Fungi Associated with Dying Buckthorn in North America
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation
2.2. Molecular Identification
2.3. Lifestyle Classification
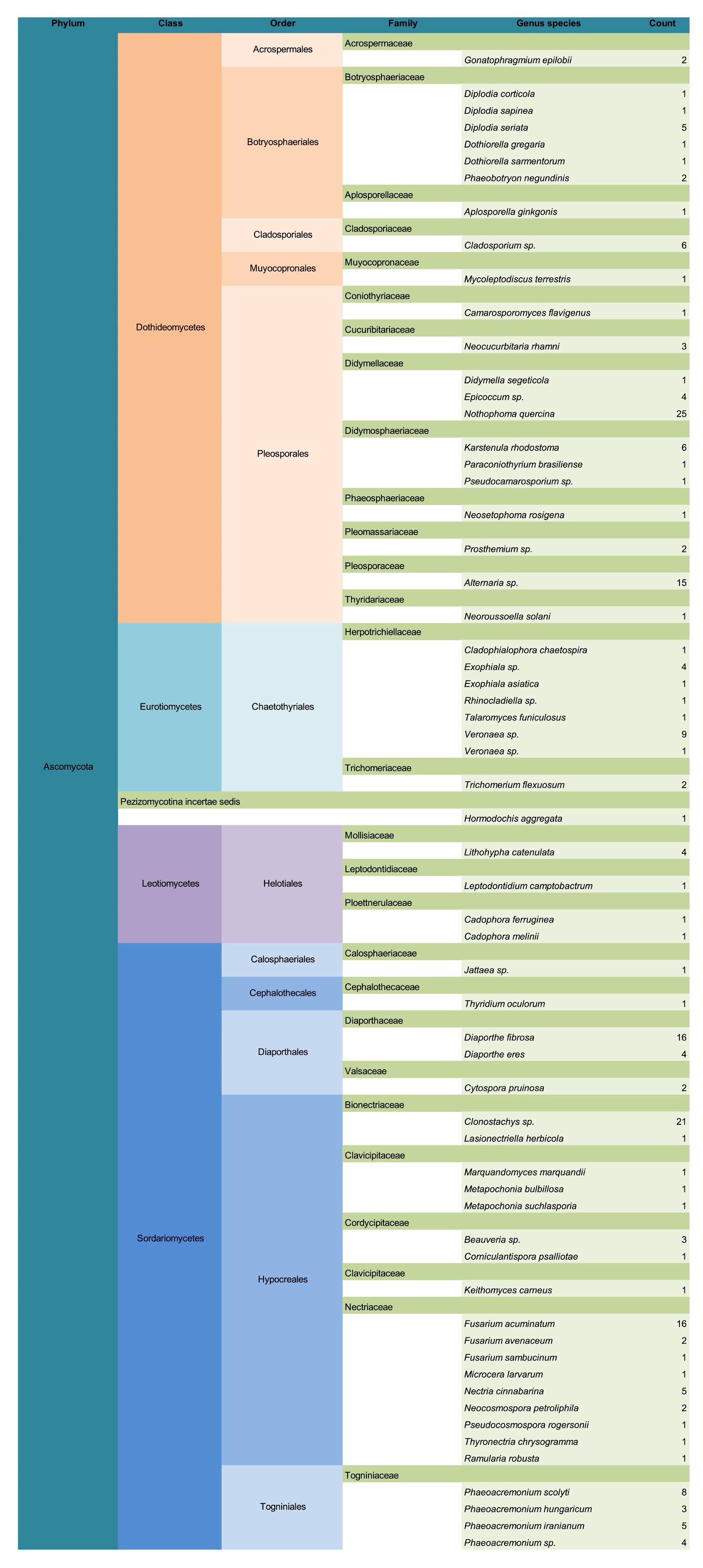
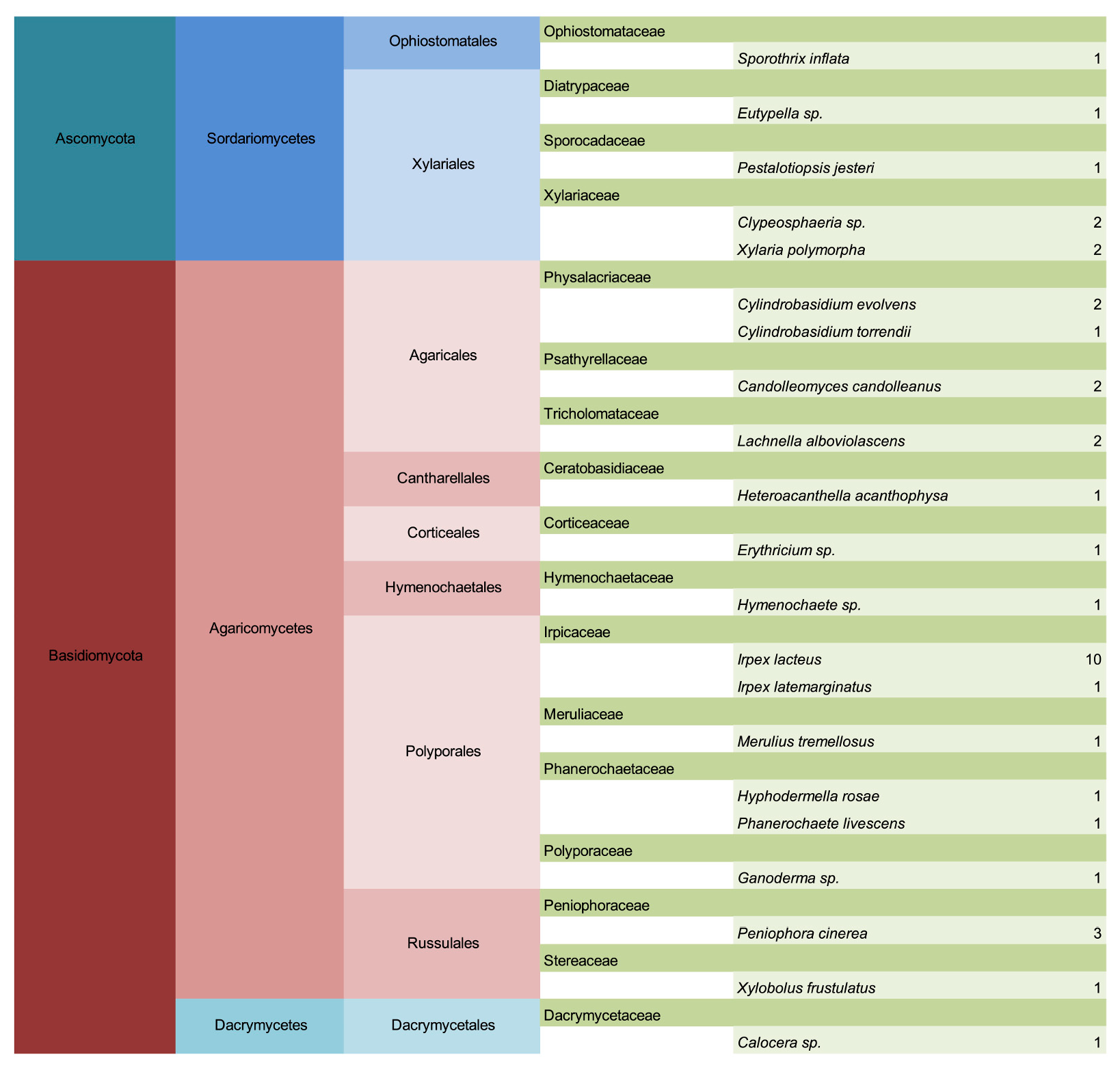
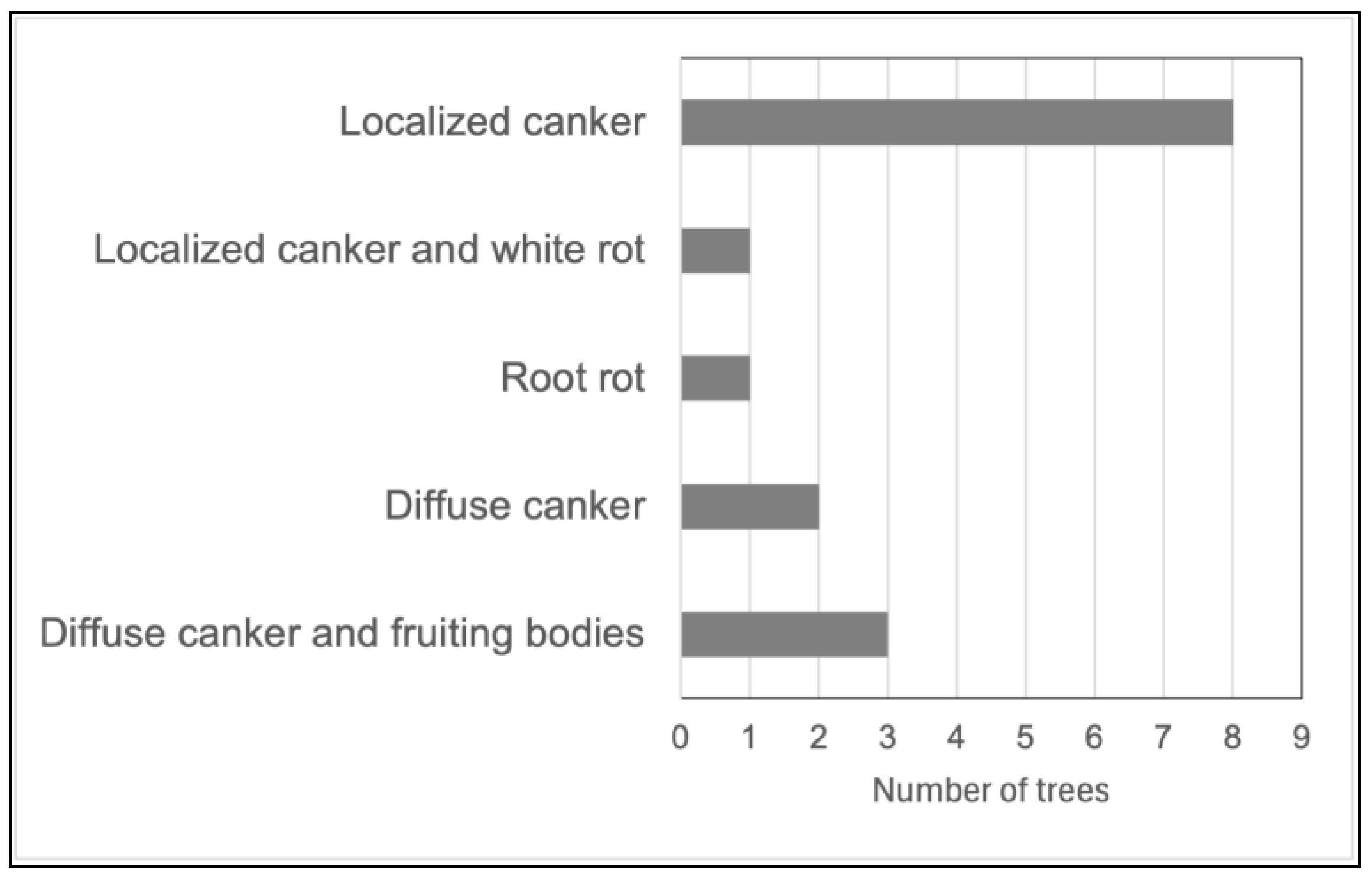
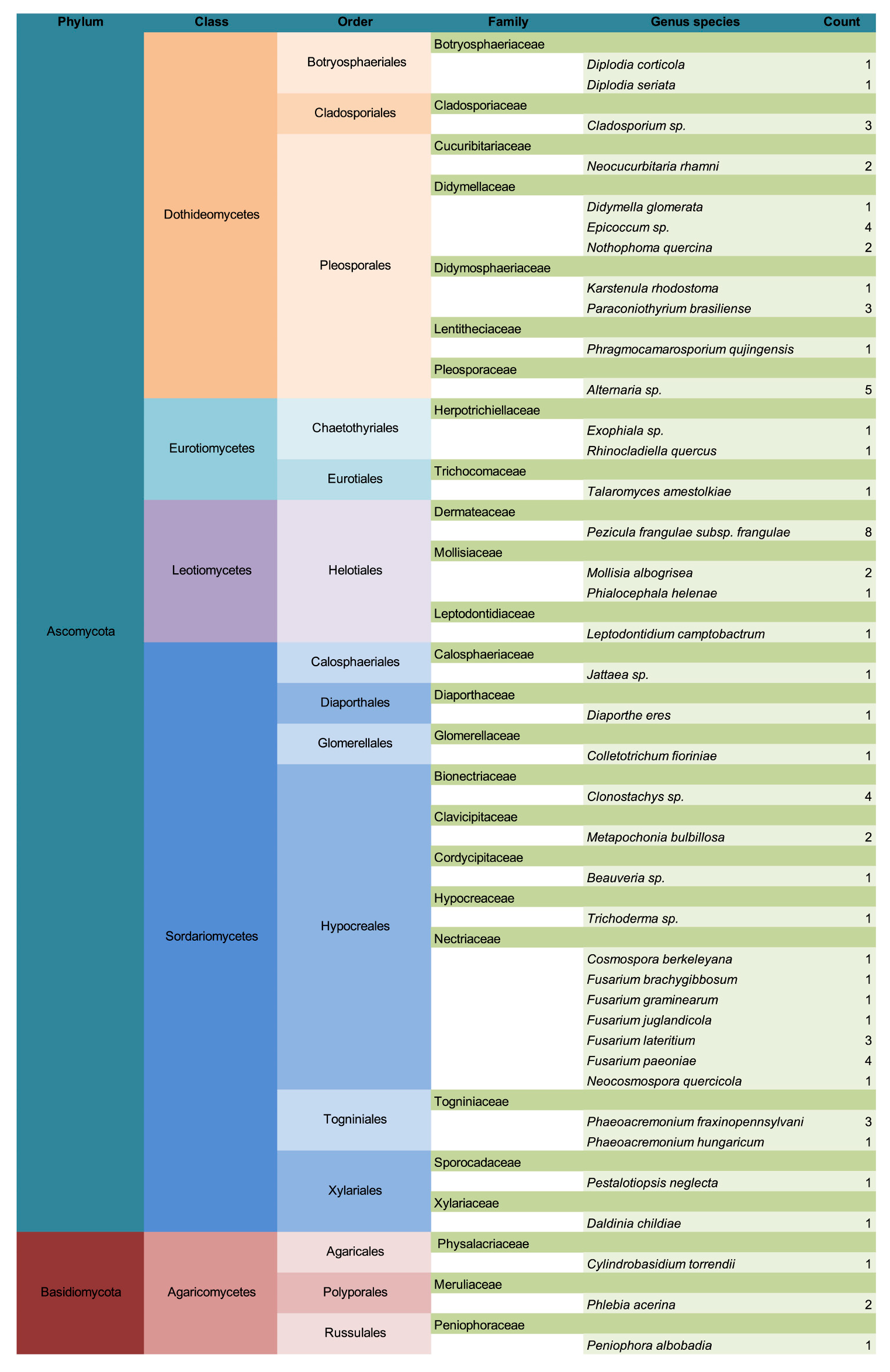
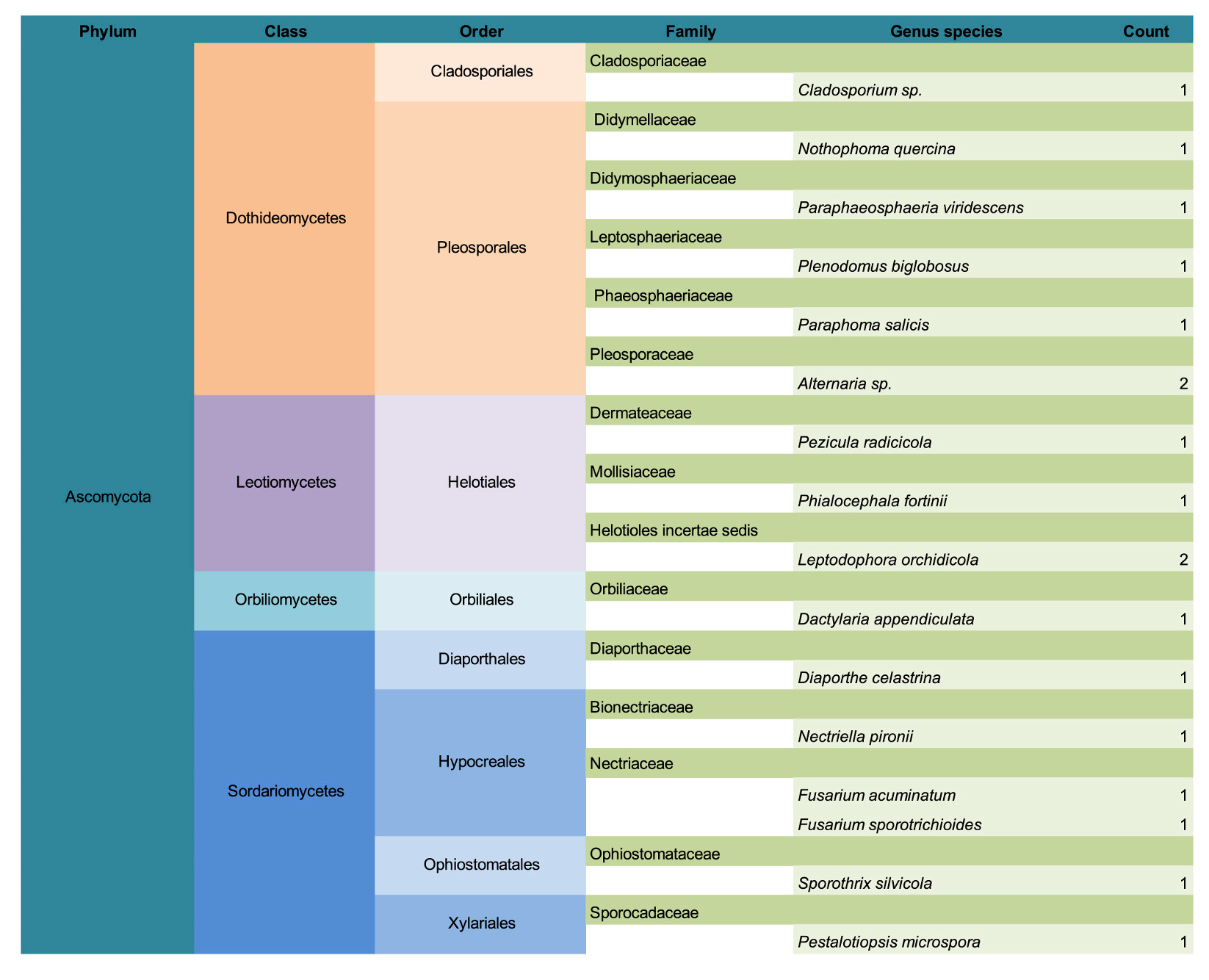
3. Results
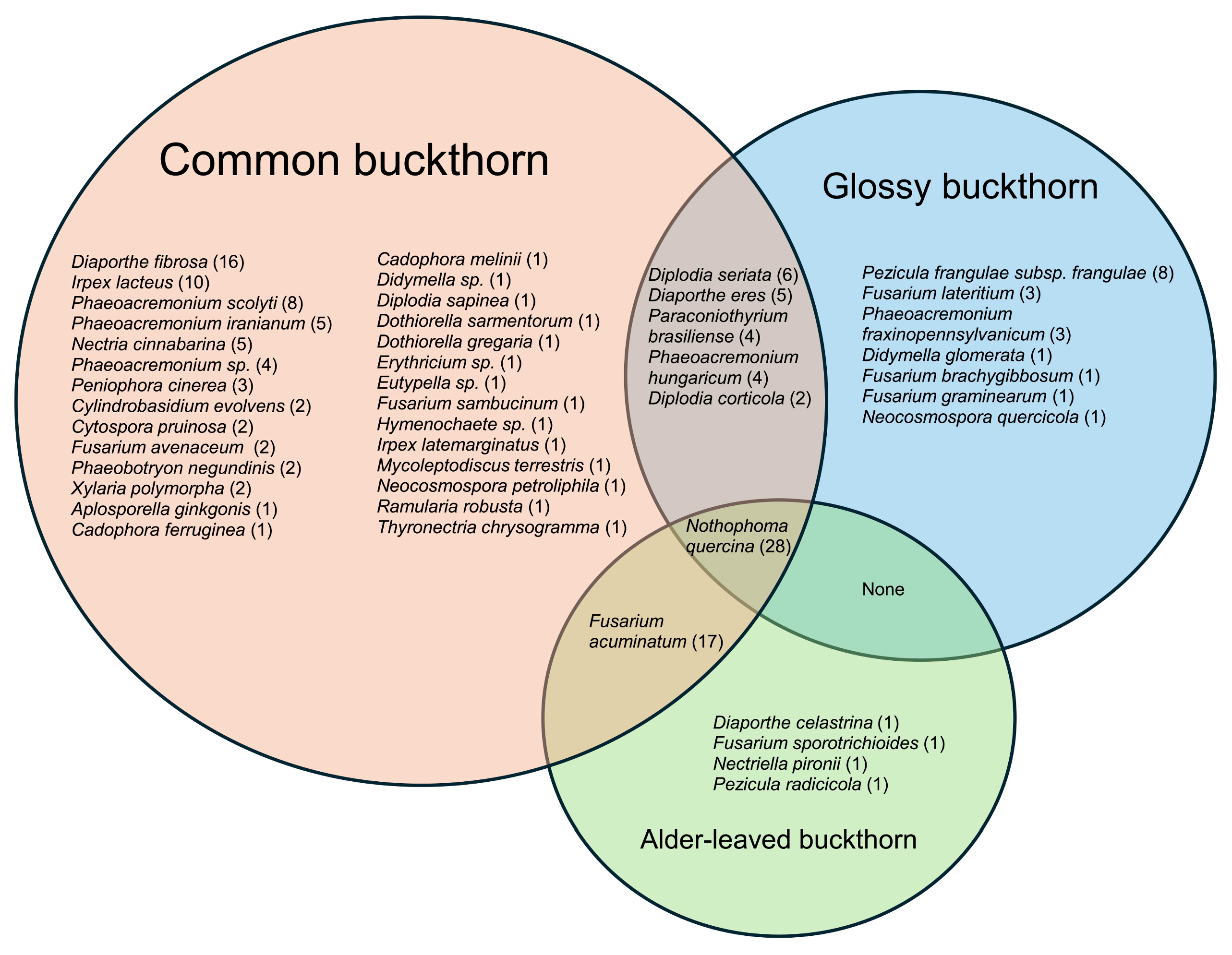
3.1. Common Buckhorn
3.2. Glossy Buckhorn
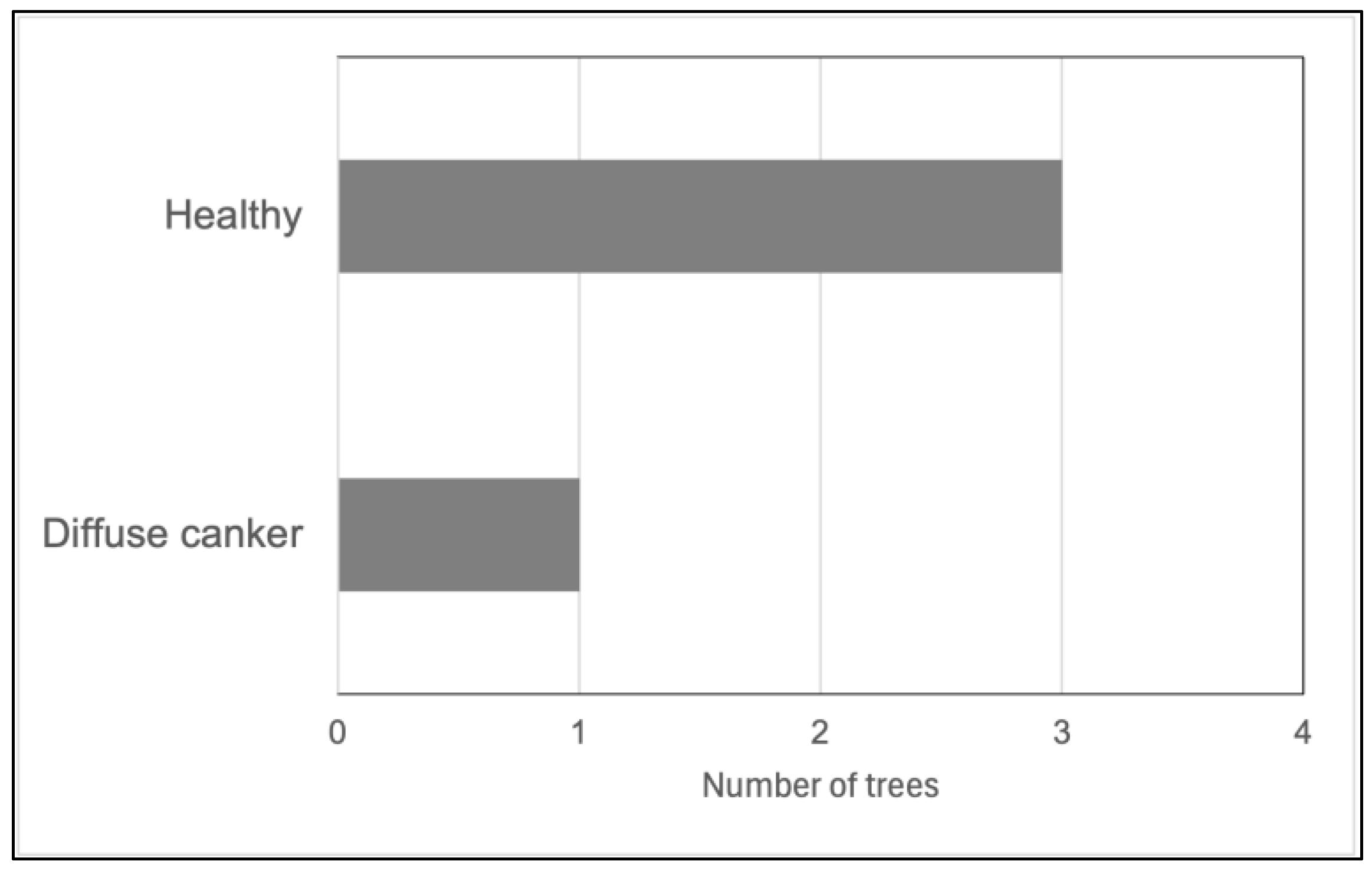
3.3. Alder-Leaved Buckhorn
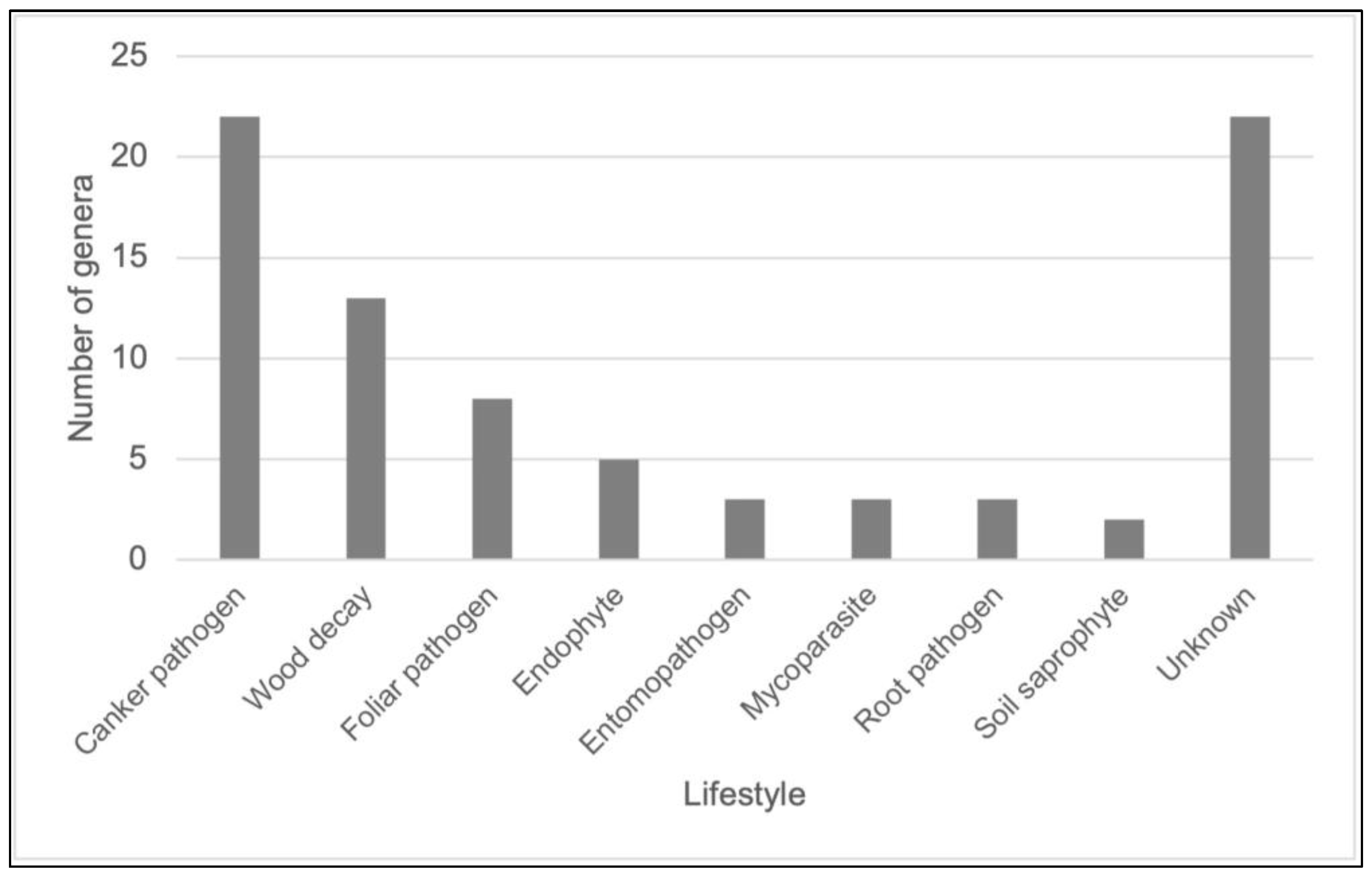
3.4. Lifestyle Distribution and Potential Pathogens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kurylo, J.; Endress, A.G. Rhamnus cathartica: Notes on its early history in North America. Northeast. Nat. 2012, 19, 601–610. [Google Scholar] [CrossRef]
- Apfelbaum, S.L.; Haney, A. Management of Degraded Oak Savanna Remnants in the Upper Midwest: Preliminary Results from Three Years of Study. In Proceedings of the Oak Woods Management Workshop; Ebinger, J., Ed.; Eastern Illinois University: Peoria, IL, USA, 1993; pp. 81–89. [Google Scholar]
- Alsum, E.M. Fifty years later: An Assessment of the Influence of Common Buckthorn (Rhamnus cathartica L.) and of Change in Overstory Vegetation in Several Floodplain Forests of the Lower Wisconsin State Riverway. Master’s Thesis, University of Wisconsin, Madison, WI, USA, 2003. [Google Scholar]
- Knight, K.S.; Kurylo, J.S.; Endress, A.G.; Stewart, J.R.; Reich, P.B. Ecology and ecosystem impacts of common buckthorn (Rhamnus cathartica): A review. Biol. Invasions 2007, 9, 925–937. [Google Scholar] [CrossRef]
- Roth, A.M.; Whitfeld, T.J.; Lodge, A.G.; Eisenhauer, N.; Frelich, L.E.; Reich, P.B. Invasive earthworms interact with abiotic conditions to influence the invasion of common buckthorn (Rhamnus cathartica). Oecologia 2015, 178, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Heimpel, G.E.; Frelich, L.E.; Landis, D.A.; Hopper, K.R.; Hoelmer, K.A.; Sezen, Z.; Asplen, M.K.; Wu, K. European buckthorn and Asian soybean aphid as components of an extensive invasional meltdown in North America. Biol. Invasions 2010, 12, 2913–2931. [Google Scholar] [CrossRef]
- Archibold, W.; Brooks, D.; Delanoy, L. An investigation of the invasive shrub European Buckthorn, Rhamnus cathartica L., near Saskatoon, Saskatchewan. Can. Field-Nat. 1997, 111, 617–621. [Google Scholar] [CrossRef]
- Croft, E.M. Non-Chemical European Buckthorn (Rhamnus cathartica) Removal Effectiveness and Costs Compared to Chemical Removal Methods. Master’s Thesis, University of Wisconsin-Stout, Menomonie, WI, USA, 2022. [Google Scholar]
- Schuster, M.J.; Bockenstedt, P.; Wragg, P.D.; Reich, P.B. Fosamine ammonium impacts on the targeted invasive shrub Rhamnus cathartica and non-target herbs. Invasive Plant Sci. Manag. 2020, 13, 210–215. [Google Scholar] [CrossRef]
- Gassmann, A.; Tosevski, I. Biological control of Rhamnus cathartica: Is it feasible? A review of work done in 2002–2012. J. Appl. Entomol. 2014, 138, 1–13. [Google Scholar] [CrossRef]
- Morin, L. Progress in biological control of weeds with plant pathogens. Annu. Rev. Phytopathol. 2020, 58, 201–223. [Google Scholar] [CrossRef]
- Pile Knapp, L.S.; Rebbeck, J.; Hutchinson, T.; Fraser, J.; Pinchot, C.C. Controlling an invasive tree with a native fungus: Inoculating Ailanthus altissima (Tree-of-Heaven) with Verticillium nonalfalfae in highly disturbed Appalachian forests of Ohio. J. For. 2022, 120, 558–574. [Google Scholar] [CrossRef]
- Au, R.C.F.; Tuchscherer, K. Efficacy of biological and chemical herbicides on non-native buckthorn during three seasonal periods. Nat. Areas J. 2014, 34, 92–98. [Google Scholar] [CrossRef]
- Dolinski, L.F.; Bal, T.L.; Webster, C.R.; Resh, S.C. Assessing the utility of a native pathogenic fungus as a biocontrol alternative to herbicide on invasive buckthorns in forests of upper Michigan. Nat. Areas J. 2024, 44, 57–64. [Google Scholar] [CrossRef]
- Mitchell, C.E.; Power, A.G. Release of invasive plants from fungal and viral pathogens. Nature 2003, 421, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Weerasuriya, N.M. Fungi Associated with Common Buckthorn (Rhamnus cathartica) in Southern Ontario. Master’s Thesis, University of Western Ontario, London, ON, Canada, 2017. [Google Scholar]
- Harrington, T.C. Cycloheximide sensitivity as a taxonomic character in Ceratocystis. Mycologia 1981, 73, 1123–1129. [Google Scholar] [CrossRef]
- Worrall, J.J. Media for selective isolation of hymenomycetes. Mycologia 1991, 83, 296–302. [Google Scholar] [CrossRef]
- Blanchette, R.A.; Held, B.W.; Hellmann, L.; Millman, L.; Büntgen, U. Arctic driftwood reveals unexpectedly rich fungal diversity. Fungal Ecol. 2016, 23, 58–65. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef]
- Hou, L.; Hernández-Restrepo, M.; Groenewald, J.Z.; Cai, L.; Crous, P.W. Citizen science project reveals high diversity in Didymellaceae (Pleosporales, Dothideomycetes). MycoKeys 2020, 65, 49–99. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Gassmann, A. Developing Biological Control of Buckthorns. In Proceedings of the Symposium on the Biology, Ecology and Management of Garlic Mustard (Alliaria petiolata) and European Buckthorn (Rhamnus cathartica); Skinner, L.C., Ed.; University of Minnesota: St Paul, MN, USA, 2005; pp. 55–57. [Google Scholar]
- Grady, K.C.; Heneghan, L.; Jabon, D. Reduced herbivory on exotic buckthorn (Rhamnus cathartica) compared to native tree species in an Illinois woodland. Bio. Notes 2012, 143, 1–6. [Google Scholar]
- Grubb, P.J.; Kollmann, J.; Lee, W.G. A garden experiment on susceptibility to rabbit-grazing, sapling growth rates, and age at first reproduction for eleven European woody species. Plant Biol. 1999, 1, 226–234. [Google Scholar] [CrossRef]
- Sweetman, H.L. Selection of woody plants as winter food by the cottontail rabbit. Ecology 1944, 25, 467–472. [Google Scholar] [CrossRef]
- Sweetman, H.L. Further studies of the winter feeding habits of cottontail rabbits. Ecology 1949, 30, 371–376. [Google Scholar] [CrossRef]
- Vanveldhuisen, M.V.; Ragsdale, D.W.; Skinner, L.C. Survey of Insect Fauna on Common Buckthorn, Rhamnus cathartica. In Proceedings of the Symposium on the Biology, Ecology and Management of Garlic Mustard (Alliaria petiolata) and European buckthorn (Rhamnus cathartica); USDA Forest Service: Morgantown, WV, USA, 2005; pp. 58–61. [Google Scholar]
- Knight, K.S. Factors that Influence Invasion Success of two Woody Invaders of Forest Understories. PhD Dissertation, University of Minnesota, Minneapolis, MN, USA, 2006. [Google Scholar]
- Flory, S.L.; Alba, C.; Clay, K.; Holt, R.D.; Goss, E.M. Emerging pathogens can suppress invaders and promote native species recovery. Biol. Invasions 2018, 20, 5–8. [Google Scholar] [CrossRef]
- Kelly, D.W.; Paterson, R.A.; Townsend, C.R.; Poulin, R.; Tompkins, D.M. Parasite spillback: A neglected concept in invasion ecology? Ecology 2009, 90, 2047–2056. [Google Scholar] [CrossRef]
- Stricker, K.B.; Harmon, P.F.; Goss, E.M.; Clay, K.; Flory, L.S. Emergence and accumulation of novel pathogens suppress an invasive species. Ecol. Lett. 2016, 19, 469–477. [Google Scholar] [CrossRef]
- Stengel, A.; Drijber, R.A.; Carr, E.; Egreja, T.; Hillman, E.; Krause, T.; Reese, S.; Herr, J.R. Rethinking the roles of pathogens and mutualists: Exploring the continuum of symbiosis in the context of microbial ecology and evolution. Phytobiomes J. 2022, 6, 108–117. [Google Scholar] [CrossRef]
- Lofgren, L.A.; LeBlanc, N.R.; Certano, A.K.; Nachtigall, J.; LaBine, K.M.; Riddle, J.; Broz, K.; Dong, Y.; Bethan, B.; Kafer, C.W.; et al. Fusarium graminearum: Pathogen or endophyte of North American grasses? New Phytol. 2018, 217, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.J.; Wood, A.R.; Den Breeÿen, A. Plant pathogens and biological control of weeds in South Africa: A review of projects and progress during the last decade. Afr. Entomol. Memoir 1999, 1, 129–137. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Franke, R.D.M.; Rajtar, N.N.; Blanchette, R.A. Fungi Associated with Dying Buckthorn in North America. Forests 2025, 16, 1148. https://doi.org/10.3390/f16071148
Franke RDM, Rajtar NN, Blanchette RA. Fungi Associated with Dying Buckthorn in North America. Forests. 2025; 16(7):1148. https://doi.org/10.3390/f16071148
Chicago/Turabian StyleFranke, Ryan D. M., Nickolas N. Rajtar, and Robert A. Blanchette. 2025. "Fungi Associated with Dying Buckthorn in North America" Forests 16, no. 7: 1148. https://doi.org/10.3390/f16071148
APA StyleFranke, R. D. M., Rajtar, N. N., & Blanchette, R. A. (2025). Fungi Associated with Dying Buckthorn in North America. Forests, 16(7), 1148. https://doi.org/10.3390/f16071148








