Single-cell and spatial omics reveal progressive loss of xylem developmental complexity across seed plants. Plant Cell 2025, 37, koaf253. [Google Scholar] [CrossRef]
- Trontin, J.-F.; Klimaszewska, K.; Morel, A.; Hargreaves, C.; Lelu-Walter, M.-A. Molecular aspects of conifer zygotic and somatic embryo development: A review of genome-wide approaches and recent insights. Tree Genet. Genomes 2016, 12, 78. [Google Scholar] [CrossRef]
- Walter, C.; Grace, L.J. Somatic embryogenesis and genetic transformation in Pinus radiata. In Protocol for Somatic Embryogenesis in Woody Plants; Forestry Sciences; Jain, S.M., Gupta, P.K., Eds.; Springer: Dordrecht, The Netherlands, 2005; Volume 77, pp. 363–384. [Google Scholar] [CrossRef]
- Charity, J.A.; Holland, L.; Grace, L.J.; Walter, C.; McInnis, S.; Klimaszewska, K.; Caron, S.; Rutledge, R.G. Regeneration of Pinus radiata plants from embryogenic tissue: Improvement of protocols. Plant Cell Rep. 2005, 24, 606–611. [Google Scholar] [CrossRef]
- Loopstra, C.A.; Stomp, A.M.; Sederoff, R.R. Agrobacterium-mediated DNA transfer in sugar pine. Plant Mol. Biol. 1990, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Sederoff, R.; Whetten, R. Regeneration of transgenic loblolly pine (Pinus taeda L.) from zygotic embryos transformed with Agrobacterium tumefaciens. Planta 2001, 213, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Germain, H.; Lachance, D.; Pelletier, G.; Fossdal, C.G.; Solheim, H.; Séguin, A. The expression pattern of the Picea glauca Defensin 1 promoter is maintained in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 785–795. [Google Scholar] [CrossRef]
- Uddenberg, D.; Akhter, S.; Ramachandran, P.; Sundström, J.F.; Carlsbecker, A. Sequenced genomes and rapidly emerging technologies pave the way for conifer evolutionary developmental biology. Front. Plant Sci. 2015, 6, 970. [Google Scholar] [CrossRef]
- Chen, K.; Chen, J.; Pi, X.; Huang, L.-J.; Li, N. Isolation, Purification, and Application of Protoplasts and Transient Expression Systems in Plants. Int. J. Mol. Sci. 2023, 24, 16892. [Google Scholar] [CrossRef]
- Sasamoto, H.; Ogita, S. Endogenous Plant Hormones in Protoplasts of Embryogenic Cells of Conifers. Prog. Biotechnol. 2001, 18, 279–288. [Google Scholar] [CrossRef]
- Poovaiah, C.; Phillips, L.; Geddes, B.; Reeves, C.; Sorieul, M.; Rhorlby, G. Genome editing with CRISPR/Cas9 in Pinus radiata (D. Don). BMC Plant Biol. 2021, 21, 363. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, J.; Gao, Y.; Zhao, R.; Zhang, J.; Kong, L. Efficient Multi-Sites Genome Editing and Plant Regeneration via Somatic Embryogenesis in Picea glauca. Front. Plant Sci. 2021, 12, 751891. [Google Scholar] [CrossRef]
- Cao, H.X.; Vu, G.T.H.; Cailing, O. From Genome Sequencing to CRISPR-Based Genome Editing for Climate-Resilient Forest Trees. Int. J. Mol. Sci. 2022, 23, 966. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9–cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, M.R. Fate of transgenes in the forest tree genome. Tree Genet. Genomes 2011, 7, 221–230. [Google Scholar] [CrossRef]
- Liu, W.; Stewart, C.N., Jr. Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 2016, 37, 36–44. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef]
- Zhong, Z.; Sretenovic, S.; Ren, Q.; Yang, L.; Bao, Y.; Qi, C.; Yuan, M.; He, Y.; Liu, S.; Liu, X.; et al. Improving Plant Genome Editing with High-Fidelity xCas9 and Non-canonical PAM-Targeting Cas9-NG. Mol. Plant 2019, 12, 1027–1036. [Google Scholar] [CrossRef]
- Zhong, R.; Lee, C.; Ye, Z.-H. Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant 2010, 3, 1087–1103. [Google Scholar] [CrossRef]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef]
- Chano, V.; Sobrino-Plata, J.; Collada, C.; Soto, A. Wood development regulators involved in apical growth in Pinus canariensis. Plant Biol. 2021, 23, 413–527. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef]
- Maleki, S.S.; Mohammadi, K.; Movahedi, A.; Wu, F.; Ji, K.S. Increase in Cell Wall Thickening and Biomass Production by Overexpression of PmCesA2 in Poplar. Front. Plant Sci. 2020, 11, 110. [Google Scholar] [CrossRef]
- Li, X.; Wu, H.X.; Dillon, S.K.; Southerton, S.G. Generation and analysis of expressed sequence tags from six developing xylem libraries in Pinus radiata D. Don. BMC Genom. 2009, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Huang, L.; Chen, M.; Zeng, W.; Feng, Z.; Huang, S.; Liu, T. Integrated Analysis of the Transcriptome and Metabolome Reveals Genes Involved in Terpenoid and Flavonoid Biosynthesis in the Loblolly Pine (Pinus taeda L.). Front. Plant Sci. 2021, 12, 729161. [Google Scholar] [CrossRef] [PubMed]
- Bomal, C.; Bedon, F.; Caron, S.; Mansfield, S.D.; Levasseur, C.; Cooke, J.E.K.; Blais, S.; Tremblay, L.; Morency, M.J.; Pavy, N.; et al. Involvement of Pinus taeda MYB1 and MYB8 in Phenylpropanoid Metabolism and Secondary Cell Wall Biogenesis: A Comparative in Planta Analysis. J. Exp. Bot. 2008, 59, 3925–3939. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Koutaniemi, S.; Malmberg, H.A.; Simola, L.K.; Terri, T.T.; Kärkönen, A. Norway spruce (Picea abies) laccases: Characterization of a laccase in a lignin-forming tissue culture. J. Integr. Plant Biol. 2015, 57, 341–348. [Google Scholar] [CrossRef]
- Sato, Y.; Wuli, B.; Sederoff, R.; Whetten, R. Molecular Cloning and Expression of Eight Laccase cDNAs in Loblolly Pine (Pinus taeda). J. Plant Res. 2001, 114, 147–155. [Google Scholar] [CrossRef]
- Herrero, J.; Esteban-Carrasco, A.; Zapata, J.M. Looking for Arabidopsis thaliana peroxidases involved in lignin biosynthesis. Plant Physiol. Biochem. 2013, 67, 77–86. [Google Scholar] [CrossRef]
- Uggla, C.; Mellerowicz, E.J.; Sundberg, B. Indole-3-Acetic Acid Controls Cambial Growth in Scots Pine by Positional Signaling. Plant Physiol. 1998, 117, 113–121. [Google Scholar] [CrossRef]
- Eriksson, M.E.; Israelsson, M.; Olsson, O.; Moritz, T. Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat. Biotechnol. 2000, 18, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Wan, C.; Fu, X.; Hu, J.; Tao, Y.; Luo, K.; Xu, C. Locally biosynthesized gibberellins in Populus stems are involved in the regulation of wood development. For. Res. 2025, 5, e005. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Yamamoto, F. An Overview of the Biology of Reaction Wood Formation. J. Integr. Plant Biol. 2007, 49, 131–143. [Google Scholar] [CrossRef]
- Seyfferth, C.; Wessels, B.; Jokipii-Lukkari, S.; Sundell, D.; Delhomme, N.; Nilsson, O.; Kangasjärvi, J.; Teeri, T.H.; Street, N.R.; Hvidsten, T.R.; et al. Ethylene-related gene expression networks in wood formation. Front. Plant Sci. 2018, 9, 272. [Google Scholar] [CrossRef]
- Lachaud, S. Participation of phytohormones in the regulation of cambial activity and xylogenesis. Ann. Sci. For. 1989, 46, 273s–276s. [Google Scholar] [CrossRef]
- Fan, F.; Zhou, Z.; Qin, H.; Tan, J.; Ding, G. Exogenous Brassinosteroid Facilitates Xylem Development in Pinus massoniana Seedlings. Int. J. Mol. Sci. 2021, 22, 7615. [Google Scholar] [CrossRef]
- Zhou, F.; Hu, B.; Li, J.; Yan, H.; Liu, Q.; Zeng, B.; Fan, C. Exogenous applications of brassinosteroids promote secondary xylem differentiation in Eucalyptus grandis. Peer J. 2024, 12, e16250. [Google Scholar] [CrossRef]
- Timell, T.E. Compression Wood in Gymnosperms; Springer: Berlin/Heidelberg, Germany, 1986. [Google Scholar]
- Mast, S.; Peng, L.; Jordan, T.W.; Flint, H.; Phillips, L.; Donaldson, L.; Strabala, T.J.; Wagner, A. Proteomic analysis of membrane preparations from developing Pinus radiata compression wood. Tree Physiol. 2010, 30, 1456–1468. [Google Scholar] [CrossRef]
- Yamashita, S.; Yoshida, M.; Yamamoto, H. Relationship between development of compression wood and gene expression. Plant Sci. 2009, 176, 729–735. [Google Scholar] [CrossRef]
- Donaldson, L.A.; Grace, J.; Dewnes, G.M. Within-tree variation in anatomical properties of compression wood in radiata pine. IAWA J. 2004, 25, 253–271. [Google Scholar] [CrossRef]
- Sundell, D.; Street, N.R.; Kumar, M.; Mellerowicz, E.J.; Kucukoglu, M.; Johnsson, C.; Kumar, V.; Mannapperuma, C.; Delhomme, N.; Nilsson, O.; et al. AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 2017, 29, 1585–1604. [Google Scholar] [CrossRef]
- Duval, I.; Lachance, D.; Giguère, I.; Bomal, C.; Morency, M.-J.; Pelletier, G.; Boyle, B.; MacKay, J.J.; Séguin, A. Large-scale screening of transcription factor–promoter interactions in spruce reveals a transcriptional network involved in vascular development. J. Exp. Bot. 2014, 65, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Farmer, A.; Thibivilliers, S.; Ryu, K.H.; Schiefelbein, J.; Libault, M. Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol. Plant. 2021, 14, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, A.; Osmolak, A.; Morales, P.; Crow, J. Seasonal changes in abundance and phosphorylation status of photosynthetic proteins in eastern white pine and balsam fir. Tree Physiol. 2008, 29, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Vanhakylä, S.; Salminen, J.-P. Seasonal Variation in Plant Polyphenols and Related Bioactivities across Three Years in Ten Tree Species as Visualized by Mass Spectrometric Fingerprint Mapping. Molecules 2023, 28, 6093. [Google Scholar] [CrossRef]
- Pascual, J.; Canal, M.J.; Escandon, M.; Meijon, M.; Weckwerth, W.; Valledor, L. Integrated Physiological, Proteomic, and Metabolomic Analysis of Ultra Violet (UV) Stress Responses and Adaptation Mechanisms in Pinus radiata. Mol. Cell. Proteom. 2017, 16, 485–501. [Google Scholar] [CrossRef]
- Uddenberg, D.; Reimegård, J.; Clapham, D.; Almqvist, C.; Arnold, S.v.; Emanuelsson, O.; Sundström, J.F. Early Cone Setting in Picea abies acrocona Is Associated with Increased Transcriptional Activity of a MADS Box Transcription Factor. Plant Physiol. 2013, 161, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Umarov, R.K.; Solovyev, V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE 2017, 12, e0171410. [Google Scholar] [CrossRef]
- Raju, S.K.K. Machine learning models reveal the importance of time-point specific cis-regulatory elements in Arabidopsis thaliana wounding response. Plant Cell 2022, 34, 716–717. [Google Scholar] [CrossRef]
- Zhou, J.; Troyanskaya, O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 2015, 12, 931–934. [Google Scholar] [CrossRef]
- Park, E.J.; Lee, W.Y.; Kurepin, L.V.; Zhang, R.; Janzen, L.; Pharis, R.P. Plant hormone-assisted early family selection in Pinus densiflora via a retrospective approach. Tree Physiol. 2015, 35, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P.; Gubler, F. Molecular Mechanism of Gibberellin Signaling in Plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef] [PubMed]
- MacKay, J.J.; O’Malley, D.M.; Presnell, T.; Booker, F.L.; Campbell, M.M.; Whetten, R.W.; Sederoff, R.R. Inheritance, gene expression, and lignin characterization in a mutant loblolly pine (Pinus taeda L.). Proc. Natl. Acad. Sci. USA 1997, 94, 8255–8260. [Google Scholar] [CrossRef]
- Wagner, A.; Tobimatsu, Y.; Phillips, L.; Flint, H.; Geddes, B.; Lu, F.; Ralph, J. Syringyl Lignin Production in Conifers: Proof of Concept in a Pine Tracheary Element System. Proc. Natl. Acad. Sci. USA 2015, 112, 6218–6223. [Google Scholar] [CrossRef]
- Brodribb, T.J.; McAdam, S.A.M.; Jordan, G.J.; Martins, S.C.V. Conifer species adapt to low-rainfall climates by following two distinct evolutionary pathways in hydraulic and stomatal traits. Proc. Natl. Acad. Sci. USA 2014, 111, 14389–14394. [Google Scholar] [CrossRef]
- Brodribb, T.J.; Cochard, H. Hydraulic Failure Defines the Recovery and Point of Death in Water-Stressed Conifers. Plant Physiol. 2008, 149, 575–584. [Google Scholar] [CrossRef]
- Bhatnagar-Mathur, P.; Vades, V.; Sharma, K.K. Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Rep. 2008, 27, 411–424. [Google Scholar] [CrossRef]
- Chang, C.Y.Y.; Bräutigam, K.; Hüner, N.P.A.; Ensminger, I. Champions of winter survival: Cold acclimation and molecular regulation of cold hardiness in evergreen conifers. New Phytol. 2021, 229, 675–691. [Google Scholar] [CrossRef]
- Strimbeck, G.R.; Schaberg, P.G.; Fossdal, C.G.; Schröder, W.P.; Kjellsen, T.D. Extreme low temperature tolerance in woody plants. Front. Plant Sci. 2015, 6, 884. [Google Scholar] [CrossRef]
- Dusenge, M.E.; Warren, J.M.; Reich, P.B.; Ward, E.J.; Murphy, B.K.; Stefanski, A.; Bermudez, R.; Cruz, M.; McLennan, D.A.; King, A.W.; et al. Boreal conifers maintain carbon uptake with warming despite failure to track optimal temperatures. Nat. Commun. 2023, 14, 4667. [Google Scholar] [CrossRef]
- Forest Research. How Does Replanting Productive Conifers Affect Carbon Sequestration on Shallow Peat Soils. Forest Research Report. 2024. Available online: https://www.forestresearch.gov.uk/news/how-does-replanting-productive-conifers-affect-carbon-sequestration-on-shallow-peat-soils/ (accessed on 27 December 2025).
- Aono, A.H.; Bajay, S.K.; Francisco, F.R.; de Souza, A.P. Genome-wide association reveals novel insights into the molecular mechanisms regulating stem volume in Pinus taeda. Tree Genet. Genomes 2025, 21, 16. [Google Scholar] [CrossRef]
- Resende, M.F.R.; Muñoz, P.; Resende, M.D.V.; Garrick, D.J.; Fernando, R.L.; Davis, J.M.; Jokela, E.J.; A Martin, T.; Peter, G.F.; Kirst, M. Accuracy of Genomic Selection Methods in a Standard Data Set of Loblolly Pine (Pinus taeda L.). Genetics 2012, 190, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Bibik, J.D.; Sahu, A.; Kim, B.; Unda, F.; Andersen, T.B.; Mansfield, S.D.; Maravelias, C.T.; Sharkey, T.D.; Hamberger, B.R. Engineered poplar for bioproduction of the triterpene squalene. Plant Biotechnol. J. 2024, 22, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Hoengenaert, L.; Boerjan, W. Accelerating wood domestication in forest trees through genome editing: Advances and prospects. Curr. Opin. Plant Biol. 2023, 71, 102329. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, J.; Zhao, J.; Niu, S. Genetic transformation in conifers: Current status and future prospects. For. Res. 2024, 4, e010. [Google Scholar] [CrossRef] [PubMed]
| Pinus Species | Genome Size (Gbp) | Assembly Quality | Transcriptome/ Epigenome Resources | References |
|---|---|---|---|---|
| P. taeda (loblolly pine) | ~22 | Improved Assembly (v2.0) with long reads | RNA-seq (xylem, needles, roots), stress/lignin datasets, methylome, ATAC-seq resources | [14,15,18,26] |
| P. lambertiana (sugar pine) | ~31 | Draft genome assembly | RNA-seq (needles, developing xylem), Megagenomic data, SNP array and GBS datasets | [19,27,28] |
| P. tabuliformis (Chinese red pine) | 25.4 | Chromosome-level Hi-C assembly | RNA-seq (seasonal/tissue-specific; hormone and infection treatments), methylome datasets | [28,29,30,31,32,33] |
| P. densiflora (Korean red pine) | ~21.7 | Haplotype-resolved HiFi + Hi-C assembly | RNA-seq (seasonal xylem/cambium), NAC/LAC family expression datasets | [10,12,17] |
| P. radiata (Monterey pine) | ~25 | Partial draft assembly; gene-space coverage | RNA-seq (stress and UV response), proteome/metabolome profiles, GBS datasets | [20,33,34] |
| P. sylvestris (Scots pine) | ~24 | Gene-space draft assembly | RNA-seq (cambium, xylem), seasonal dormancy proteomics, metabolomic analysis, GBS datasets | [32,35,36,37] |
| P. contorta (Lodgepole pine) | ~23 | Draft genome assembly | Metatranscriptome, Small RNA and miRNA profiling, RNA-seq (needle, conelets) | [38,39,40,41] |
| P. pinaster (Maritime Pine) | ~24 | Draft assembly (Illumina + long-read scaffolding) | RNA-seq (seedlings, embryogenic tissues), SNP array resources | [41,42,43] |
| P. monticola (Western White Pine) | 27–28 | Draft genome assembly | RNA-seq (needles), SNP array datasets | [44,45,46] |
| P. massoniana (Masson’s pine) | ~21.9 | Chromosome-level assembly | RNA-seq (hormone, abiotic/biotic stress, resin-producing tissues), multi-omics datasets | [47,48,49,50,51] |
| P. albicaulis (Whitebark pine) | ~27.6 | Assembly in progress | Genomic resources under development, RNA-seq (disease), metabolome profiles | [52,53] |
| P. yunnanensis (Yunnan pine) | Not reported | Draft genome assembly | RNA-seq (seedlings, axillary buds, trunk spiral-grain tissues) | [54,55,56] |
| P. koraiensis (Korean pine) | 25~30 | Draft genome assembly | RNA-seq (MeJA-treated seedlings; male/female cone development), metabolomic datasets | [57,58,59] |
| P. halepensis (Aleppo pine) | Not reported | Not reported | RNA-seq (drought/fire-adapted accessions; drought stress from needles of six physiological stages) | [60,61] |
| Gene/Pathway | Pinus Species | Functional Roles | Expression Context | References |
|---|---|---|---|---|
| NAC–MYB cascade | P. densiflora | Core transcriptional network guiding tracheid differentiation and secondary cell wall (SCW) formation | Highly expressed in developing xylem (DX) of both opposite wood (OW) and compression wood (CW) | [70] |
| PdeNAC2, PdeNAC9, PdeNAC38, PdeNAC41 | P. densiflora | VND/NST/SND-like transcription factor (TF) regulating tracheid differentiation | Preferentially expressed in DX of both OW and CW | [70] |
| PtMYB1, PtMYB4, PtMYB8 | P. taeda | Activate SCW biosynthetic genes | Upregulated in DX; responsible for SCW formation in both OW and CW | [64,73] |
| PmMYB4 | P. massoniana | SCW regulator; lignin/cellulose biosynthesis. | Highly expressed in lignifying tissues such as bark and semi-lignified stem in wood | [47] |
| PdeMYB46 (MYB46) | P. densiflora | Master regulator controlling downstream SCW TFs and biosynthetic pathways | Strongly expressed in DX of both OW and CW | [68] |
| PrCesA3, PrCesA11 PmCesA2 | P. radiata P. massoniana | Core cellulose synthase subunits required for SCW cellulose assembly | Upregulated in CW with enhanced cellulose synthesis in the S2 layer | [96,100,101] |
| PdeLAC28 | P. densiflora | Catalyzes monolignol oxidation; promotes lignin deposition and SCW thickening | Exclusively upregulated in CW during the growing season in spring and summer | [21] |
| PrC4H (Cinnamate 4- Hydroxylase) | P. radiata | Early phenylpropanoid enzyme producing p-coumaric acid | Upregulated in both OW and CW | [101] |
| COMT (Caffeic O-methyl- transferase) | P. pinaster | Methylates monolignol intermediates during lignification | Upregulated in both OW and CW | [41] |
| ACC oxidase (1-amino cyclopropane- 1-carboxylate oxidase) | P. pinaster | Ethylene-producing enzyme influencing CW-related signaling | Upregulated in both OW and CW | [41] |
| SAM synthase (S-adenosyl-L- Methionine synthase) | P. pinaster | Provides SAM for transmethylation in lignin and ethylene biosynthesis | Upregulated in both OW and CW | [41] |
| PpNAC1 | P. pinaster | Regulates phenylalanine biosynthesis genes | Expression in developing vascular tissues | [102] |
| HD-ZIP III TFs | P. canariensis | Control vascular patterning and wood formation | Expressed during secondary growth in wood | [98] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Jang, H.-A.; Pyo, S.-W.; Choi, Y.-I.; Lee, H.; Bae, E.-K.; Ko, J.-H. Expanding the Research Frontiers of Pinus Species in Wood Biology. Forests 2026, 17, 48. https://doi.org/10.3390/f17010048
Jang H-A, Pyo S-W, Choi Y-I, Lee H, Bae E-K, Ko J-H. Expanding the Research Frontiers of Pinus Species in Wood Biology. Forests. 2026; 17(1):48. https://doi.org/10.3390/f17010048
Chicago/Turabian StyleJang, Hyun-A, Seung-Won Pyo, Young-Im Choi, Hyoshin Lee, Eun-Kyung Bae, and Jae-Heung Ko. 2026. "Expanding the Research Frontiers of Pinus Species in Wood Biology" Forests 17, no. 1: 48. https://doi.org/10.3390/f17010048
APA StyleJang, H.-A., Pyo, S.-W., Choi, Y.-I., Lee, H., Bae, E.-K., & Ko, J.-H. (2026). Expanding the Research Frontiers of Pinus Species in Wood Biology. Forests, 17(1), 48. https://doi.org/10.3390/f17010048






