Phylogenetic Diversity in Forests: Insights into Evolutionary Patterns and Conservation Strategies
Abstract
1. Introduction
Literature Search and Methodology
2. Concepts and Metrics of Phylogenetic Diversity
2.1. Core Concepts
2.2. Quantitative Metrics and Formulas
| Metric | Formula/Basis | Description | Strengths | Limitations | References |
|---|---|---|---|---|---|
| PD (Faith’s Phylogenetic Diversity) | ∑ (branch lengths connecting all species in a sample) | Measures total evolutionary history represented in a community | Simple, widely adopted, scales with richness | Sensitive to species richness and tree topology | [18] |
| MPD (Mean Pairwise Distance) | Mean of all phylogenetic distances between species pairs | Reflects average relatedness across a community; emphasizes deep divergence | Detects phylogenetic clustering and overdispersion | Biased by richness; influenced by basal divergence | [59,60] |
| MNTD (Mean Nearest Taxon Distance) | Mean phylogenetic distance to each species’ closest relative | Captures tip-level evolutionary divergence across a community | Useful for identifying recent radiations | Sensitive to rare taxa and sample completeness | [18,61] |
| NTI (Nearest Taxon Index) | −(MNTDobs − MNTDrand)/ StDev(MNTDrand) | Measures degree of tip-level clustering relative to null model | Indicates recent evolutionary filtering | Depends strongly on null model and taxon resolution | [59] |
| NRI (Net Relatedness Index) | −(MPDobs − MPDrand)/StDev(MPDrand) | Quantifies overall community clustering against random expectations | Captures deep phylogenetic structure | Impacted by species pool and branch length distributions | [60,61] |
| PE (Phylogenetic Endemism) | PD with each taxon’s branch length weighted by the inverse of their range size | Measures geographically restricted evolutionary history | Highlights endemism hotspots and refugia | Requires detailed range and tree data | [62,63] |
| RPD (Relative Phylogenetic Diversity) | Observed PD/the PD expected from null model | Compares the observed PD to a null expectation | Indicates over- or under-dispersion | Interpretation depends on null model | [63] |
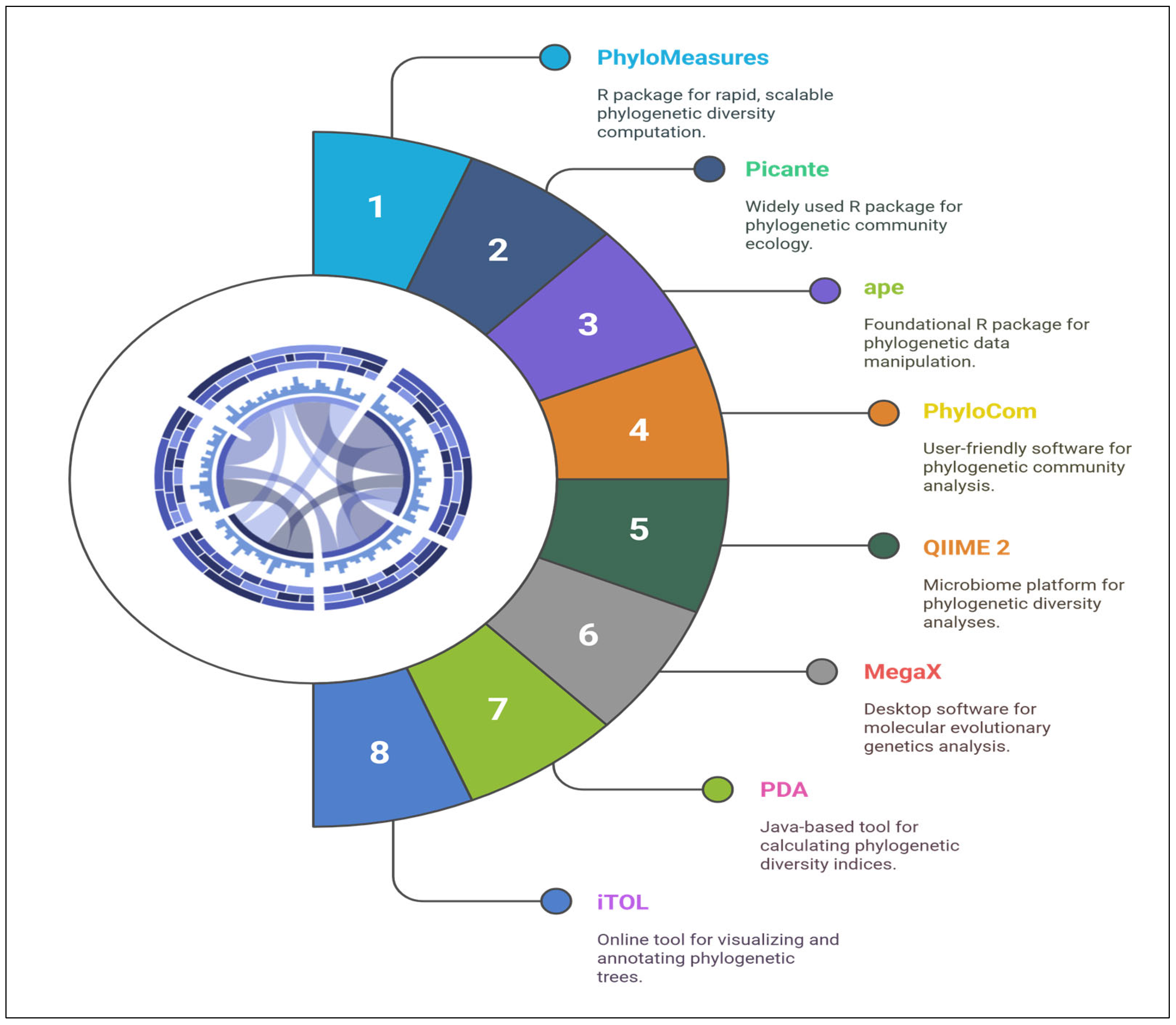
2.3. Tools and Software
3. Patterns of Phylogenetic Diversity in Forest Ecosystems
3.1. Global Patterns
3.2. Elevational and Latitudinal Gradients
3.3. Case Studies
4. Drivers of Phylogenetic Structure
4.1. Environmental Filters
4.2. Disturbance and Land-Use Change
4.3. Historical Biogeography and Speciation
5. Phylogenetic Diversity and Conservation Priorities
5.1. Integrating PD into Conservation Planning
5.2. Applications in Protected Area Design
5.3. Surrogacy and Ecosystem Services
| Study | Region | PD ↔ FD | PD ↔ Services | Conclusion |
|---|---|---|---|---|
| Rapacciuolo, et al. [126] | Americas | Strong congruence | Good proxy in terrestrial vertebrates | PD is a useful surrogate for conserving FD and services in large-scale prioritization |
| Mazel, et al. [52] | Global | Variable (−85% to +92%) | Unreliable overall | PD does not consistently capture FD; risk in assuming surrogate strength |
| Thompson, et al. [128] | Canada | Positive correlation | Linked to trophic function | Both PD and FD improve ecosystem function across trophic levels |
| Wang, et al. [33] | China (semi-arid grassland) | Positive relationship | Enhances multifunctionality | Both FD and PD are more predictive than species richness for ecosystem functions |
| Doxa, et al. [129] | Southern France | Weak congruence | Spatial mismatch | PD and FD vary independently; be cautious if using PD as a sole metric in landscape conservation |
| Qin, et al. [130] | China (restoration sites) | Consistent congruence | Functional restoration | PD tracks ecosystem recovery, making it useful for restoration assessment |
| Trindade-Filho, et al. [131] | Brazil | Good alignment (for birds) | Partial | Indicator groups can represent FD and PD, but this varied by taxon |
| Montaño-Centellas, et al. [132] | Global (raptors) | High congruence | Strong relevance | PD correlates well with functional traits and conservation priority |
6. Emerging Trends and Methodological Advances
6.1. Genomic and Phylogenomic Advances
6.2. Remote Sensing and Phylogenetics
6.3. Trait–Phylogeny Integration
7. Future Directions
7.1. Data Gaps and Standardization
7.2. Regional Focus and Collaboration
7.3. Science–Policy Interface
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wudu, K.; Abegaz, A.; Ayele, L.; Ybabe, M. The impacts of climate change on biodiversity loss and its remedial measures using nature based conservation approach: A global perspective. Biodivers. Conserv. 2023, 32, 3681–3701. [Google Scholar] [CrossRef]
- Frei, K.; E-Vojtkó, A.; Tölgyesi, C.; Vojtkó, A.; Farkas, T.; Erdős, L.; Li, G.; Lőrincz, Á.; Bátori, Z. Topographic complexity drives trait composition as well as functional and phylogenetic diversity of understory plant communities in microrefugia: New insights for conservation. For. Ecosyst. 2025, 12, 100278. [Google Scholar] [CrossRef]
- Karger, D.N.; Kessler, M.; Lehnert, M.; Jetz, W. Limited protection and ongoing loss of tropical cloud forest biodiversity and ecosystems worldwide. Nat. Ecol. Evol. 2021, 5, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Aerts, R.; Honnay, O. Forest restoration, biodiversity and ecosystem functioning. BMC Ecol. 2011, 11, 29. [Google Scholar] [CrossRef]
- Kumar, R.; Kumar, A.; Saikia, P. Deforestation and forests degradation impacts on the environment. In Environmental Degradation: Challenges and Strategies for Mitigation; Springer: Cham, Switzerland, 2022; pp. 19–46. [Google Scholar]
- Maurya, M.J.; Vivek, M. Deforestation: Causes, Consequences and Possible Solutions. Ideal. J. Adv. Res. Progress. Spectr. (IJARPS) 2025, 4, 70–76. [Google Scholar]
- Stephenson, P.J.; Londoño-Murcia, M.C.; Borges, P.A.V.; Claassens, L.; Frisch-Nwakanma, H.; Ling, N.; McMullan-Fisher, S.; Meeuwig, J.J.; Unter, K.M.M.; Walls, J.L. Measuring the impact of conservation: The growing importance of monitoring fauna, flora and funga. Diversity 2022, 14, 824. [Google Scholar] [CrossRef]
- Baker, M.; Yesilonis, I.; Templeton, L.; Shobe, B.; Bos, J.; Sonti, N.F.; Lautar, K. Distributed urban forest patch sampling detects edge effects and woodland condition for monitoring and management. Ecosphere 2025, 16, e70236. [Google Scholar] [CrossRef]
- Norris, K.; Harper, N. Extinction processes in hot spots of avian biodiversity and the targeting of pre–emptive conservation action. Proc. R. Soc. Lond. Ser. Biol. Sci. 2004, 271, 123–130. [Google Scholar] [CrossRef]
- Winfree, R.; Fox, W.J.; Williams, N.M.; Reilly, J.R.; Cariveau, D.P. Abundance of common species, not species richness, drives delivery of a real-world ecosystem service. Ecol. Lett. 2015, 18, 626–635. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Colwell, R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001, 4, 379–391. [Google Scholar] [CrossRef]
- Purvis, A.; Hector, A. Getting the measure of biodiversity. Nature 2000, 405, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, S.A.; Hovick, S.M.; Dibble, C.J.; Rasmussen, N.L.; Van Allen, B.G.; Maitner, B.S.; Ahern, J.R.; Bell-Dereske, L.P.; Roy, C.L.; Meza-Lopez, M.; et al. Does phylogeny matter? Assessing the impact of phylogenetic information in ecological meta-analysis. Ecol. Lett. 2012, 15, 627–636. [Google Scholar] [CrossRef] [PubMed]
- El-Barougy, R.F.; Bersier, L.-F.; Gray, S.M.; El-Keblawy, A.; Galal, T.; Ullah, F.; Elgamal, I.A.; Dakhil, M.A. Shaping beta diversity in arid landscape through native plant species contributions: Synergy of climate, soil, and species traits. Front. Plant Sci. 2025, 16, 1521596. [Google Scholar] [CrossRef] [PubMed]
- Zaman, W.; Ayaz, A.; Park, S. Integrating morphological and molecular data in plant taxonomy. Pak. J. Bot. 2025, 57, 4. [Google Scholar] [CrossRef]
- Anderegg, L.D.L.; Griffith, D.M.; Cavender-Bares, J.; Riley, W.J.; Berry, J.A.; Dawson, T.E.; Still, C.J. Representing plant diversity in land models: An evolutionary approach to make “Functional Types” more functional. Glob. Change Biol. 2022, 28, 2541–2554. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Schneider, F.D.; Santos, M.J.; Armstrong, A.; Carnaval, A.; Dahlin, K.M.; Fatoyinbo, L.; Hurtt, G.C.; Schimel, D.; Townsend, P.A. Integrating remote sensing with ecology and evolution to advance biodiversity conservation. Nat. Ecol. Evol. 2022, 6, 506–519. [Google Scholar] [CrossRef]
- De Azevedo, B.P.; Rother, D.C. Assessing phylogenetic diversity metrics in terrestrial ecological restoration: Global trends and gaps. Restor. Ecol. 2025, 33, e14292. [Google Scholar] [CrossRef]
- Lv, T.; Wang, N.; Xie, L.; Chen, S.; Zhao, R.; Feng, Y.; Li, Y.; Ding, H.; Fang, Y. Environmental heterogeneity affecting community assembly patterns and phylogenetic diversity of three forest communities at Mt. Huangshan, China. Forests 2022, 13, 133. [Google Scholar] [CrossRef]
- Roy, A.; Tripathi, S.K.; Basu, S.K. Formulating diversity vector for ecosystem comparison. Ecol. Model. 2004, 179, 499–513. [Google Scholar] [CrossRef]
- Saqib, S.; Liu, Y.; Ye, J.; Omollo, W.O.; Lu, L.; Hu, H.; Zhang, Q.; Liu, B.; Ahmad, M.; Shinwari, Z.K. Identifying conservation priority areas using spatial phylogenetic approaches in west Himalaya. Pak. J. Bot. 2024, 56, 1029–1039. [Google Scholar] [CrossRef]
- Shipley, B.R.; McGuire, J.L. Interpreting and integrating multiple endemism metrics to identify hotspots for conservation priorities. Biol. Conserv. 2022, 265, 109403. [Google Scholar] [CrossRef]
- Santo-Silva, E.E.; Benchimol, M.; Peres, C.A. Phylogenetic homogenization of Amazonian tree assemblages in forest islands after 26 years of isolation. Appl. Veg. Sci. 2021, 24, e12601. [Google Scholar] [CrossRef]
- Mori, A.S.; Lertzman, K.P.; Gustafsson, L. Biodiversity and ecosystem services in forest ecosystems: A research agenda for applied forest ecology. J. Appl. Ecol. 2017, 54, 12–27. [Google Scholar] [CrossRef]
- Erickson, D.L.; Jones, F.A.; Swenson, N.G.; Pei, N.; Bourg, N.A.; Chen, W.; Davies, S.J.; Ge, X.-j.; Hao, Z.; Howe, R.W. Comparative evolutionary diversity and phylogenetic structure across multiple forest dynamics plots: A mega-phylogeny approach. Front. Genet. 2014, 5, 358. [Google Scholar] [CrossRef]
- Kylafis, G.; Loreau, M. Niche construction in the light of niche theory. Ecol. Lett. 2011, 14, 82–90. [Google Scholar] [CrossRef]
- González-Caro, S.; Umaña, M.N.; Álvarez, E.; Stevenson, P.R.; Swenson, N.G. Phylogenetic alpha and beta diversity in tropical tree assemblages along regional-scale environmental gradients in northwest South America. J. Plant Ecol. 2014, 7, 145–153. [Google Scholar] [CrossRef]
- Sirami, C.; Caplat, P.; Popy, S.; Clamens, A.; Arlettaz, R.; Jiguet, F.; Brotons, L.; Martin, J.-L. Impacts of global change on species distributions: Obstacles and solutions to integrate climate and land use. Glob. Ecol. Biogeogr. 2017, 26, 385–394. [Google Scholar] [CrossRef]
- McNamara, J.M.; Barta, Z.; Klaassen, M.; Bauer, S. Cues and the optimal timing of activities under environmental changes. Ecol. Lett. 2011, 14, 1183–1190. [Google Scholar] [CrossRef]
- Feng, G.; Svenning, J.-C.; Mi, X.; Jia, Q.; Rao, M.; Ren, H.; Bebber, D.P.; Ma, K. Anthropogenic disturbance shapes phylogenetic and functional tree community structure in a subtropical forest. For. Ecol. Manag. 2014, 313, 188–198. [Google Scholar] [CrossRef]
- Culmsee, H.; Leuschner, C. Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 2013, 40, 1997–2010. [Google Scholar] [CrossRef]
- Jones, D.A. Phylogenetic Regionalization of the World’s Turtles Reveals Concentrations of Neo and Paleo Endemism. Master’s Thesis, San Diego State University, San Diego, CA, USA, 2024. [Google Scholar]
- Wang, Q.; Huang, J.; Zang, R.; Li, Z.; El-Kassaby, Y.A. Centres of neo-and paleo-endemism for Chinese woody flora and their environmental features. Biol. Conserv. 2022, 276, 109817. [Google Scholar] [CrossRef]
- Isaac, N.J.B.; Pearse, W.D. The Use of EDGE (Evolutionary Distinct Globally Endangered) and EDGE-Like Metrics to Evaluate Taxa for Conservation. In Phylogenetic Diversity: Applications and Challenges in Biodiversity Science; Scherson, R.A., Faith, D.P., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 27–39. [Google Scholar]
- Wang, T.; Zuo, Y.; Manda, T.; Hwarari, D.; Yang, L. Harnessing Artificial Intelligence, Machine Learning and Deep Learning for Sustainable Forestry Management and Conservation: Transformative Potential and Future Perspectives. Plants 2025, 14, 998. [Google Scholar] [CrossRef] [PubMed]
- Tolisano, J. Artists as the New Naturalists: A Response and Expansion to Jacobson et al. Conserv. Biol. 2007, 21, 1135–1136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guardiola, M.; Pino, J.; Rodà, F. Patch history and spatial scale modulate local plant extinction and extinction debt in habitat patches. Divers. Distrib. 2013, 19, 825–833. [Google Scholar] [CrossRef]
- Asad, F.; Adil, M.; Zhu, H.; Büntgen, U.; Hamayun, M.; Alrefaei, A.F.; Ali, S. A tree ring-based spring temperature reconstruction for the Hindu Kush region in northern Pakistan. Trees For. People 2024, 16, 100541. [Google Scholar] [CrossRef]
- Chisika, S.N.; Yeom, C. The implication of the changing forest management paradigms in formulating forestry policies in Kenya. Forestist 2024, 74, 278–288. [Google Scholar] [CrossRef]
- Tuanmu, M.-N.; Jetz, W. A global, remote sensing-based characterization of terrestrial habitat heterogeneity for biodiversity and ecosystem modelling. Glob. Ecol. Biogeogr. 2015, 24, 1329–1339. [Google Scholar] [CrossRef]
- Mace, G.M.; Norris, K.; Fitter, A.H. Biodiversity and ecosystem services: A multilayered relationship. Trends Ecol. Evol. 2012, 27, 19–26. [Google Scholar] [CrossRef]
- Faith, D.P. Biodiversity and evolutionary history: Useful extensions of the PD phylogenetic diversity assessment framework. Ann. N. Y. Acad. Sci. 2013, 1289, 69–89. [Google Scholar] [CrossRef]
- Sajjad, S.; Islam, M.; Muhammad, K.; Ghafoor, S.-u.; Ullah, I.; Khan, A.; Siraj, M.; Alrefaei, A.F.; Shah, J.A.; Ali, S. Comprehensive Evaluation of Cryptic Juglans Genotypes: Insight from Molecular Markers and Phylogenetic Analysis. Genes 2024, 15, 1417. [Google Scholar] [CrossRef]
- Worth, J.R.P.; Kikuchi, S.; Kanetani, S.; Takahashi, D.; Aizawa, M.; Marchuk, E.A.; Choi, H.J.; Polezhaeva, M.A.; Sheiko, V.V.; Ueno, S. Chloroplast genome-based genetic resources via genome skimming for the subalpine forests of Japan and adjacent regions. Ecol. Evol. 2024, 14, e11584. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, D.S.; Cadotte, M.W.; MacDonald, A.A.M.; Marushia, R.G.; Mirotchnick, N. Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 2012, 15, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Vellend, M.; Cornwell, W.K.; Magnuson-Ford, K.; Mooers, A.Ø. Measuring phylogenetic biodiversity. Biol. Divers. Front. Meas. Assess. 2011, 14, 194–207. [Google Scholar]
- Faith, D.P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61, 1–10. [Google Scholar] [CrossRef]
- Miller, J.T.; Jolley-Rogers, G.; Mishler, B.D.; Thornhill, A.H. Phylogenetic diversity is a better measure of biodiversity than taxon counting. J. Syst. Evol. 2018, 56, 663–667. [Google Scholar] [CrossRef]
- Sandel, B.; Weigelt, P.; Kreft, H.; Keppel, G.; van der Sande, M.T.; Levin, S.; Smith, S.; Craven, D.; Knight, T.M. Current climate, isolation and history drive global patterns of tree phylogenetic endemism. Glob. Ecol. Biogeogr. 2020, 29, 4–15. [Google Scholar] [CrossRef]
- Mishler, B.D.; Guralnick, R.; Soltis, P.S.; Smith, S.A.; Soltis, D.E.; Barve, N.; Allen, J.M.; Laffan, S.W. Spatial phylogenetics of the North American flora. J. Syst. Evol. 2020, 58, 393–405. [Google Scholar] [CrossRef]
- Swenson, N.G. The role of evolutionary processes in producing biodiversity patterns, and the interrelationships between taxonomic, functional and phylogenetic biodiversity. Am. J. Bot. 2011, 98, 472–480. [Google Scholar] [CrossRef]
- Mazel, F.; Pennell, M.W.; Cadotte, M.W.; Diaz, S.; Dalla Riva, G.V.; Grenyer, R.; Leprieur, F.; Mooers, A.O.; Mouillot, D.; Tucker, C.M. Prioritizing phylogenetic diversity captures functional diversity unreliably. Nat. Commun. 2018, 9, 2888. [Google Scholar] [CrossRef]
- Faith, D.P. Phylogenetic diversity and conservation evaluation: Perspectives on multiple values, indices, and scales of application. In Phylogenetic Diversity: Applications and Challenges in Biodiversity Science; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–26. [Google Scholar]
- Islam, T.; Rasray, B.A.; Wani, S.A.; Dar, F.A.; Nawchoo, I.A.; Khuroo, A.A. Elevational Patterns of Plant Taxonomic, Functional and Phylogenetic Diversity in the Himalaya. In Functional and Phylogenetic Diversity in the Himalaya; Springer Nature: Singapore, 2025. [Google Scholar]
- Roeder, M.; McLeish, M.; Beckschaefer, P.; de Blecourt, M.; Paudel, E.; Harrison, R.D.; Slik, F. Phylogenetic clustering increases with succession for lianas in a Chinese tropical montane rain forest. Ecography 2015, 38, 832–841. [Google Scholar] [CrossRef]
- Swenson, N.G. Phylogenetic resolution and quantifying the phylogenetic diversity and dispersion of communities. PLoS ONE 2009, 4, e4390. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-X.; Zhang, M.-L. Spatial patterns of species diversity and phylogenetic structure of plant communities in the Tianshan Mountains, arid Central Asia. Front. Plant Sci. 2017, 8, 2134. [Google Scholar] [CrossRef] [PubMed]
- Uchida, K.; Hiraiwa, M.K.; Cadotte, M.W. Non-random loss of phylogenetically distinct rare species degrades phylogenetic diversity in semi-natural grasslands. J. Appl. Ecol. 2019, 56, 1419–1428. [Google Scholar] [CrossRef]
- Yang, J.; Wu, F. Impacts of the attributes of patches and the surrounding matrix on the phylogenetic diversity of tree species in urban remnant forests. Land Degrad. Dev. 2024, 35, 3007–3017. [Google Scholar] [CrossRef]
- Santo-Silva, E.E.; Santos, B.A.; Arroyo-Rodríguez, V.; Melo, F.P.L.; Faria, D.; Cazetta, E.; Mariano-Neto, E.; Hernández-Ruedas, M.A.; Tabarelli, M. Phylogenetic dimension of tree communities reveals high conservation value of disturbed tropical rain forests. Divers. Distrib. 2018, 24, 776–790. [Google Scholar] [CrossRef]
- Liu, W.; Ling, N.; Guo, J.; Ruan, Y.; Zhu, C.; Shen, Q.; Guo, S. Legacy effects of 8-year nitrogen inputs on bacterial assemblage in wheat rhizosphere. Biol. Fert. Soils 2020, 56, 583–596. [Google Scholar] [CrossRef]
- Rosauer, D.A.N.; Laffan, S.W.; Crisp, M.D.; Donnellan, S.C.; Cook, L.G. Phylogenetic endemism: A new approach for identifying geographical concentrations of evolutionary history. Mol. Ecol. 2009, 18, 4061–4072. [Google Scholar] [CrossRef]
- Mishler, B.D.; Knerr, N.; González-Orozco, C.E.; Thornhill, A.H.; Laffan, S.W.; Miller, J.T. Phylogenetic measures of biodiversity and neo-and paleo-endemism in Australian Acacia. Nat. Commun. 2014, 5, 4473. [Google Scholar] [CrossRef]
- Nosrati, H.; Mirtajadini, S.M.; Jahanshahi, M. Phylogenetic structure of plant community, and its relationship with environmental components. Authorea 2023, 29657431. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Gastauer, M.; Meira-Neto, J.A.A. Avoiding inaccuracies in tree calibration and phylogenetic community analysis using Phylocom 4.2. Ecol. Inf. 2013, 15, 85–90. [Google Scholar] [CrossRef]
- Webb, C.O.; Ackerly, D.D.; Kembel, S.W. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 2008, 24, 2098–2100. [Google Scholar] [CrossRef] [PubMed]
- Ben-Said, M. Spatial point-pattern analysis as a powerful tool in identifying pattern-process relationships in plant ecology: An updated review. Ecol. Process. 2021, 10, 1–23. [Google Scholar] [CrossRef]
- Laffan, S.W.; Lubarsky, E.; Rosauer, D.F. Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography 2010, 33, 643–647. [Google Scholar] [CrossRef]
- Cai, L.; Kreft, H.; Taylor, A.; Schrader, J.; Dawson, W.; Essl, F.; van Kleunen, M.; Pergl, J.; Pyšek, P.; Winter, M. Phylogenetic endemism of the world’s seed plants. BioRxiv 2022. [Google Scholar] [CrossRef]
- Lemmon, E.M.; Lemmon, A.R. High-throughput genomic data in systematics and phylogenetics. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 99–121. [Google Scholar] [CrossRef]
- Morlon, H.; Schwilk, D.W.; Bryant, J.A.; Marquet, P.A.; Rebelo, A.G.; Tauss, C.; Bohannan, B.J.M.; Green, J.L. Spatial patterns of phylogenetic diversity. Ecol. Lett. 2011, 14, 141–149. [Google Scholar] [CrossRef]
- Swenson, N.; Erickson, D.; Mi, X.; Bourg, N.; Forero-Montaña, J.; Ge, X.; Howe, R.; Lake, J.; Liu, X.; Pei, N.; et al. Phylogenetic and functional alpha and beta diversity in temperate and tropical tree communities. Ecology 2012, 93, S112–S125. [Google Scholar] [CrossRef]
- Tietje, M.; Antonelli, A.; Forest, F.; Govaerts, R.; Smith, S.A.; Sun, M.; Baker, W.J.; Eiserhardt, W.L. Global hotspots of plant phylogenetic diversity. New Phytol. 2023, 240, 1636–1646. [Google Scholar] [CrossRef]
- Cardoso, D.; Särkinen, T.; Alexander, S.; Amorim, A.M.; Bittrich, V.; Celis, M.; Daly, D.C.; Fiaschi, P.; Funk, V.A.; Giacomin, L.L. Amazon plant diversity revealed by a taxonomically verified species list. Proc. Natl. Acad. Sci. USA 2017, 114, 10695–10700. [Google Scholar] [CrossRef]
- Moraes, R.M.; Correa, S.B.; Doria, C.R.C.; Duponchelle, F.; Miranda, G.; Montoya, M.; Phillips, O.; Salinas, N.; Silman, M.; Ulloa Ulloa, C. Amazonian ecosystems and their ecological functions. In Amazon Assessment Report 2021; United Nations Sustainable Development Solutions Network: New York, NY, USA, 2021; Volume 1. [Google Scholar]
- Le, H.; Mao, J.; Cavender-Bares, J.; Pinto-Ledezma, J.N.; Deng, Y.; Zhao, C.; Xiong, G.; Xu, W.; Xie, Z. Non-native plants tend to be phylogenetically distant but functionally similar to native plants under intense disturbance at the Three Gorges Reservoir Area. New Phytol. 2024, 244, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Namkhan, M.; Gale, G.A.; Savini, T.; Tantipisanuh, N. Loss and vulnerability of lowland forests in mainland Southeast Asia. Conserv. Biol. 2021, 35, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Kuhnhäuser, B.G.; Bates, C.D.; Dransfield, J.; Geri, C.; Henderson, A.; Julia, S.; Lim, J.Y.; Morley, R.J.; Rustiami, H.; Schley, R.J. Island geography drives evolution of rattan palms in tropical Asian rainforests. Science 2025, 387, 1204–1209. [Google Scholar] [CrossRef] [PubMed]
- Dani, R.S.; Divakar, P.K.; Baniya, C.B. Diversity and composition of plants species along elevational gradient: Research trends. Biodivers. Conserv. 2023, 32, 2961–2980. [Google Scholar] [CrossRef]
- Zu, K.; Zhang, C.; Chen, F.; Zhang, Z.; Ahmad, S.; Nabi, G. Latitudinal gradients of angiosperm plant diversity and phylogenetic structure in China’s nature reserves. Glob. Ecol. Conserv. 2023, 42, e02403. [Google Scholar] [CrossRef]
- Fuentes-Lillo, E.; Lembrechts, J.J.; Barros, A.; Aschero, V.; Bustamante, R.O.; Cavieres, L.A.; Clavel, J.; Herrera, I.; Jiménez, A.; Tecco, P. Going up the Andes: Patterns and drivers of non-native plant invasions across latitudinal and elevational gradients. Biodivers. Conserv. 2023, 32, 4199–4219. [Google Scholar] [CrossRef]
- Liu, Y.; Saqib, S.; Lu, L.; Lai, Y.; Hu, H.; Peng, D.; Zaman, W.; Zhao, L.; Liu, B.; Wang, Q. Phylogenetic regionalization of the Pan-Himalayan vascular flora. J. Syst. Evol. 2025, 63, 12–24. [Google Scholar] [CrossRef]
- Shi, N.; Wang, C.; Wang, J.; Wu, N.; Naudiyal, N.; Zhang, L.; Wang, L.; Sun, J.; Du, W.; Wei, Y. Biogeographic patterns and richness of the meconopsis species and their influence factors across the pan-Himalaya and adjacent regions. Diversity 2022, 14, 661. [Google Scholar] [CrossRef]
- Liu, J.-Q.; Li, J.-L.; Lai, Y.-J. Plant diversity and ecology on the Qinghai–Tibet Plateau. J. Syst. Evol. 2021, 59, 1139. [Google Scholar] [CrossRef]
- Huang, H.; Liu, H.; Ma, W.; Qin, L.; Chen, L.; Guo, H.; Xu, H.; Li, J.; Yang, C.; Hu, H.; et al. High-throughput MALDI-MSI metabolite analysis of plant tissue microarrays. Plant Biotechnol. J. 2023, 21, 2574–2584. [Google Scholar] [CrossRef]
- Khan, S.W.; Qamar Abbas, A.H.; Hussain, A.; Ali, H.; Ali, S. Assessment of Floristic Diversity in the Desert Ecosystem of Central Karakoram National Park (Karakoram Range) Gilgit-Baltistan, Pakistan. Pak. J. Bot. 2021, 53, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Hao, Z.; Zhang, J. Phylogenetic structure and phylogenetic diversity of angiosperm assemblages in forests along an elevational gradient in Changbaishan, China. J. Plant Ecol. 2014, 7, 154–165. [Google Scholar] [CrossRef]
- Li, R.; Qian, L.; Sun, H. Current progress and future prospects in phylofloristics. Plant Divers. 2018, 40, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Paz, A.; Brown, J.L.; Cordeiro, C.L.O.; Aguirre-Santoro, J.; Assis, C.; Amaro, R.C.; Raposo do Amaral, F.; Bochorny, T.; Bacci, L.F.; Caddah, M.K. Environmental correlates of taxonomic and phylogenetic diversity in the Atlantic Forest. J. Biogeogr. 2021, 48, 1377–1391. [Google Scholar] [CrossRef]
- Potter, K.M.; Woodall, C.W. Trends over time in tree and seedling phylogenetic diversity indicate regional differences in forest biodiversity change. Ecol. Appl. 2012, 22, 517–531. [Google Scholar] [CrossRef]
- Mienna, I.M. Patterns and Drivers of Phylogenetic Diversity and Endemism in the Norwegian Vascular Flora. Master’s Thesis, NTNU, Trondheim, Norway, 2018. [Google Scholar]
- Guo, W.-Y.; Serra-Diaz, J.M.; Eiserhardt, W.L.; Maitner, B.S.; Merow, C.; Violle, C.; Pound, M.J.; Sun, M.; Slik, F.; Blach-Overgaard, A. Climate change and land use threaten global hotspots of phylogenetic endemism for trees. Nat. Commun. 2023, 14, 6950. [Google Scholar] [CrossRef]
- Brown, J.L.; Paz, A.; Reginato, M.; Renata, C.A.; Assis, C.; Lyra, M.; Caddah, M.K.; Aguirre-Santoro, J.; d’Horta, F.; Raposo do Amaral, F. Seeing the forest through many trees: Multi-taxon patterns of phylogenetic diversity in the Atlantic Forest hotspot. Divers. Distrib. 2020, 26, 1160–1176. [Google Scholar] [CrossRef]
- Anacker, B.L.; Whittall, J.B.; Goldberg, E.E.; Harrison, S.P. Origins and consequences of serpentine endemism in the California flora. Evolution 2011, 65, 365–376. [Google Scholar] [CrossRef]
- Fritz, S.A.; Rahbek, C. Global patterns of amphibian phylogenetic diversity. J. Biogeogr. 2012, 39, 1373–1382. [Google Scholar] [CrossRef]
- Safi, K.; Cianciaruso, M.V.; Loyola, R.D.; Brito, D.; Armour-Marshall, K.; Diniz-Filho, J.A.F. Understanding global patterns of mammalian functional and phylogenetic diversity. Philos. Trans. R. Soc. Biol. Sci. 2011, 366, 2536–2544. [Google Scholar] [CrossRef]
- Cheikh Albassatneh, M.; Escudero, M.; Monnet, A.C.; Arroyo, J.; Bacchetta, G.; Bagnoli, F.; Dimopoulos, P.; Hampe, A.; Leriche, A.; Médail, F. Spatial patterns of genus-level phylogenetic endemism in the tree flora of Mediterranean Europe. Divers. Distrib. 2021, 27, 913–928. [Google Scholar] [CrossRef]
- González-Caro, S.; Duivenvoorden, J.F.; Balslev, H.; Cavelier, J.; Grandez, C.; Macia, M.J.; Romero-Saltos, H.; Sanchez, M.; Valencia, R.; Duque, A. Scale-dependent drivers of the phylogenetic structure and similarity of tree communities in northwestern Amazonia. J. Ecol. 2021, 109, 888–899. [Google Scholar] [CrossRef]
- Cavender-Bares, J.; Kozak, K.H.; Fine, P.V.A.; Kembel, S.W. The merging of community ecology and phylogenetic biology. Ecol. Lett. 2009, 12, 693–715. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.M.; Cadotte, M.W.; Carvalho, S.B.; Davies, T.J.; Ferrier, S.; Fritz, S.A.; Grenyer, R.; Helmus, M.R.; Jin, L.S.; Mooers, A.O.; et al. A guide to phylogenetic metrics for conservation, community ecology and macroecology. Biol. Rev. 2017, 92, 698–715. [Google Scholar] [CrossRef]
- Faith, D.P.; Magallón, S.; Hendry, A.P.; Conti, E.; Yahara, T.; Donoghue, M.J. Evosystem services: An evolutionary perspective on the links between biodiversity and human well-being. Curr. Opin. Environ. Sustain. 2010, 2, 66–74. [Google Scholar] [CrossRef]
- Qian, H.; Kessler, M.; Deng, T.; Jin, Y. Patterns and drivers of phylogenetic structure of pteridophytes in China. Glob. Ecol. Biogeogr. 2021, 30, 1835–1846. [Google Scholar] [CrossRef]
- Cadotte, M.W.; Tucker, C.M. Should environmental filtering be abandoned? Trends Ecol. Evol. 2017, 32, 429–437. [Google Scholar] [CrossRef]
- Tu, W.-Q.; Lu, W.-X.; Gu, J.-Q.; Lou, A.-R. The species diversity and phylogenetic structure patterns of desert plant communities in the Turpan-Hami region, Xinjiang. Glob. Ecol. Conserv. 2024, 55, e03239. [Google Scholar] [CrossRef]
- Liang, J.; Ding, Z.; Lie, G.; Zhou, Z.; Zhang, Z.; Hu, H. Patterns and drivers of phylogenetic diversity of seed plants along an elevational gradient in the central Himalayas. Glob. Ecol. Conserv. 2023, 47, e02661. [Google Scholar] [CrossRef]
- Xiao, Y.; Yang, L.; Nie, X.; Li, C.; Xiong, F.; Wang, L.; Zhou, G. Examining differences in phylogenetic composition enhances understanding of the phylogenetic structure of the shrub community in the northeastern Qinghai-Tibetan Plateau. Ecol. Evol. 2020, 10, 6723–6731. [Google Scholar] [CrossRef]
- Luo, Y.; Zhou, M.; Jin, S.; Wang, Q.; Yan, D. Changes in phylogenetic structure and species composition of woody plant communities across an elevational gradient in the southern Taihang Mountains, China. Glob. Ecol. Conserv. 2023, 42, e02412. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, Z.; Shang, K.; Zhao, M.; Kong, J.; Wang, X.; Wang, Y.; Song, H.; Zhang, O.; Lv, X.; et al. A taxonomic and phylogenetic perspective on plant community assembly along an elevational gradient in subtropical forests. J. Plant Ecol. 2021, 14, 702–716. [Google Scholar] [CrossRef]
- Mori, A.S.; Ota, A.T.; Fujii, S.; Seino, T.; Kabeya, D.; Okamoto, T.; Ito, M.T.; Kaneko, N.; Hasegawa, M. Biotic homogenization and differentiation of soil faunal communities in the production forest landscape: Taxonomic and functional perspectives. Oecologia 2015, 177, 533–544. [Google Scholar] [CrossRef] [PubMed]
- de Albuquerque, F.S.; Castro-Díez, P.; Rodríguez, M.Á.; Cayuela, L. Assessing the influence of environmental and human factors on native and exotic species richness. Acta Oecologica 2011, 37, 51–57. [Google Scholar] [CrossRef]
- Kadereit, J.W.; Abbott, R.J. Plant speciation in the Quaternary. Plant Ecol. Divers. 2021, 14, 105–142. [Google Scholar] [CrossRef]
- Hofreiter, M.; Stewart, J. Ecological change, range fluctuations and population dynamics during the Pleistocene. Curr. Biol. 2009, 19, R584–R594. [Google Scholar] [CrossRef]
- Sanmartín, I. Historical biogeography: Evolution in time and space. Evol. Educ. Outreach 2012, 5, 555–568. [Google Scholar] [CrossRef]
- Alexandre, J.; Diniz-Filho, F. Phylogenetic diversity and conservation priorities under distinct models of phenotypic evolution. Conserv. Biol. 2004, 18, 698–704. [Google Scholar] [CrossRef]
- Lees, C.M.; Rutschmann, A.; Santure, A.W.; Beggs, J.R. Science-based, stakeholder-inclusive and participatory conservation planning helps reverse the decline of threatened species. Biol. Conserv. 2021, 260, 109194. [Google Scholar] [CrossRef]
- Folk, R.A.; Belitz, M.W.; Siniscalchi, C.M.; Kates, H.R.; Soltis, D.E.; Soltis, P.S.; Guralnick, R.P.; Borges, L.M. Phylogenetic diversity and regionalization of root nodule symbiosis. BioRxiv 2023. [Google Scholar] [CrossRef]
- Bhola, N.; Klimmek, H.; Kingston, N.; Burgess, N.D.; van Soesbergen, A.; Corrigan, C.; Harrison, J.; Kok, M.T.J. Perspectives on area-based conservation and its meaning for future biodiversity policy. Conserv. Biol. 2021, 35, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Gillingham, P.K.; Britton, J.R.; Jones, G.; Miller-Rushing, A.; Stafford, R.; Slater, H. Climate change adaptation for biodiversity in protected areas: An overview of actions. Biol. Conserv. 2024, 289, 110375. [Google Scholar] [CrossRef]
- Cupertino-Eisenlohr, M.; Simon, M. Evolutionary diversity gradients in neotropical tree assemblages: New insights from Non-Flooded Evergreen forests. Biotropica 2023, 55, 454–466. [Google Scholar] [CrossRef]
- Edwards, D.; Massam, M.; Haugaasen, T.; Gilroy, J. Tropical secondary forest regeneration conserves high levels of avian phylogenetic diversity. Biol. Conserv. 2017, 209, 432–439. [Google Scholar] [CrossRef]
- Rodrigues, A.; Gaston, K. Maximising phylogenetic diversity in the selection of networks of conservation areas. Biol. Conserv. 2002, 105, 103–111. [Google Scholar] [CrossRef]
- Woinarski, J.; Price, O.; Faith, D. Application of a taxon priority system for conservation planning by selecting areas which are most distinct from environments already reserved. Biol. Conserv. 1996, 76, 147–159. [Google Scholar] [CrossRef]
- Cardillo, M. Phylogenetic diversity in conservation: A brief history, critical overview, and challenges to progress. Camb. Prism. Extinction 2023, 1, e11. [Google Scholar] [CrossRef]
- Rapacciuolo, G.; Graham, C.H.; Marin, J.; Behm, J.E.; Costa, G.C.; Hedges, S.B.; Helmus, M.R.; Radeloff, V.C.; Young, B.E.; Brooks, T.M. Species diversity as a surrogate for conservation of phylogenetic and functional diversity in terrestrial vertebrates across the Americas. Nat. Ecol. Evol. 2019, 3, 53–61. [Google Scholar] [CrossRef]
- Wang, M.; Lu, N.; An, N.; Fu, B. Plant functional and phylogenetic diversity regulate ecosystem multifunctionality in semi-arid grassland during succession. Front. Environ. Sci. 2022, 9, 791801. [Google Scholar] [CrossRef]
- Thompson, P.L.; Davies, T.J.; Gonzalez, A. Ecosystem functions across trophic levels are linked to functional and phylogenetic diversity. PLoS ONE 2015, 10, e0117595. [Google Scholar] [CrossRef]
- Doxa, A.; Devictor, V.; Baumel, A.; Pavon, D.; Medail, F.; Leriche, A. Beyond taxonomic diversity: Revealing spatial mismatches in phylogenetic and functional diversity facets in Mediterranean tree communities in southern France. For. Ecol. Manag. 2020, 474, 118318. [Google Scholar] [CrossRef]
- Qin, H.; Wang, Y.; Zhang, F.; Chen, J.; Zhang, G.; Dong, G. Application of species, phylogenetic and functional diversity to the evaluation on the effects of ecological restoration on biodiversity. Ecol Inf. 2016, 32, 53–62. [Google Scholar] [CrossRef]
- Trindade-Filho, J.; Sobral, F.L.; Cianciaruso, M.V.; Loyola, R.D. Using indicator groups to represent bird phylogenetic and functional diversity. Biol. Conserv. 2012, 146, 155–162. [Google Scholar] [CrossRef]
- Montaño-Centellas, F.; Baiser, B.; McGrew, A.; Trotta, L.; Li, D. Global patterns and drivers of raptor phylogenetic and functional diversity. Glob. Ecol. Biogeogr. 2023, 32, 281–294. [Google Scholar] [CrossRef]
- Scher, C.L.; Karimi, N.; Glasenhardt, M.C.; Tuffin, A.; Cannon, C.H.; Scharenbroch, B.C.; Hipp, A.L. Application of remote sensing technology to estimate productivity and assess phylogenetic heritability. Appl. Plant Sci. 2020, 8, e11401. [Google Scholar] [CrossRef]
- Guo, C.; Luo, Y.; Gao, L.M.; Yi, T.S.; Li, H.T.; Yang, J.B.; Li, D.Z. Phylogenomics and the flowering plant tree of life. J. Integr. Plant Biol. 2023, 65, 299–323. [Google Scholar] [CrossRef]
- Smith, S.A.; Walker-Hale, N.; Walker, J.F.; Brown, J.W. Phylogenetic conflicts, combinability, and deep phylogenomics in plants. Syst. Biol. 2020, 69, 579–592. [Google Scholar] [CrossRef]
- Marcondes, R. Realistic scenarios of missing taxa in phylogenetic comparative methods and their effects on model selection and parameter estimation. PeerJ 2019, 7, e7917. [Google Scholar] [CrossRef]
- Zaharias, P.; Pante, E.; Gey, D.; Fedosov, A.; Puillandre, N. Data, time and money: Evaluating the best compromise for inferring molecular phylogenies of non-model animal taxa. Mol. Phylogenetics Evol. 2020, 142, 106660. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Wu, P.; Cheng, T.; Yu, J.; Zhou, S.; Hong, D. Resolving the systematic positions of enigmatic taxa: Manipulating the chloroplast genome data of Saxifragales. Mol. Phylogenetics Evol. 2018, 126, 321–330. [Google Scholar] [CrossRef]
- Thomson, R.; Shedlock, A.; Edwards, S.; Shaffer, H.; Thomson, R. Developing markers for multilocus phylogenetics in non-model organisms: A test case with turtles. Mol. Phylogenetics Evol. 2008, 49, 514–525. [Google Scholar] [CrossRef] [PubMed]
- Melin, M.; Hill, R.A.; Bellamy, P.E.; Hinsley, S.A. On bird species diversity and remote sensing—Utilizing lidar and hyperspectral data to assess the role of vegetation structure and foliage characteristics as drivers of avian diversity. IEEE J. Sel. Top. Appl. Earth 2019, 12, 2270–2278. [Google Scholar] [CrossRef]
- Kamoske, A.G.; Dahlin, K.M.; Read, Q.D.; Record, S.; Stark, S.C.; Serbin, S.P.; Zarnetske, P.L. Towards mapping biodiversity from above: Can fusing lidar and hyperspectral remote sensing predict taxonomic, functional, and phylogenetic tree diversity in temperate forests? Glob. Ecol. Biogeogr. 2022, 31, 1440–1460. [Google Scholar] [CrossRef]
- Bae, S.; Müller, J.; Lee, D.; Vierling, K.T.; Vogeler, J.C.; Vierling, L.A.; Hudak, A.T.; Latifi, H.; Thorn, S. Taxonomic, functional, and phylogenetic diversity of bird assemblages are oppositely associated to productivity and heterogeneity in temperate forests. Remote Sens. Env. 2018, 215, 145–156. [Google Scholar] [CrossRef]
- Coverdale, T.C.; Davies, A.B. Unravelling the relationship between plant diversity and vegetation structural complexity: A review and theoretical framework. J. Ecol. 2023, 111, 1378–1395. [Google Scholar] [CrossRef]
- Dunn, P.C.; Blesius, L. Modeling Insolation, Multi-Spectral Imagery and LiDAR Point-Cloud Metrics to Predict Plant Diversity in a Temperate Montane Forest. Geographies 2021, 1, 79–103. [Google Scholar] [CrossRef]
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding forest health with remote sensing-part I—A review of spectral traits, processes and remote-sensing characteristics. Remote Sens. 2016, 8, 1029. [Google Scholar] [CrossRef]
- Baraloto, C.; Hardy, O.J.; Paine, C.E.T.; Dexter, K.G.; Cruaud, C.; Dunning, L.T.; Gonzalez, M.A.; Molino, J.F.; Sabatier, D.; Savolainen, V. Using functional traits and phylogenetic trees to examine the assembly of tropical tree communities. J. Ecol. 2012, 100, 690–701. [Google Scholar] [CrossRef]
- Swenson, N.G. Phylogenetic imputation of plant functional trait databases. Ecography 2014, 37, 105–110. [Google Scholar] [CrossRef]
- Shi, W.; Wang, Y.-Q.; Xiang, W.; Li, X.; Cao, K. Environmental filtering and dispersal limitation jointly shaped the taxonomic and phylogenetic beta diversity of natural forests in southern China. Ecol. Evol. 2021, 11, 8783–8794. [Google Scholar] [CrossRef]
- Saqib, S.; Ullah, F.; Omollo, W.O.; Liu, Y.; Tao, H.-Y.; Zaman, W.; Temur, A.; Liu, B.; Lai, Y.; Chen, Z.; et al. Identifying hotspots and climate drivers of alien plant species for conservation prioritization across the Pan-Himalaya. Biol. Conserv. 2025, 302, 110994. [Google Scholar] [CrossRef]
- Rajarajan, K.; Vishnu, R.; Priyadarshini, E.; Arunachalam, P.; Ramanan, S.S. Phylogenetics in the Context of Tree Diversity and Conservation. In Molecular Genetics and Genomics Tools in Biodiversity Conservation; Springer: Singapore, 2022. [Google Scholar]
- Palmer, C.; Fischer, B. Should Global Conservation Initiatives Prioritize Phylogenetic Diversity? Philosophia 2021, 50, 2283–2302. [Google Scholar] [CrossRef]
- Chao, A.; Henderson, P.; Chiu, C.H.; Moyes, F.; Hu, K.-H.; Dornelas, M.; Magurran, A. Measuring temporal change in alpha diversity: A framework integrating taxonomic, phylogenetic and functional diversity and the iNEXT.3D standardization. Methods Ecol. Evol. 2021, 12, 1926–1940. [Google Scholar] [CrossRef]
- Pigot, A.; Phillimore, A.; Phillimore, A.; Owens, I.; Owens, I.; Orme, C. The shape and temporal dynamics of phylogenetic trees arising from geographic speciation. Syst. Biol. 2010, 59, 660–673. [Google Scholar] [CrossRef]
- Singer, B.; Di Nardo, A.; Hein, J.; Ferretti, L. Comparing Phylogeographies to Reveal Incompatible Geographical Histories within Genomes. Mol. Biol. Evol. 2024, 41, msae126. [Google Scholar] [CrossRef]
- Davies, S.; Abiem, I.; Salim, K.; Aguilar, S.; Allen, D.; Alonso, A.; Anderson-Teixeira, K.; Anderson-Teixeira, K.; Andrade, A.; Arellano, G.; et al. ForestGEO: Understanding forest diversity and dynamics through a global observatory network. Biol. Conserv. 2021, 253, 108907. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, J.-J.; Li, H.-N.; Liu, J.; Burgess, K.; Ge, X. The effects of evolutionary and environmental variance on estimates of phylogenetic diversity in temperate forest plots. J. Plant Ecol. 2020, 14, 96–107. [Google Scholar] [CrossRef]
- Schmitt, S.; Maréchaux, I.; Chave, J.; Fischer, F.; Piponiot, C.; Traissac, S.; Hérault, B. Functional diversity improves tropical forest resilience: Insights from a long-term virtual experiment. J. Ecol. 2020, 108, 831–843. [Google Scholar] [CrossRef]





| Region | Forest Type | PD Values | PE Values | Notes | References |
|---|---|---|---|---|---|
| Pan-Himalayan region | Mixed coniferous | Moderate | High | Glacial refugia preserve ancient lineages, resulting in relictual endemism | [89,90] |
| Amazon Basin | Tropical rainforest | High | Moderate | Exceptional diversification, some ancient lineages, others rapidly radiated | [49,91] |
| Eastern Europe | Temperate deciduous | Low | Low | Reflects recent post-glacial recolonization with few endemic clades | [92,93] |
| Atlantic Forest (Brazil) | Tropical/subtropical forest | Moderate | High | High levels of both paleo- and neo-endemism; threatened hotspot | [94,95] |
| California Floristic Prov. | Mediterranean woodland | High | High | Ancient serpentine flora with extreme niche conservatism | [96] |
| Southeast Asia | Lowland tropical rainforest | Very high | Moderate | Extremely high richness and deep evolutionary history | [97] |
| Central Africa | Congo rainforest | High | Low | Continuous habitat has allowed diversification, but less spatial isolation | [49] |
| Northern Scandinavia | Boreal forest | Low | Low | Dominated by a few lineages, cold-filtered communities | [93] |
| Australia (SW) | Eucalyptus woodlands | Moderate | High | Gondwanan relicts with strong habitat specialization | [98] |
| Mediterranean Europe | Evergreen sclerophyll | Moderate | Moderate | Diverse genera with moderate endemicity; historical isolation a contributing factor | [99] |
| Environmental Variable | Expected Effect | Impact on PD Metrics | Explanation | References |
|---|---|---|---|---|
| Precipitation | Increased PD | PD ↑, NTI ↓ | Promotes niche heterogeneity and speciation, reducing phylogenetic clustering | [19,64] |
| Aridity | Phylogenetic clustering | NTI ↑, MPD ↓ | Acts as an abiotic filter, allowing survival of only drought-tolerant lineages | [57,106] |
| Soil pH | Non-linear effect | PD peaks at neutral pH | Influences microbial symbiosis and nutrient availability, affecting clade composition | [107,108] |
| Elevation | Varies by gradient | PD ↓, NTI ↑ at high altitudes | Harsher conditions restrict the community to more closely related species | [109] |
| Temperature | Moderate increases → PD ↑ | PD ↑, MPD ↑ | Warmer zones support diverse taxa but may experience homogenization under extreme conditions | [110] |
| Nutrient availability | Depends on limitation | MPD varies, NTI ↓ in rich soils | High-nutrient soils can support more distantly related species with varied strategies | [64] |
| Disturbance regime | Dispersal-driven assembly | PD ↑, NTI ↓ or ↑, depending on context | Can promote the coexistence of disparate lineages or clade filtering, depending on the disturbance type | [108] |
| Study | Remote Sensing Method | Application | Forest Type | Key Insights |
|---|---|---|---|---|
| Kamoske, et al. [141] | LiDAR + Hyperspectral (NDVI) | Predict tree PD, FD, and taxonomic diversity | Temperate deciduous forest (USA) | Combined data provides accurate biodiversity mapping across taxa, specifically linking PD to structural and functional diversity. |
| Bae, et al. [142] | LiDAR + NDVI | Model bird phylogenetic & functional diversity | Temperate forest (North America) | NDVI is linked to productivity; LiDAR captures structural heterogeneity, which affects community traits and PD. |
| Melin, et al. [140] | LiDAR + RENDVI | Assess vegetation structure–diversity link | Mixed woodlands (UK) | High canopy structural diversity (LiDAR) predicts avian PD more effectively than NDVI alone, emphasizing the connection between canopy structure and PD. |
| Coverdale and Davies [143] | LiDAR | Structural complexity–diversity theory | Global review | LiDAR is crucial in linking vertical complexity to community assembly and evolutionary history |
| Dunn and Blesius [144] | LiDAR + Multispectral (NDVI) | Predict PD in montane forests | Temperate montane (California) | Elevation, solar insolation, and NDVI/structure data can be combined to model PD |
| Lausch, et al. [145] | NDVI + Spectral Time Series | Monitor forest health & structure | Multi-biome/global | NDVI seasonality can be used as a proxy for forest phenology influencing PD estimates, helping to link phenology to evolutionary processes. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Amin, A.; Akhtar, M.S.; Zaman, W. Phylogenetic Diversity in Forests: Insights into Evolutionary Patterns and Conservation Strategies. Forests 2025, 16, 1004. https://doi.org/10.3390/f16061004
Ali S, Amin A, Akhtar MS, Zaman W. Phylogenetic Diversity in Forests: Insights into Evolutionary Patterns and Conservation Strategies. Forests. 2025; 16(6):1004. https://doi.org/10.3390/f16061004
Chicago/Turabian StyleAli, Sajid, Adnan Amin, Muhammad Saeed Akhtar, and Wajid Zaman. 2025. "Phylogenetic Diversity in Forests: Insights into Evolutionary Patterns and Conservation Strategies" Forests 16, no. 6: 1004. https://doi.org/10.3390/f16061004
APA StyleAli, S., Amin, A., Akhtar, M. S., & Zaman, W. (2025). Phylogenetic Diversity in Forests: Insights into Evolutionary Patterns and Conservation Strategies. Forests, 16(6), 1004. https://doi.org/10.3390/f16061004








