Genome-Wide Identification of the PRP Gene Family Members of the Dove Tree (Davidia involucrata Baill.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of the DiPRP Genes and Predicted Subcellular Localization
2.3. Analysis of the Expression Patterns of the DiPRPs
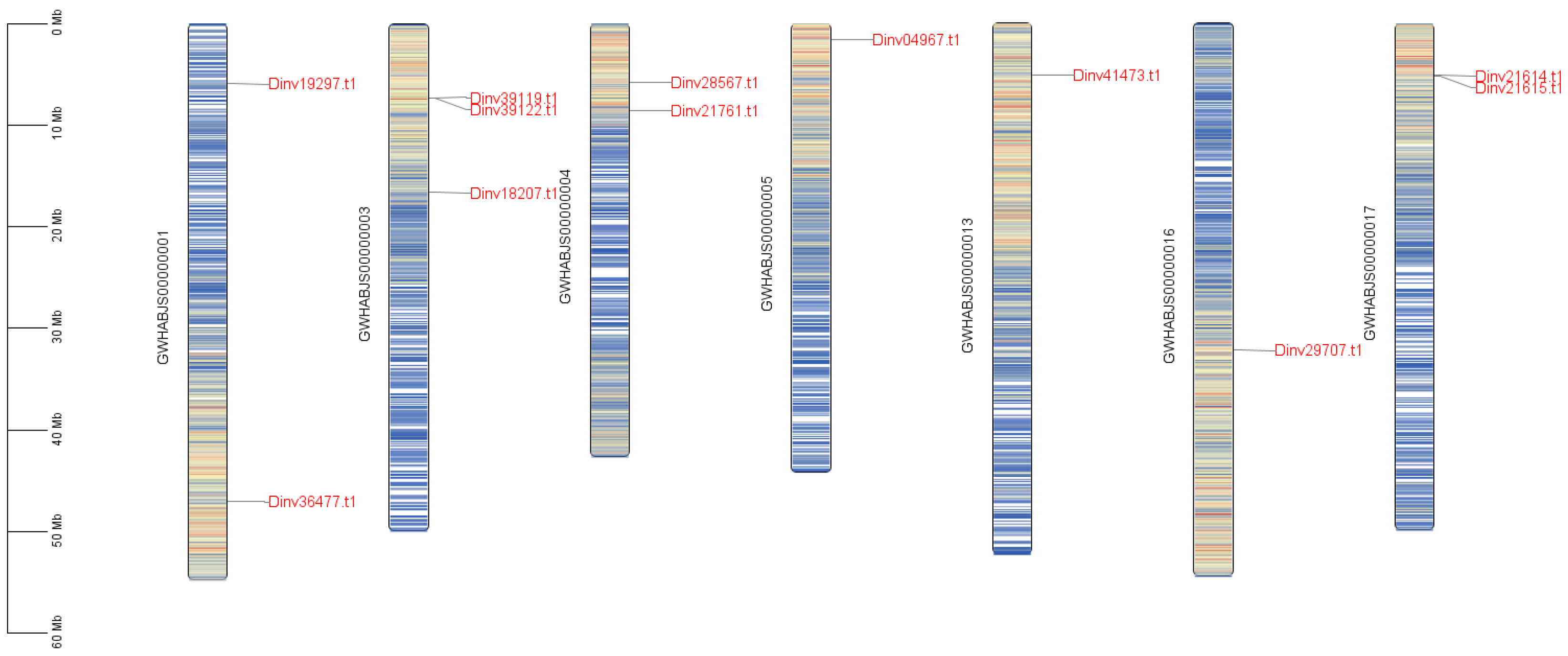
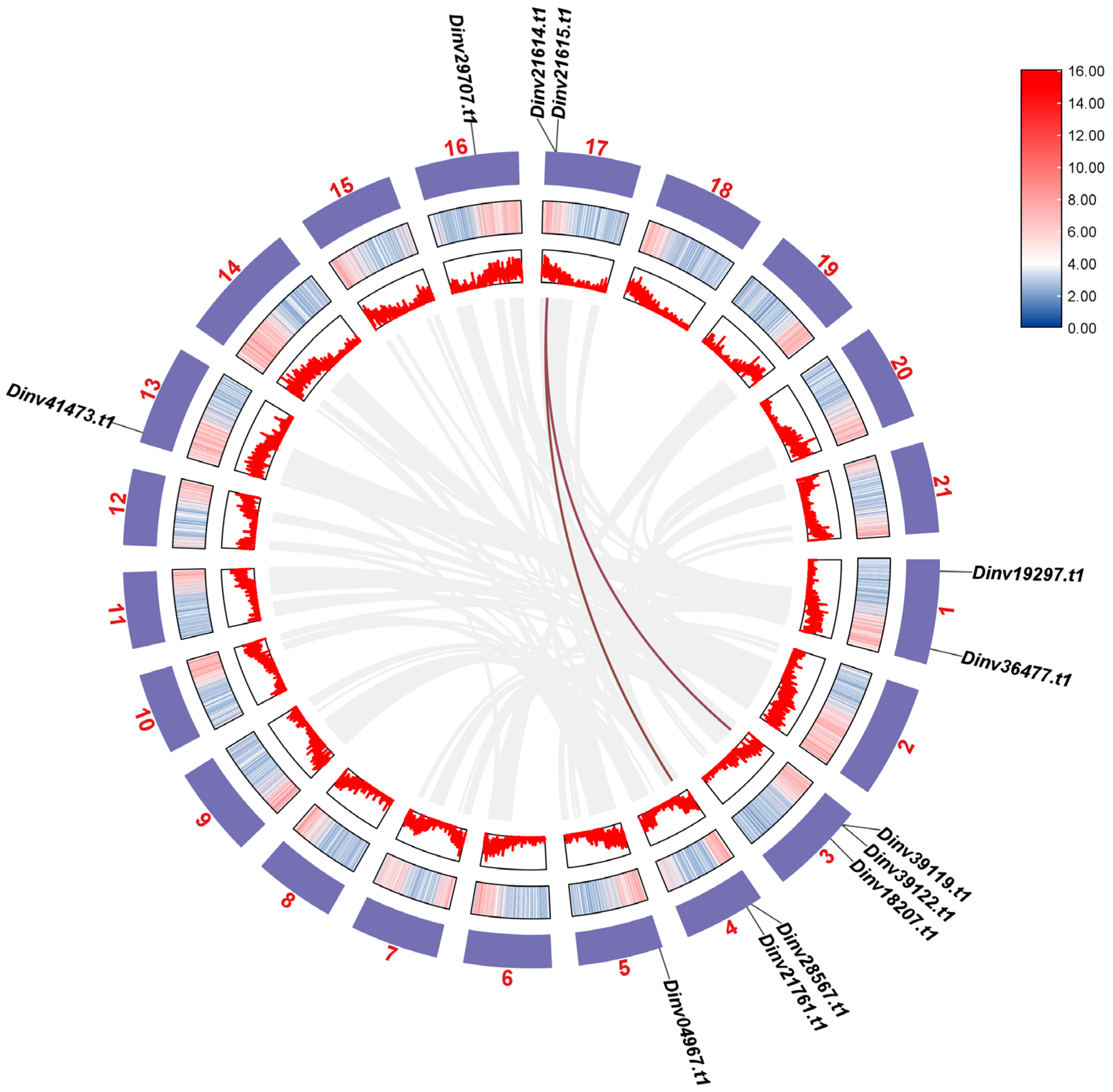
2.4. Chromosome Localization
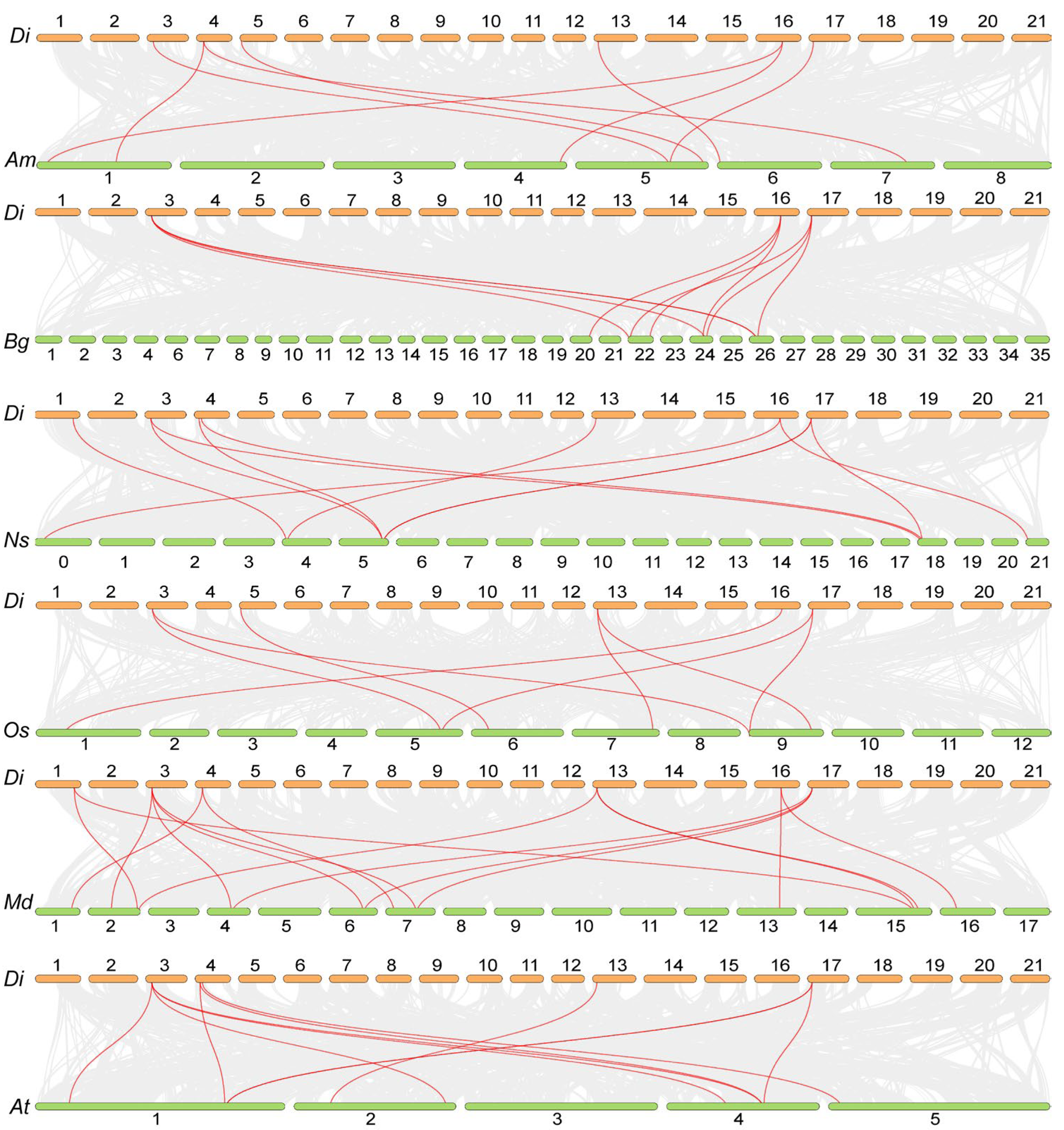
2.5. Inter-Species Collinearity Analysis
2.6. Identification of Conserved Motifs and Cis-Elements in the Promoter Regions of the DiPRP Genes
2.7. RNA Extraction and qRT-PCR
2.8. Vector Construction and Arabidopsis Transformation
2.9. Statistical Analysis
3. Result
3.1. The PRP Genes in Davidia Genome
3.2. Analysis of Collinearity of the PRPs Between Davidia and Other Species
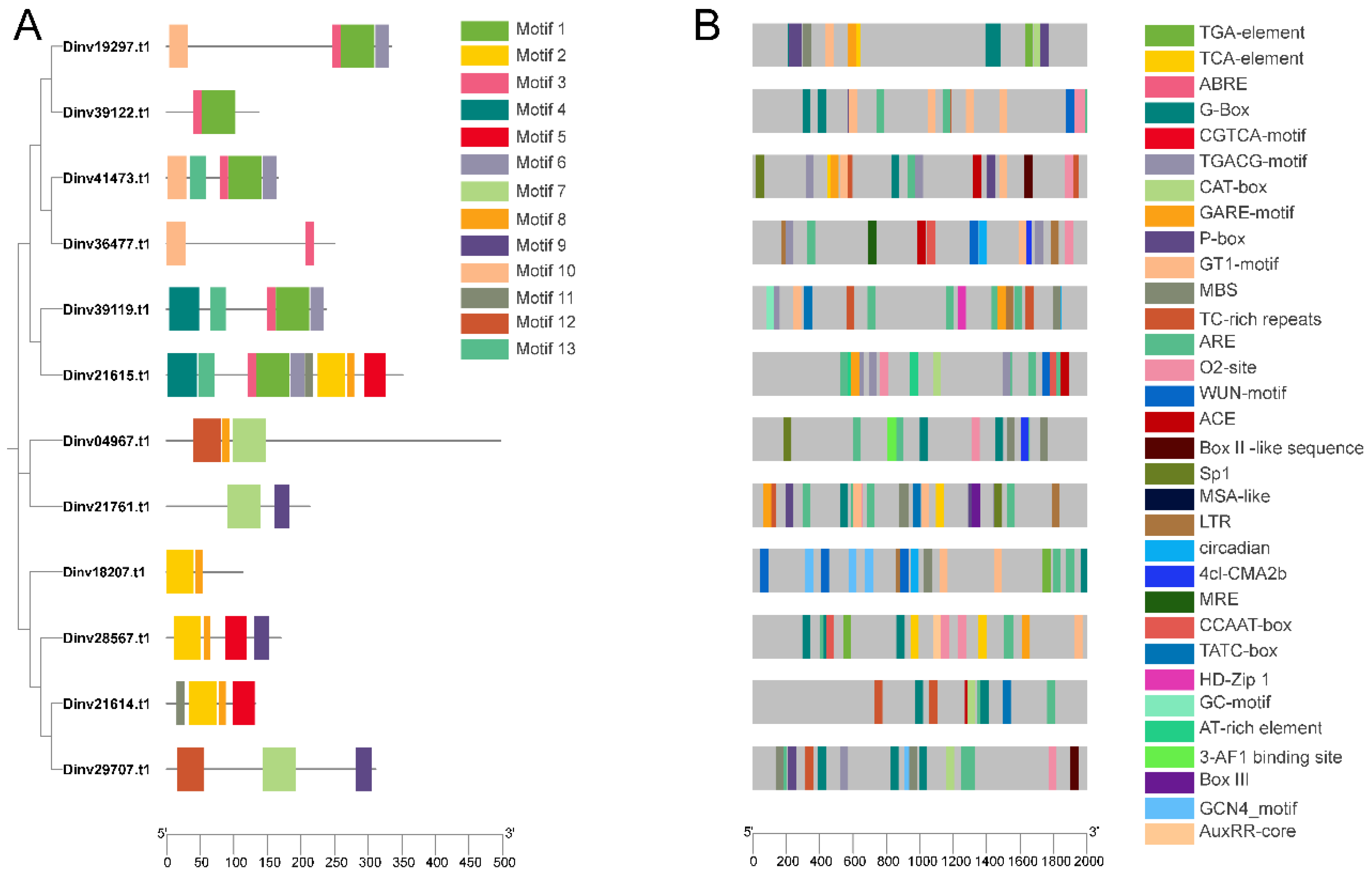
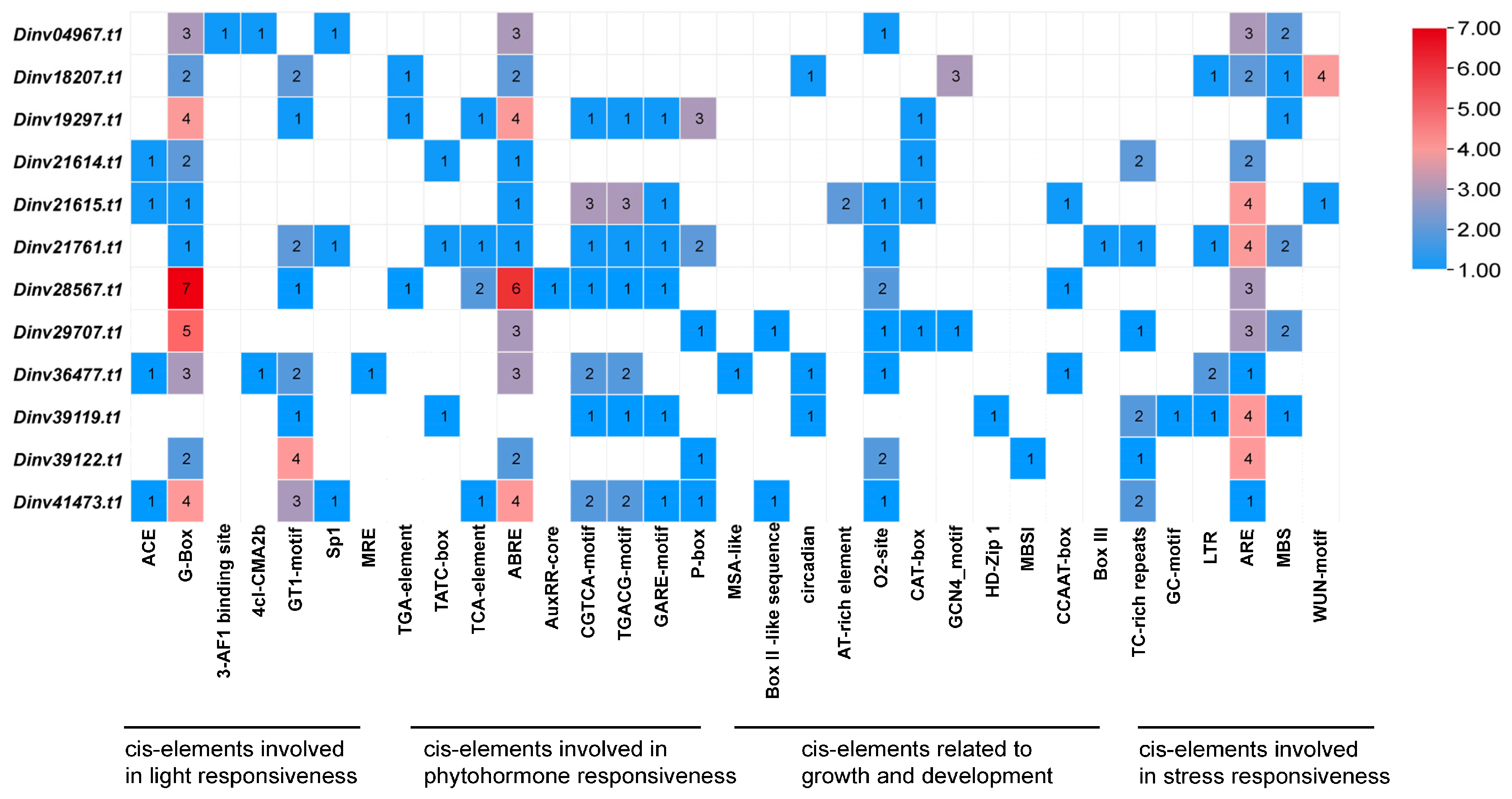
3.3. Conserved Motifs of DiPRPs and Cis-Elements of ProDiPRPs
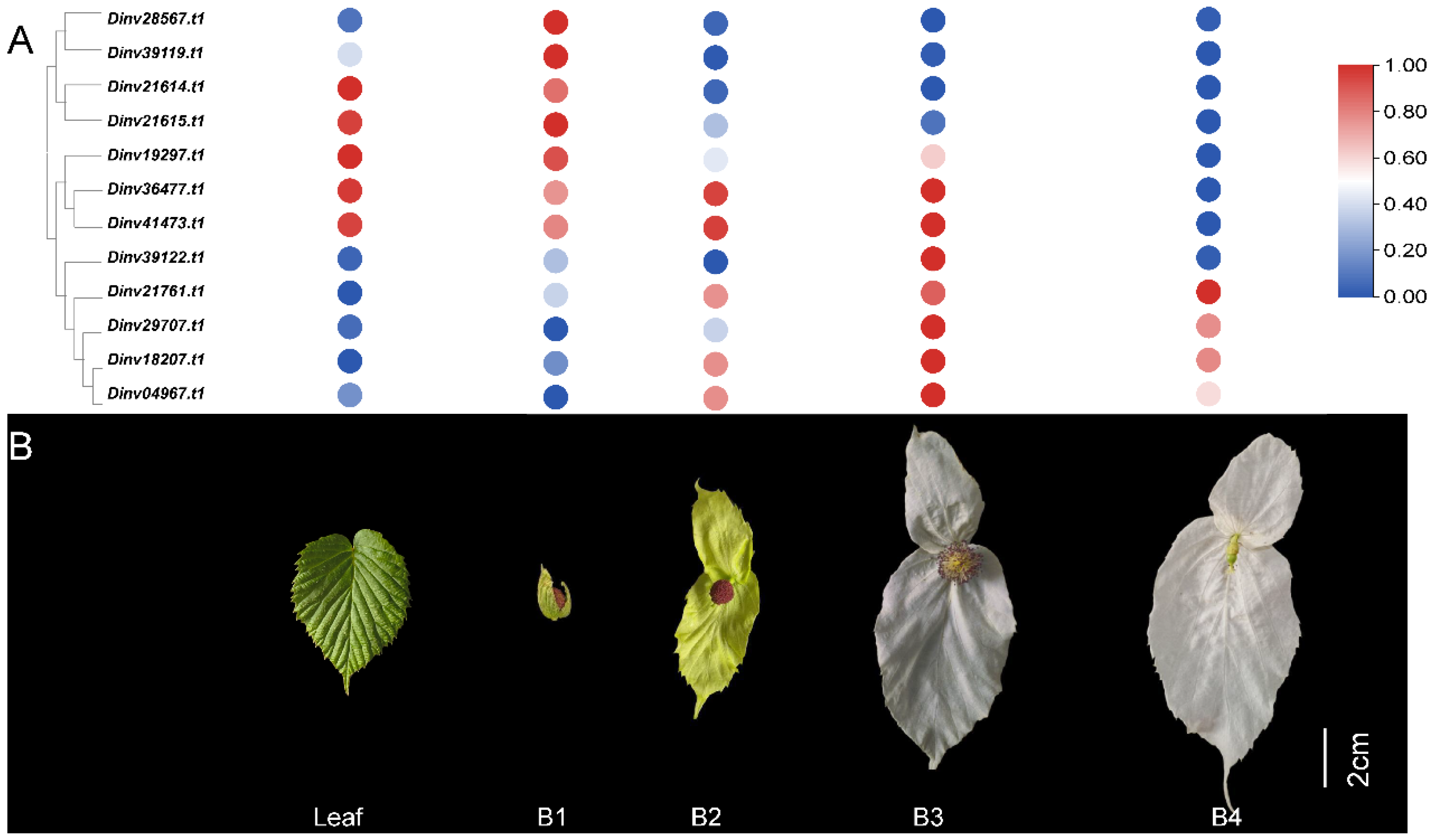
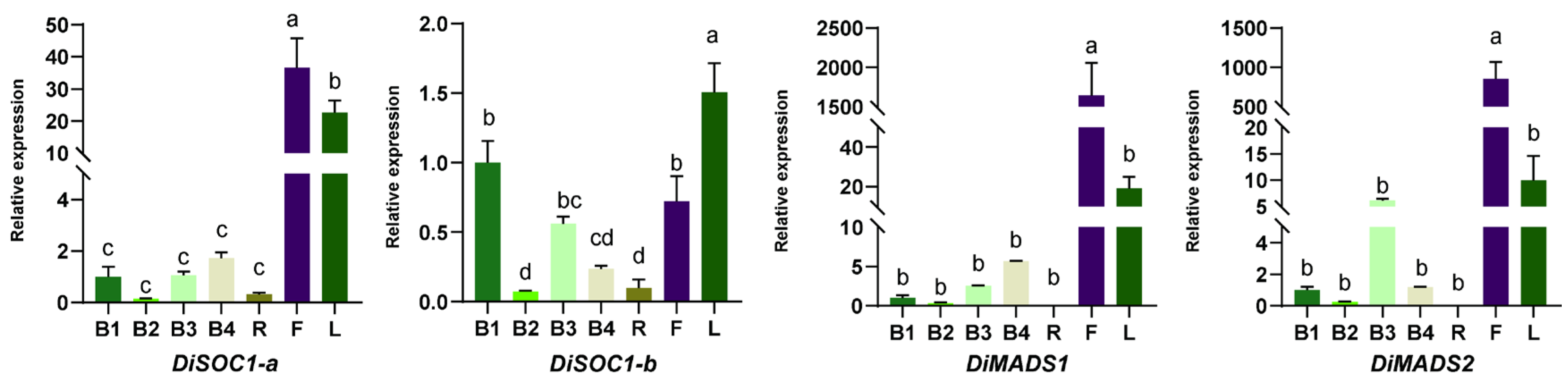
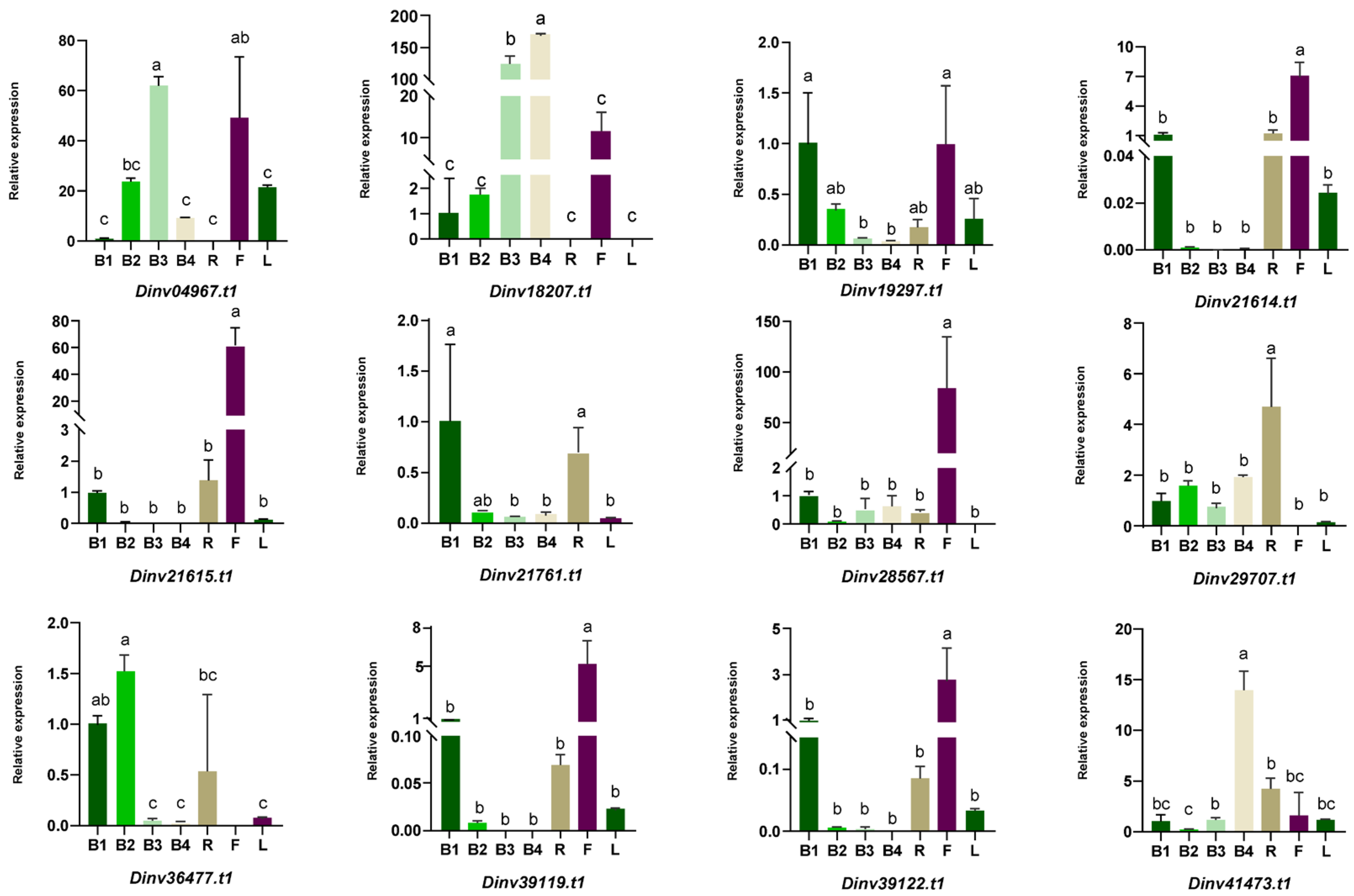
3.4. The Expression Patterns of DiPRPs During Bract Development
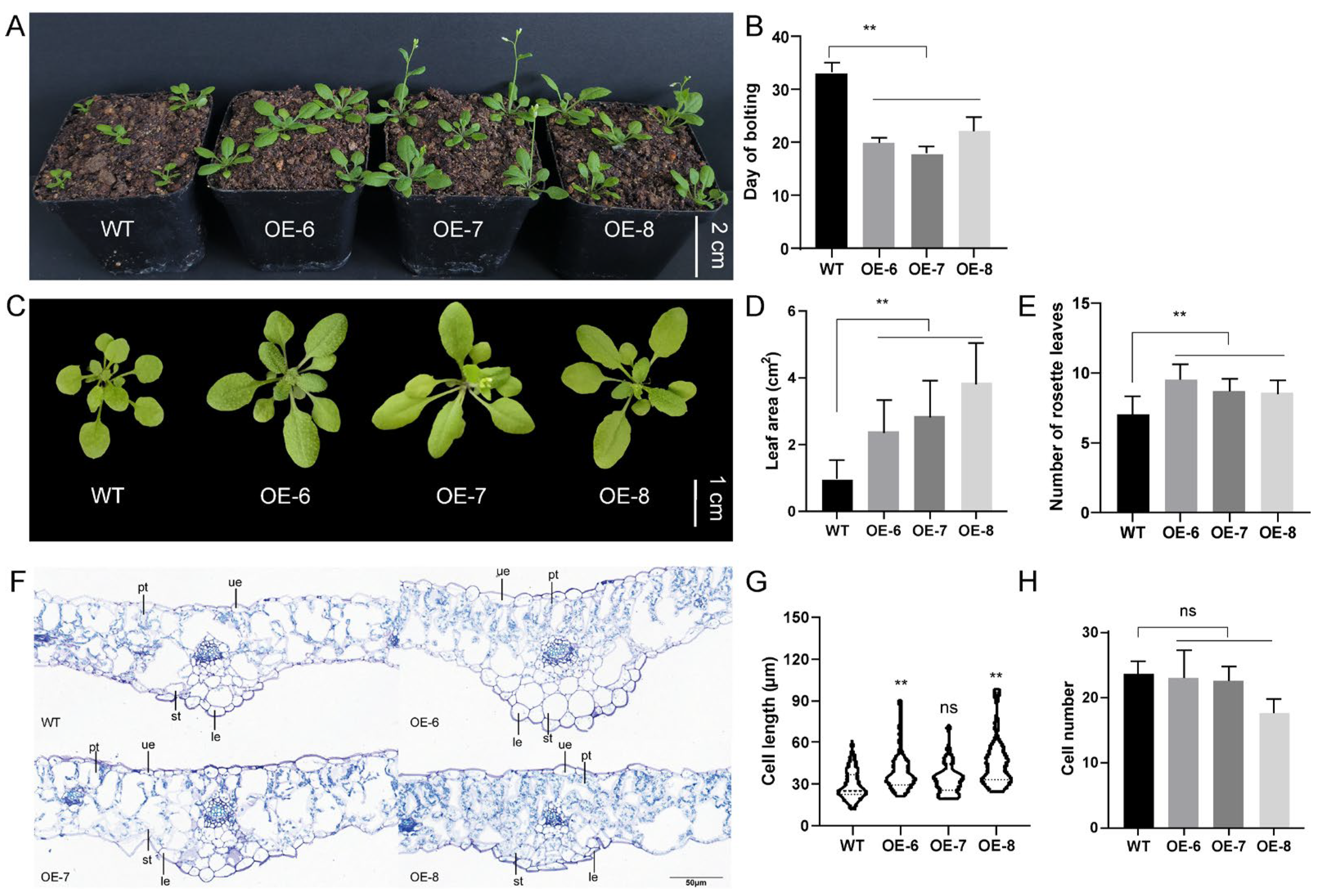
3.5. Functional Validation of a Dinv18207.t1 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Causier, B.; Schwarz-Sommer, Z.; Davies, B. Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 2010, 21, 73–79. [Google Scholar] [CrossRef]
- Sun, J.; Gong, Y.; Renner, S.S.; Huang, S. Multifunctional Bracts in the Dove Tree Davidia involucrata (Nyssaceae: Cornales): Rain Protection and Pollinator Attraction. Am. Nat. 2008, 171, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, W.S.; Antonsen, L.; Pélabon, C. Phenotypic selection on Dalechampia blossoms: Honest signaling affects pollination success. Ecology 2005, 86, 3323–3333. [Google Scholar] [CrossRef]
- Keasar, T.; Sadeh, A.; Gerchman, Y.; Shmida, A. The signaling function of an extra-floral display: What selects for signal development? Oikos 2009, 118, 1752–1759. [Google Scholar] [CrossRef]
- Higginson, A.D.; Gilbert, F.S.; Reader, T.; Barnard, C.J. Reduction of visitation rates by honeybees (Apis mellifera) to individual inflorescences of lavender (Lavandula stoechas) upon removal of coloured accessory bracts (Hymenoptera: Apidae). Èntomol. Gen. 2007, 29, 165–178. [Google Scholar] [CrossRef]
- Song, B.; Zhang, Z.-Q.; Stöcklin, J.; Yang, Y.; Niu, Y.; Chen, J.-G.; Sun, H. Multifunctional bracts enhance plant fitness during flowering and seed development in Rheum nobile (Polygonaceae), a giant herb endemic to the high Himalayas. Oecologia 2013, 172, 359–370. [Google Scholar] [CrossRef]
- Omori, Y.; Ohba, H. Pollen development of Rheum nobile Hook.f. & Thomson (Polygonaceae), with reference to its sterility induced by bract removal. Bot. J. Linn. Soc. 1996, 122, 269–278. [Google Scholar] [CrossRef]
- Song, B.; Gao, Y.; Stöcklin, J.; Song, M.; Sun, L.; Sun, H. Ultraviolet screening increases with elevation in translucent bracts of Rheum nobile (Polygonaceae), an alpine ‘glasshouse’ plant from the high Himalayas. Bot. J. Linn. Soc. 2020, 193, 276–286. [Google Scholar] [CrossRef]
- He, J.; Lin, J.; Chen, W. The current status of endemic and endangered species Davidia involucrata and the preserving strategies. Biodivers. Sci. 1995, 3, 213–221. [Google Scholar] [CrossRef]
- Xu, W.; Cao, F.; Li, M.; Liu, Z.; Wu, Y.; Wu, X.; Zhang, X. Research on the changes of endogenous hormones in the development of Davidia involucrata bracts. J. Cent. S. Univ. For. Technol. 2017, 37, 103–109. [Google Scholar] [CrossRef]
- Vekemans, D.; Viaene, T.; Caris, P.; Geuten, K. Transference of function shapes organ identity in the dove tree inflorescence. New Phytol. 2011, 193, 216–228. [Google Scholar] [CrossRef]
- Li, Y. Cloning and Analyzing of Bract Differentially Expressed Genes of Davidia involucrata Baill. Ph.D. Thesis, Sichuan University, Chengdu, China, 2002. [Google Scholar]
- Li, G.; Cao, C.; Yang, H.; Wang, J.; Wei, W.; Zhu, D.; Gao, P.; Zhao, Y. Molecular cloning and potential role of DiSOC1s in flowering regulation in Davidia involucrata Baill. Plant Physiol. Biochem. 2020, 157, 453–459. [Google Scholar] [CrossRef]
- Sun, M.; Zhu, L.; Zeng, L.; Yang, H.; Cao, C.; Wang, R.; Zhao, Y.; Wei, W. Analysis and Characterization of MADS-box Genes from Davidia involucrata Baill. and Regulation of Flowering Time in Arabidopsis. Russ. J. Plant Physiol. 2022, 69, 62. [Google Scholar] [CrossRef]
- Niu, J. Identification of Key Genes Involved in Bract Development and Functional Validation of a DiPRP4 Gene in the Dove Tree (Davidia involucrata Baill.). Master’s Thesis, Center South University of Foresty and Technolgy, Changsha, China, 2024. [Google Scholar]
- Esen, A.; Bietz, J.A.; Paulis, J.W.; Wall, J.S. Tandem repeats in the N-terminal sequence of a proline-rich protein from corn endosperm. Nature 1982, 296, 678–679. [Google Scholar] [CrossRef]
- Chen, J.; Varner, J.E. Isolation and characterization of cDNA clones for carrot extensin and a proline-rich 33-kDa protein. Proc. Natl. Acad. Sci. USA 1985, 82, 4399–4403. [Google Scholar] [CrossRef]
- Josè-Estanyol, M.; Puigdomènech, P. Plant cell wall glycoproteins and their genes. Plant Physiol. Biochem. 2000, 38, 97–108. [Google Scholar] [CrossRef]
- José-Estanyol, M.; Gomis-Rüth, F.; Puigdomènech, P. The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 2004, 42, 355–365. [Google Scholar] [CrossRef]
- Josè, M.; Puigdomènech, P. Structure and expression of genes coding for structural proteins of the plant cell wall. New Phytol. 1993, 125, 259–282. [Google Scholar] [CrossRef]
- Su, L.; Li, W.; Chen, X.; Wang, P.; Liu, D. Proline-rich protein PRPL1 enhances Panax notoginseng defence against Fusarium solani by regulating reactive oxygen species balance and strengthening the cell wall barrier. Plant Cell Environ. 2024, 47, 2375–2393. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Y.; Ding, C.; Gao, X.; Wang, R.; Mo, W.; Lv, F.; Wang, S.; Liu, L.; Tang, Z.; et al. Proline-rich protein gene PdPRP regulates secondary wall formation in poplar. J. Plant Physiol. 2019, 233, 58–72. [Google Scholar] [CrossRef]
- Chen, Y.; Ma, T.; Zhang, L.; Kang, M.; Zhang, Z.; Zheng, Z.; Sun, P.; Shrestha, N.; Liu, J.; Yang, Y. Genomic analyses of a “living fossil”: The endangered dove-tree. Mol. Ecol. Resour. 2020, 20, 756–769. [Google Scholar] [CrossRef]
- Niu, J. “Transcriptome of Davidia involucrata Baill.” the National Center for Biological Information. Available online: https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA008410 (accessed on 15 July 2025).
- Gabella, C.; Duvaud, S.; Durinx, C. Managing the life cycle of a portfolio of open data resources at the SIB Swiss Institute of Bioinformatics. Brief. Bioinform. 2021, 23, 1477–4054. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ren, R.; Dai, P.H.; Li, M.; Liu, Z.; Cao, F. Selection and stability evaluation of reference genes for real-time quantitative PCR in dove tree (Davidia involucrata). Plant Physiol. J. 2016, 52, 1565–1575. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.-S.; Niu, Q.-W.; Chua, N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Yang, J. Improvement of traditional paraffin section preparation methods. J. Biol. 2006, 1, 45–46. [Google Scholar] [CrossRef]
- Baum, D.A.; Day, C.D. Cryptic Bracts Exposed: Insights into the Regulation of Leaf Expansion. Dev. Cell 2004, 6, 318–319. [Google Scholar] [CrossRef][Green Version]
- Whipple, C.J.; Hall, D.H.; DeBlasio, S.; Taguchi-Shiobara, F.; Schmidt, R.J.; Jackson, D.P. A Conserved Mechanism of Bract Suppression in the Grass Family. Plant Cell 2010, 22, 565–578. [Google Scholar] [CrossRef]
- Wang, L.; Yin, H.; Qian, Q.; Yang, J.; Huang, C.; Hu, X.; Luo, D. NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 2009, 19, 598–611. [Google Scholar] [CrossRef] [PubMed]
- Fowler, T.J.; Bernhardt, C.; Tierney, M.L. Characterization and Expression of Four Proline-Rich Cell Wall Protein Genes in Arabidopsis Encoding Two Distinct Subsets of Multiple Domain Proteins. Plant Physiol. 1999, 121, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, R.E.; Nagao, R.T.; Key, J.L. Patterns of soybean proline-rich protein gene expression. Plant Cell 1992, 4, 99–110. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Menke, U.; Renault, N.; Mueller-Roeber, B. StGCPRP, a Potato Gene Strongly Expressed in Stomatal Guard Cells, Defines a Novel Type of Repetitive Proline-Rich Proteins. Plant Physiol. 2000, 122, 677–686. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, J.; Ning, J.; Liu, Y.; Xu, J.; Tian, S.; Zhang, L.; Sun, M. NtProRP1, a novel proline-rich protein, is an osmotic stress-responsive factor and specifically functions in pollen tube growth and early embryogenesis in Nicotiana tabacum. Plant Cell Environ. 2014, 37, 499–511. [Google Scholar] [CrossRef]
- Zhang, X.; Gong, X.; Li, D.; Yue, H.; Qin, Y.; Liu, Z.; Li, M.; Ma, F. Genome-Wide Identification of PRP Genes in Apple Genome and the Role of MdPRP6 in Response to Heat Stress. Int. J. Mol. Sci. 2021, 22, 5942. [Google Scholar] [CrossRef]
- Rajasheker, G.; Nagaraju, M.; Varghese, R.P.; Jalaja, N.; Somanaboina, A.K.; Singam, P.; Ramakrishna, C.; Penna, S.; Sreenivasulu, N.; Kishor, P.B.K. Identification and analysis of proline-rich proteins and hybrid proline-rich proteins super family genes from Sorghum bicolor and their expression patterns to abiotic stress and zinc stimuli. Front. Plant Sci. 2022, 13, 952732. [Google Scholar] [CrossRef]
- Kapoor, R.; Kumar, G.; Arya, P.; Jaswal, R.; Jain, P.; Singh, K.; Sharma, T.R. Genome-Wide Analysis and Expression Profiling of Rice Hybrid Proline-Rich Proteins in Response to Biotic and Abiotic Stresses, and Hormone Treatment. Plants 2019, 8, 343. [Google Scholar] [CrossRef]
- Priyanka, B.; Sekhar, K.; Reddy, V.D.; Rao, K.V. Expression of pigeonpea hybrid-proline-rich protein encoding gene (CcHyPRP) in yeast and Arabidopsis affords multiple abiotic stress tolerance. Plant Biotechnol. J. 2010, 8, 76–87. [Google Scholar] [CrossRef]
- Zhai, Y.; Lei, T.-T.; Yan, F.; Huang, K.-M.; Li, X.-W.; Zhang, Q.-L.; Zhang, H.-J.; Su, L.-T.; Sun, X.; Wang, Y.; et al. Cloning and Expression of a Stress-induced GmPRP Gene in Soybean (Glycine max). Acta Agron. Sin. 2011, 37, 2152–2157. [Google Scholar] [CrossRef]
- Zhang, X.; Gong, X.; Yu, H.; Su, X.; Cheng, S.; Huang, J.; Lei, Z.; Li, M.; Ma, F. The proline-rich protein MdPRP6 confers tolerance to salt stress in transgenic apple (Malus domestica). Sci. Hortic. 2022, 308, 111581. [Google Scholar] [CrossRef]
- Liu, L.; Xu, W.; Hu, X.; Liu, H.; Lin, Y. W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 2016, 6, 20881. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Kobayashi, K.; Obayashi, T.; Masuda, T. Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal. Behav. 2012, 7, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of Transcription Factor HY5 Genomic Binding Sites Revealed Its Hierarchical Role in Light Regulation of Development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Wang, P.; Korkaric, M.; Capper, R.G.; Saunders, N.J.; Langdale, J.A. GLK Transcription Factors Coordinate Expression of the Photosynthetic Apparatus in Arabidopsis. Plant Cell 2009, 21, 1109–1128. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef]
- An, J.; Zhang, X.; Bi, S.; You, C.; Wang, X.; Hao, Y. MdbHLH93, an apple activator regulating leaf senescence, is regulated by ABA and MdBT2 in antagonistic ways. New Phytol. 2019, 222, 735–751. [Google Scholar] [CrossRef]
- Wu, X.; Gao, R.; Mao, R.; Lin, Y.; Yang, Z.; Li, J.; Cao, F.; Li, M. Inducing bract-like leaves in Arabidopsis through ectopically expressing an ASR gene from the dove tree. Ind. Crop. Prod. 2022, 180, 114796. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4- and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Tang, W.; Pan, X.; Huang, A.; Gao, X.; Anderson, C.T.; Yang, Z. Arabidopsis pavement cell morphogenesis requires FERONIA binding to pectin for activation of ROP GTPase signaling. Curr. Biol. 2022, 32, 497–507.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Kanyuka, K.; Papp-Rupar, M. Pectin: A critical component in cell-wall-mediated immunity. Trends Plant Sci. 2023, 28, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Sun, G.; Yu, Q.; Gao, T.; Zhu, Q.; Wang, R.; Huang, S.; Han, Z.; Cervone, F.; Yin, H.; et al. A plant mechanism of hijacking pathogen virulence factors to trigger innate immunity. Science 2024, 383, 732–739. [Google Scholar] [CrossRef]
- Seo, S.-G.; Jeon, S.B.; Kim, J.-S.; Shin, J.-M.; Kim, J.-B.; Kang, S.-W.; Lee, G.-P.; Kim, S.-H. Characterization and expression pattern of IbPRP1 and IbPRP2 stress-related genes from sweetpotato. Genes Genom. 2010, 32, 487–497. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, T.; Zhong, G.; Zheng, Y. Overexpression of a new proline-rich protein encoding Gene CsPRP4 increases starch accumulation in Citrus. Sci. Hortic. 2020, 260, 108744. [Google Scholar] [CrossRef]
- Liu, D.Q.; Han, Q.; Shah, T.; Chen, C.Y.; Wang, Q.; Tang, B.F.; Ge, F. A hybrid proline-rich cell-wall protein gene JsPRP1 from Juglans sigillata Dode confers both biotic and abiotic stresses in transgenic tobacco plants. Trees 2018, 32, 1199–1209. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Wang, X.; Wang, W.; Li, Z.; Wu, J.; Wang, G.; Wu, L.; Zhang, G.; Ma, Z. HyPRP1 performs a role in negatively regulating cotton resistance to V. dahliae via the thickening of cell walls and ROS accumulation. BMC Plant Biol. 2018, 18, 339. [Google Scholar] [CrossRef]
- Hörtensteiner, S. Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 2009, 14, 155–162. [Google Scholar] [CrossRef]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC Transcription Factor Promotes Leaf Senescence via ABA Biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- Gothandam, K.M.; Nalini, E.; Karthikeyan, S.; Shin, J.S. OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol. Biol. 2010, 72, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Telfer, A.; Bollman, K.M.; Poethig, R.S. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 1997, 124, 645–654. [Google Scholar] [CrossRef]
- Chien, J.C.; Sussex, I.M. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 111, 1321–1328. [Google Scholar] [CrossRef]
- Dvořáková, L.; Srba, M.; Opatrny, Z.; Fischer, L. Hybrid proline-rich proteins: Novel players in plant cell elongation? Ann. Bot. 2011, 109, 453–462. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Zhang, X.; Luo, Y.; Niu, J.; Li, J.; Li, M. Genome-Wide Identification of the PRP Gene Family Members of the Dove Tree (Davidia involucrata Baill.). Forests 2025, 16, 1425. https://doi.org/10.3390/f16091425
Fan Y, Zhang X, Luo Y, Niu J, Li J, Li M. Genome-Wide Identification of the PRP Gene Family Members of the Dove Tree (Davidia involucrata Baill.). Forests. 2025; 16(9):1425. https://doi.org/10.3390/f16091425
Chicago/Turabian StyleFan, Yanling, Xiyi Zhang, Yanxian Luo, Jie Niu, Jia Li, and Meng Li. 2025. "Genome-Wide Identification of the PRP Gene Family Members of the Dove Tree (Davidia involucrata Baill.)" Forests 16, no. 9: 1425. https://doi.org/10.3390/f16091425
APA StyleFan, Y., Zhang, X., Luo, Y., Niu, J., Li, J., & Li, M. (2025). Genome-Wide Identification of the PRP Gene Family Members of the Dove Tree (Davidia involucrata Baill.). Forests, 16(9), 1425. https://doi.org/10.3390/f16091425





