Genome-Wide Identification of the UGT Gene Family in Poplar Populus euphratica and Functional Analysis of PeUGT110 Under Drought Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of PeUGT Proteins
2.2. Analyses of Phylogenetic Relationship, Chromosome Location, and Synteny
2.3. Analyses of Gene Structures, Conserved Motifs and Promoter Cis-Acting Element of PeUGT Genes
2.4. Expression Patterns of PeUGT Genes in Different Tissues
2.5. Quantitative Real-Time PCR
2.6. Arabidopsis Transformation and Drought Treatment
2.7. Plant Materials and Drought Treatments
3. Results
3.1. Identification and Characteristics of PeUGTs
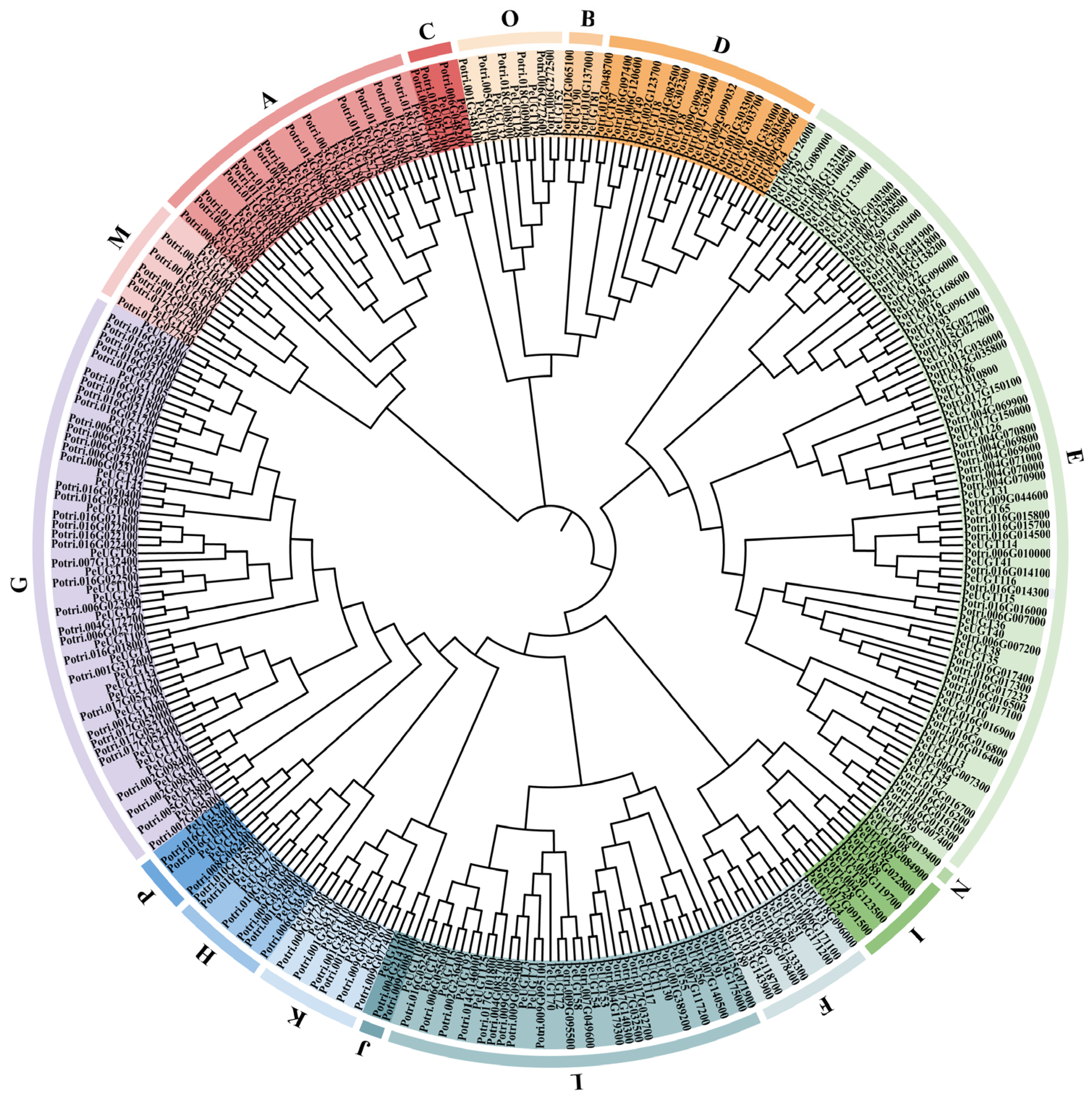
3.2. Phylogenetic Analysis of UGT Genes
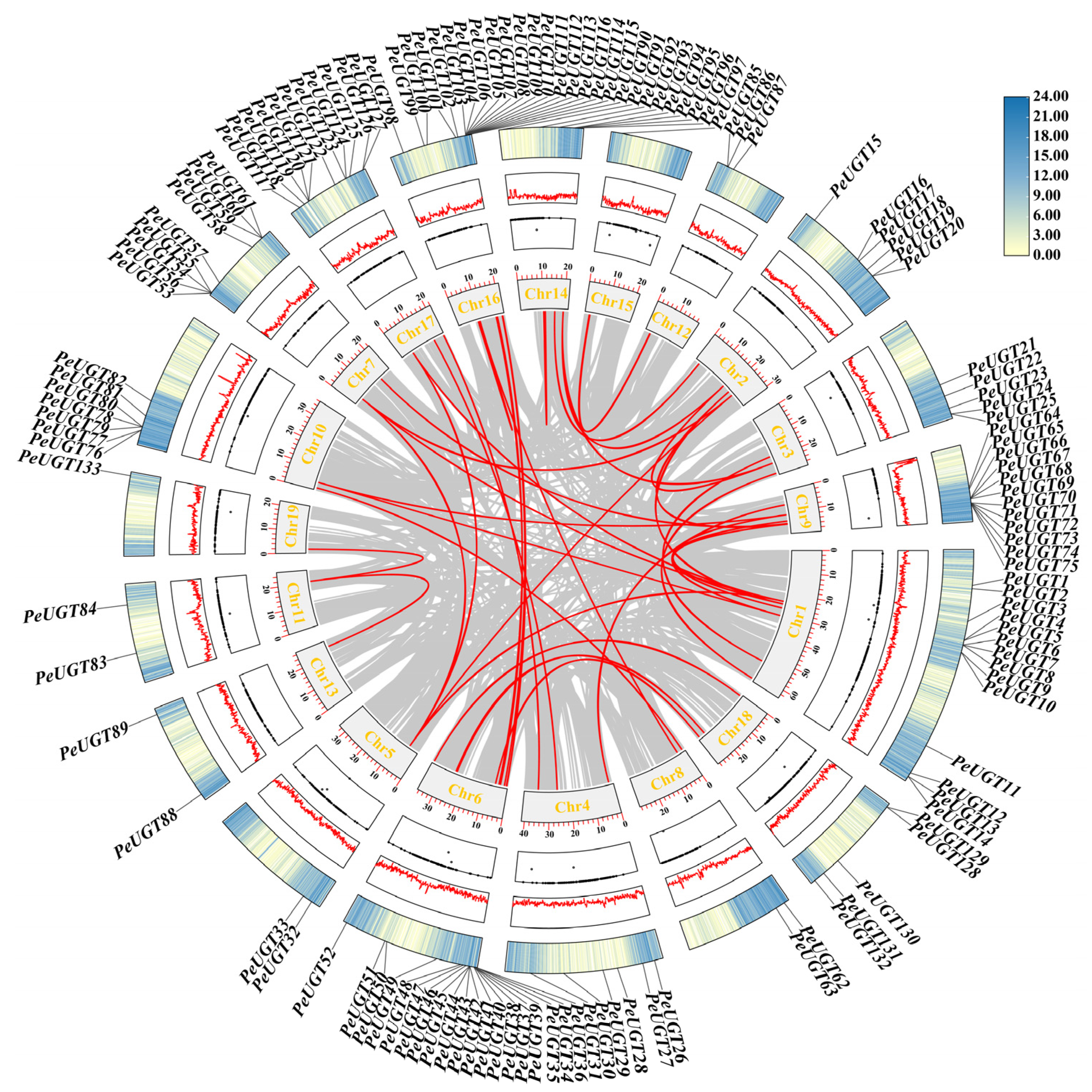
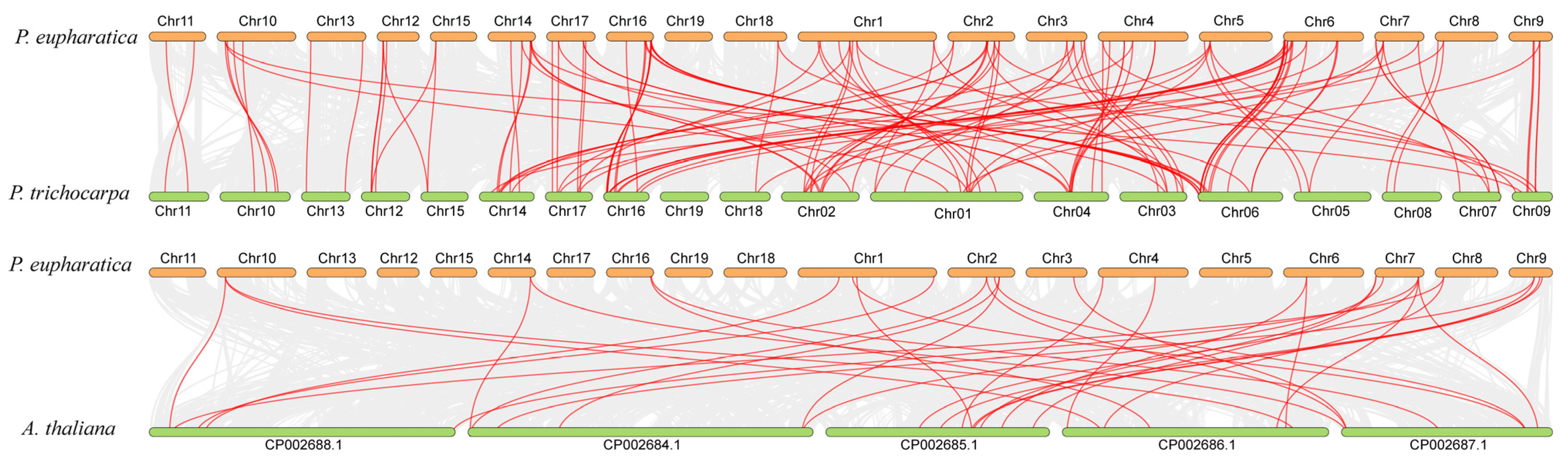
3.3. Chromosome Localization and Gene Duplication Analysis of UGT Gene in P. euphratica
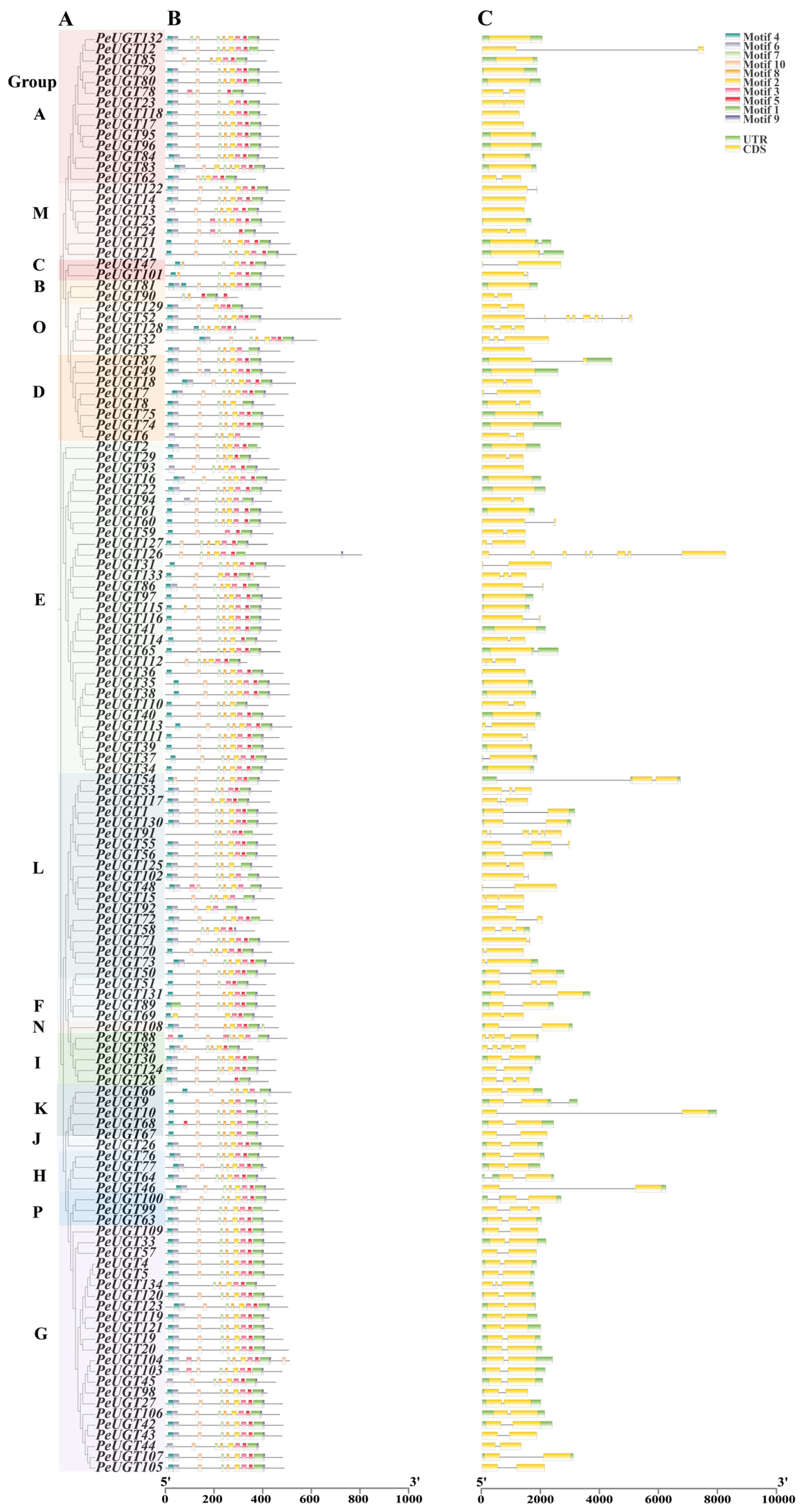
3.4. Structure and Conserved Motif Analysis of PeUGT Genes
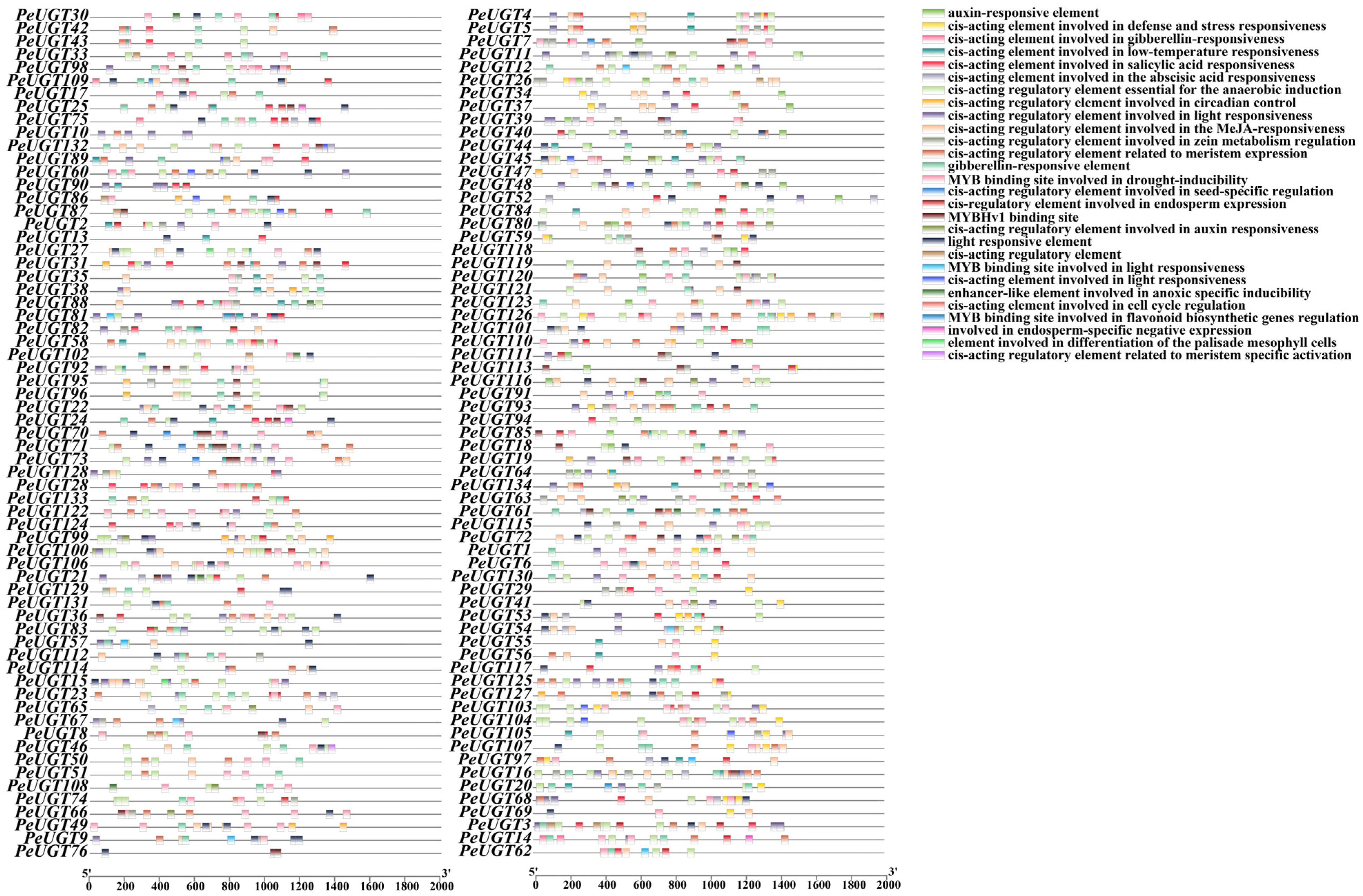
3.5. Cis-Acting Elements Analysis of PeUGT Genes
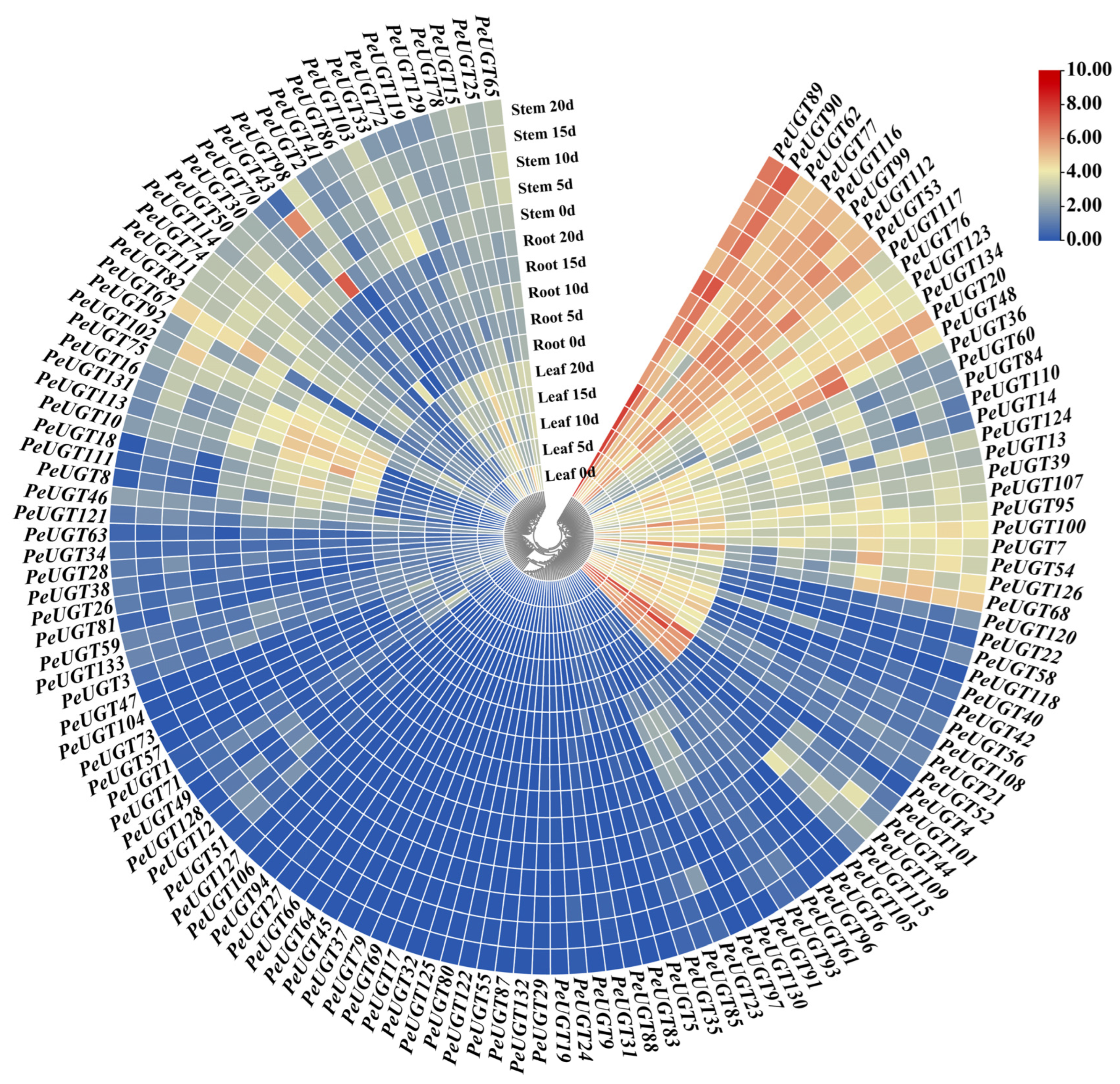
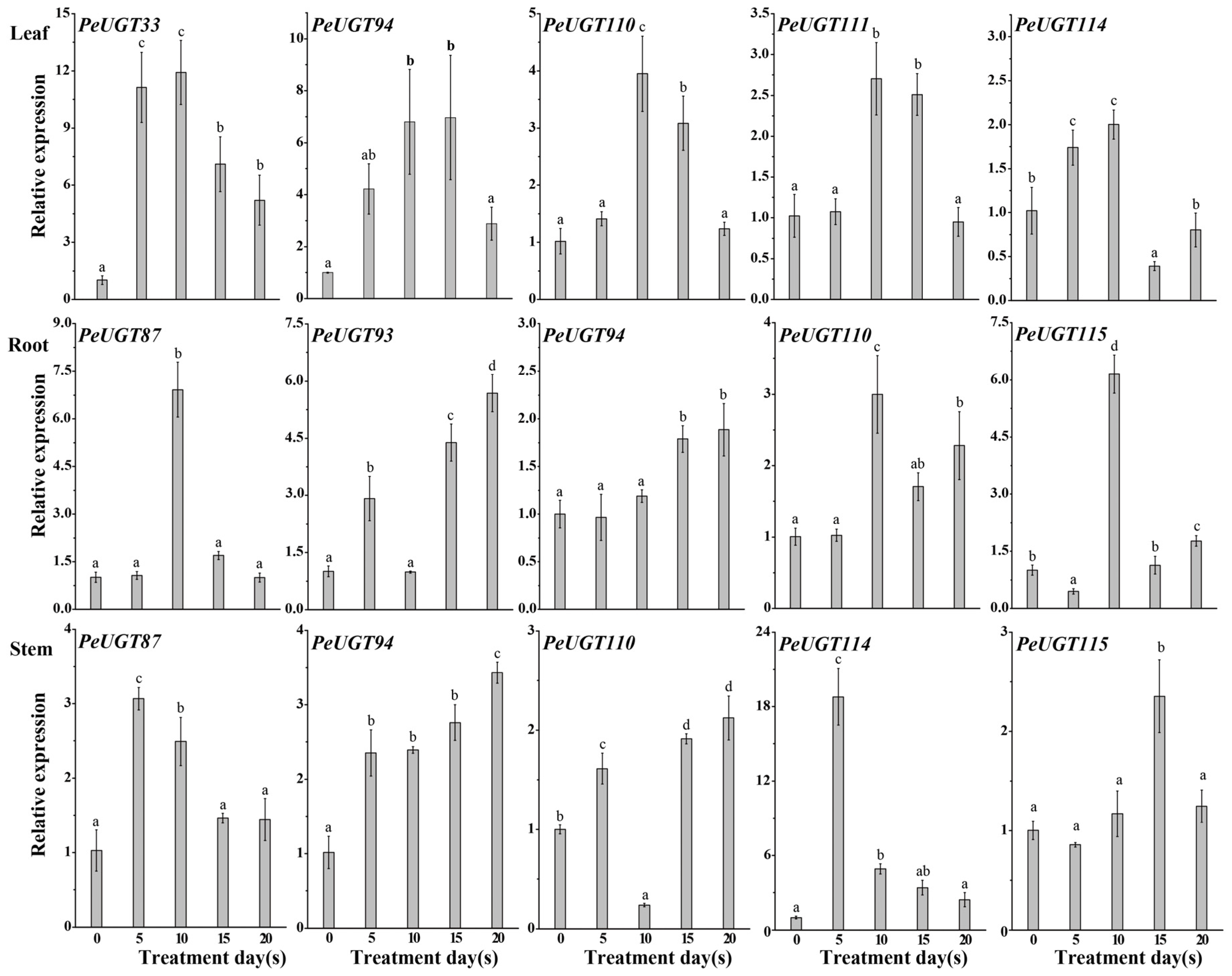
3.6. Expression of PeUGT Genes in Different Tissues Under Drought Stress
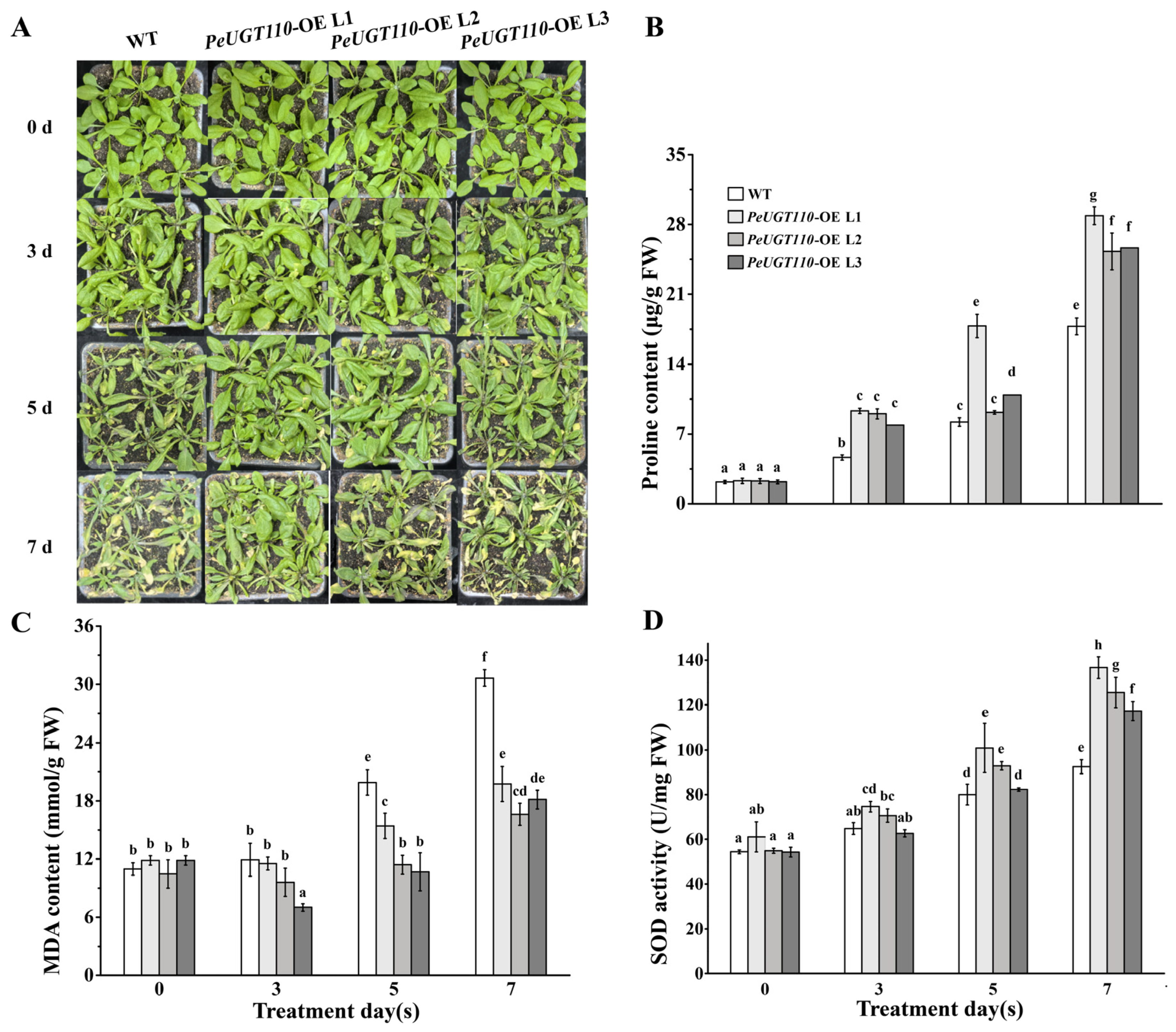
3.7. Overexpression of the PeUGT110 Gene Improves the Drought Tolerance in Arabidopsis
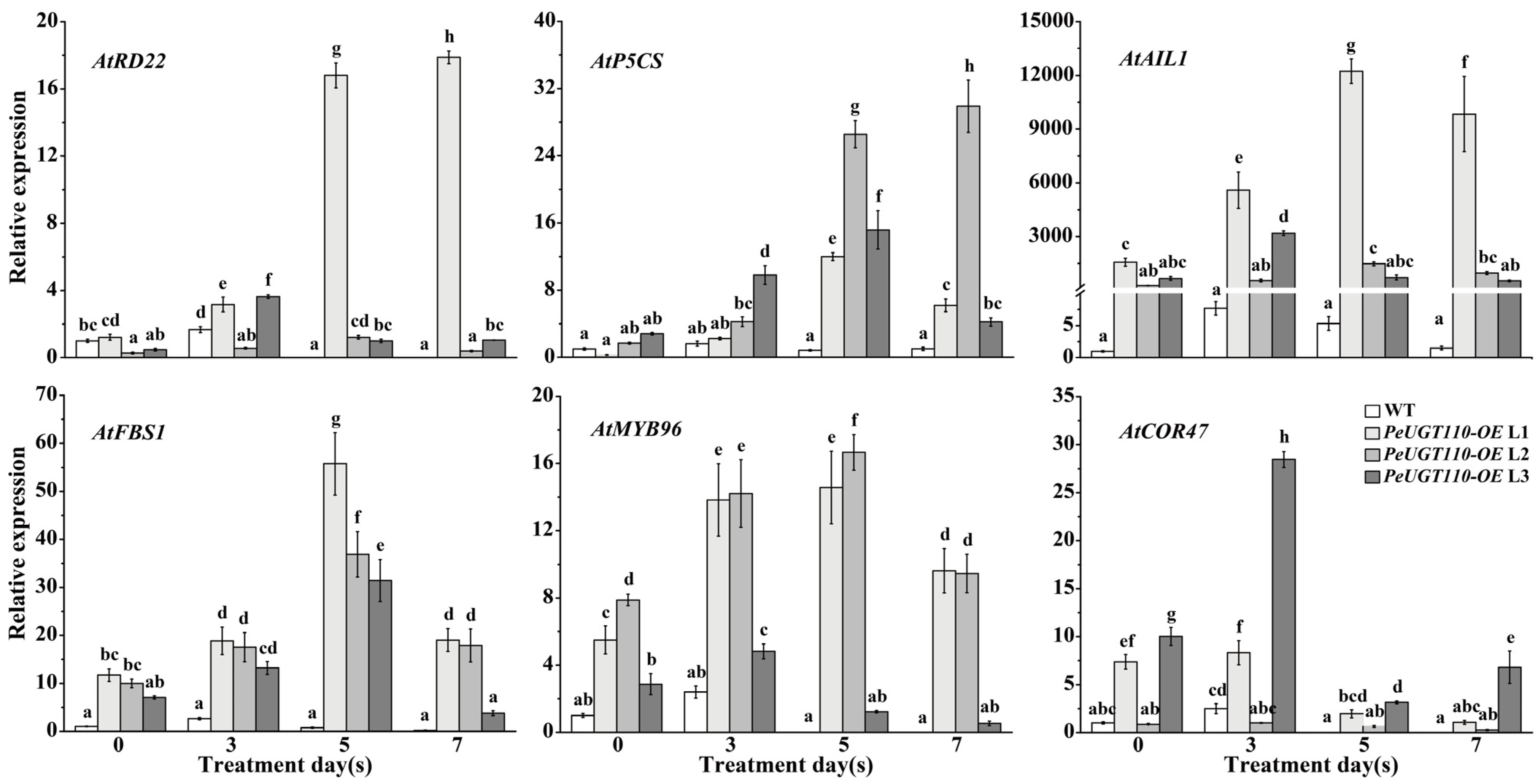
3.8. Expression Profiling of Drought Stress-Related Genes in Arabidopsis Overexpressing PeUGT110
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K. Drought stress in plants: An overview. Plant responses to drought stress: From morphological to molecular features. Int. J. Agric. Biol. 2012, 11, 100–105. [Google Scholar]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of climate change on crops adaptation and strategies to tackle its outcome: A review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M. Plant drought stress: Effects, mechanisms and management. In Sustainable Agriculture; Springer: Dordrecht, The Netherlands, 2009; pp. 153–188. [Google Scholar]
- Fathi, A.; Tari, D.B. Effect of drought stress and its mechanism in plants. Int. J. Life Sci. 2016, 10, 1–6. [Google Scholar] [CrossRef]
- Kaur, G.; Asthir, B. Molecular responses to drought stress in plants. Biol. Plant. 2017, 61, 201–209. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.-K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Paquette, S.; Møller, B.L.; Bak, S. On the origin of family 1 plant glycosyltransferases. Phytochemistry 2003, 62, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Osmani, S.A.; Bak, S.; Møller, B.L. Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 2009, 70, 325–347. [Google Scholar] [CrossRef] [PubMed]
- Offen, W.; Martinez-Fleites, C.; Yang, M.; Kiat-Lim, E.; Davis, B.G.; Tarling, C.A.; Ford, C.M.; Bowles, D.J.; Davies, G.J. Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 2006, 25, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Hanada, K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011, 66, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Gachon, C.M.; Langlois-Meurinne, M.; Saindrenan, P. Plant secondary metabolism glycosyltransferases: The emerging functional analysis. Trends Plant Sci. 2005, 10, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.E.; Tian, L. Phylogenomic analysis of UDP-dependent glycosyltransferases provides insights into the evolutionary landscape of glycosylation in plant metabolism. Plant J. 2019, 100, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.; Li, Y.; Lim, E.-K.; Bowles, D.J. Higher plant glycosyltransferases. Genome Biol. 2001, 2, reviews3004.1. [Google Scholar] [CrossRef] [PubMed]
- Barvkar, V.T.; Pardeshi, V.C.; Kale, S.M.; Kadoo, N.Y.; Gupta, V.S. Phylogenomic analysis of UDP glycosyltransferase 1 multigene family in Linum usitatissimum identified genes with varied expression patterns. BMC Genom. 2012, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Pang, C.; Fan, S.; Song, M.; Yu, J.; Wei, H.; Ma, Q.; Li, L.; Zhang, C.; Yu, S. Genome-wide analysis of the family 1 glycosyltransferases in cotton. Mol. Genet. Genom. 2015, 290, 1805–1818. [Google Scholar] [CrossRef] [PubMed]
- Kamal, K.A.; Shah, F.A.; Zhao, Y.; Chen, Z.; Fu, S.; Zhu, Z.; Ren, J.; Liu, H. Genome-wide identification of the UGT genes family in Acer rubrum and role of ArUGT52 in anthocyanin biosynthesis under cold stress. BMC Plant Biol. 2025, 25, 288. [Google Scholar] [CrossRef] [PubMed]
- Rehman, H.M.; Khan, U.M.; Nawaz, S.; Saleem, F.; Ahmed, N.; Rana, I.A.; Atif, R.M.; Shaheen, N.; Seo, H. Genome wide analysis of family-1 UDP glycosyltransferases in Populus trichocarpa specifies abiotic stress responsive glycosylation mechanisms. Genes 2022, 13, 1640. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Yan, Q.; Wu, F.; Wang, Y.; Wang, S.; Zong, X.; Zhou, P.; Zhang, J. Genome-wide analysis of the UDP-glycosyltransferase family reveals its roles in coumarin biosynthesis and abiotic stress in Melilotus albus. Int. J. Mol. Sci. 2021, 22, 10826. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Song, J.; Sheng, S.; Wang, D.; Wang, T.; Wang, N.; Chen, A.; Wang, L.; Peng, Y.; Ma, Y. Genome-wide characterization of Solanum tuberosum UGT gene family and functional analysis of StUGT178 in salt tolerance. BMC Genom. 2024, 25, 1206. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dong, G.; Zhao, S.; Zhang, F.; Ma, X.; Liu, C.; Ding, Y.; Hou, B. Arabidopsis glycosyltransferase UGT86A1 promotes plant adaptation to salt and drought stresses. Physiol. Plant. 2025, 177, e70050. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Yu, H.M.; Meng, X.F.; Lin, J.S.; Li, Y.J.; Hou, B.K. Ectopic expression of glycosyltransferase UGT76E11 increases flavonoid accumulation and enhances abiotic stress tolerance in Arabidopsis. Plant Biol. 2018, 20, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Commun. 2020, 11, 2629. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wang, X.; Yu, H.; Dong, N.; Li, J.; Chang, X.; Wang, J.; Jiang, C.; Liu, J.; Chi, X. Genome-wide analysis of UDP-glycosyltransferases family and identification of UGT genes involved in drought stress of Platycodon grandiflorus. Front. Plant Sci. 2024, 15, 1363251. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Chen, X.; Hou, H.; Fan, Y.; Wang, L.; Zeng, H.; Chen, X.; Ding, Y.; Hu, X.; Yan, Q. Genome-wide characterization of the tomato UDP-glycosyltransferase gene family and functional identification of SlUDPGT52 in drought tolerance. Hortic. Adv. 2023, 1, 14. [Google Scholar] [CrossRef]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; Van De Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, X.M.; Kojima, M.; Sakakibara, H.; Hou, B.K. N-glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 2200–2213. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Yan, H.; Arndt, S.K. Leaf and whole tree adaptations to mild salinity in field grown Populus euphratica. Tree Physiol. 2009, 29, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Xu, J.; Shao, X.; Luo, W.; Niu, Z.; Gao, C.; Wan, D. A novel gene coding γ-aminobutyric acid transporter may improve the tolerance of Populus euphratica to adverse environments. Front. Plant Sci. 2019, 10, 1083. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Ma, J.; Niu, Z.; Bai, X.; Lei, W.; Shao, X.; Chen, N.; Zhou, F.; Wan, D. Tissue-specific transcriptome analysis reveals multiple responses to salt stress in Populus euphratica seedlings. Genes 2017, 8, 372. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Li, G.; Hu, H.; Lv, J.; Zheng, Q.; Liu, J.; Wan, D. A gene that underwent adaptive evolution, LAC2 (LACCASE), in Populus euphratica improves drought tolerance by improving water transport capacity. Hortic. Res. 2021, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, Y.; Zhang, J.; Ma, X.; Li, Y.; Li, M.; Wang, D.; Kang, M.; Wu, H.; Yang, Y. Improved genome assembly provides new insights into genome evolution in a desert poplar (Populus euphratica). Mol. Ecol. Resour. 2020, 20, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Su, Y.; Chen, N.; Shen, S. Genome-wide analysis of the UGT gene family and identification of flavonoids in Broussonetia papyrifera. Molecules 2021, 26, 3449. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, X.; Lu, M.; Chen, J.; Shi, T. Gene expression and evolution of Family-1 UDP-glycosyltransferases—Insights from an aquatic flowering plant (sacred lotus). Aquat. Bot. 2020, 166, 103270. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, Z.; Shen, M.; Li, Q.; Wang, S.; Jiang, J.; Zeng, W. Identification and functional verification of the glycosyltransferase gene family involved in flavonoid synthesis in Rubus Chingii Hu. Plants 2024, 13, 1390. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Feng, Y.; Ke, D.; Teng, Y.; Chen, Y.; Langjia, R. Molecular identification and characterization of UDP-glycosyltransferase (UGT) multigene family in pomegranate. Horticulturae 2023, 9, 540. [Google Scholar] [CrossRef]
- Cui, L.; Yao, S.; Dai, X.; Yin, Q.; Liu, Y.; Jiang, X.; Wu, Y.; Qian, Y.; Pang, Y.; Gao, L. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis). J. Exp. Bot. 2016, 67, 2285–2297. [Google Scholar] [CrossRef] [PubMed]
- Caputi, L.; Malnoy, M.; Goremykin, V.; Nikiforova, S.; Martens, S. A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J. 2012, 69, 1030–1042. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, Y.; Qu, X.; Wu, F.; Li, X.; Ren, M.; Tong, Y.; Wu, X.; Yang, A.; Chen, Y. Genome-wide analysis of UDP-glycosyltransferases family and identification of UGT genes involved in abiotic stress and flavonol biosynthesis in Nicotiana tabacum. BMC Plant Biol. 2023, 23, 204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Guo, Y.; Zhang, Y.; Guo, J.; Li, K.; Fu, W.; Jia, Z.; Li, W.; Tran, L.-S.P.; Jia, K.-P. Genome-wide identification, characterization and expression profiles of the CCD gene family in Gossypium species. 3 Biotech 2021, 11, 249. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Lu, Q.; Liu, R.; Gong, J.; Gong, W.; Liu, A.; Ge, Q.; Li, J.; Shang, H.; Li, P. Genome-wide characterization of the UDP-glycosyltransferase gene family in upland cotton. 3 Biotech 2019, 9, 453. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Admas, T.; Cheng, B.; Meng, Y.; Pan, R.; Zhang, W. UGT gene family identification and functional analysis of HvUGT1 under drought stress in wild barley. Physiol. Mol. Biol. Plants 2024, 30, 1225–1238. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Dai, J.; Chen, Z.; Tie, W.; Yan, Y.; Yang, H.; Zeng, J.; Hu, W. Comprehensive analysis and expression profiles of cassava UDP-glycosyltransferases (UGT) family reveal their involvement in development and stress responses in cassava. Genomics 2021, 113, 3415–3429. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Liu, Y.; Yao, R.; He, D.; Yan, L.; Chen, Y.; Huai, D.; Wang, Z.; Yu, B.; Kang, Y. Genome-wide analysis of UDP-glycosyltransferase gene family and identification of a flavonoid 7-O-UGT (AhUGT75A) enhancing abiotic stress in peanut (Arachis hypogaea L.). BMC Plant Biol. 2023, 23, 626. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.; Kim, S.R.; Zhao, G.; Yi, J.; Yoo, Y.; Jin, P.; Lee, S.W.; Jung, K.h.; Zhang, D.; An, G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013, 161, 663–675. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhao, L.; Huang, H.; Zhang, Y.; Wang, J.; Lu, X.; Wang, S.; Wang, D.; Chen, X.; Chen, C. Genome-wide identification, evolution and function analysis of UGTs superfamily in cotton. Front. Mol. Biosci. 2022, 9, 965403. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Qian, P.; Ye, M.; Mu, K.; Wang, S.; Chen, M.; Ma, H. GmUGT73F4 plays important roles in enhancing seed vitality and tolerance to abiotic stresses in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 2022, 150, 313–328. [Google Scholar] [CrossRef]
- Li, H.; Yue, H.; Xie, J.; Bu, J.; Li, L.; Xin, X.; Zhao, Y.; Zhang, H.; Yang, L.; Wang, J. Transcriptomic profiling of the high-vigour maize (Zea mays L.) hybrid variety response to cold and drought stresses during seed germination. Sci. Rep. 2021, 11, 19345. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
An, J.; He, Q.; Xi, J.; Li, J.; Wang, G. Genome-Wide Identification of the UGT Gene Family in Poplar Populus euphratica and Functional Analysis of PeUGT110 Under Drought Stress. Forests 2025, 16, 1214. https://doi.org/10.3390/f16081214
An J, He Q, Xi J, Li J, Wang G. Genome-Wide Identification of the UGT Gene Family in Poplar Populus euphratica and Functional Analysis of PeUGT110 Under Drought Stress. Forests. 2025; 16(8):1214. https://doi.org/10.3390/f16081214
Chicago/Turabian StyleAn, Jilong, Qing He, Jinfeng Xi, Jing Li, and Gaini Wang. 2025. "Genome-Wide Identification of the UGT Gene Family in Poplar Populus euphratica and Functional Analysis of PeUGT110 Under Drought Stress" Forests 16, no. 8: 1214. https://doi.org/10.3390/f16081214
APA StyleAn, J., He, Q., Xi, J., Li, J., & Wang, G. (2025). Genome-Wide Identification of the UGT Gene Family in Poplar Populus euphratica and Functional Analysis of PeUGT110 Under Drought Stress. Forests, 16(8), 1214. https://doi.org/10.3390/f16081214





