Revealing the Bacteriome in Crop–Livestock–Forest Integration Systems in the Cerrado of MATOPIBA, Brazil
Abstract
1. Introduction
2. Materials and Methods
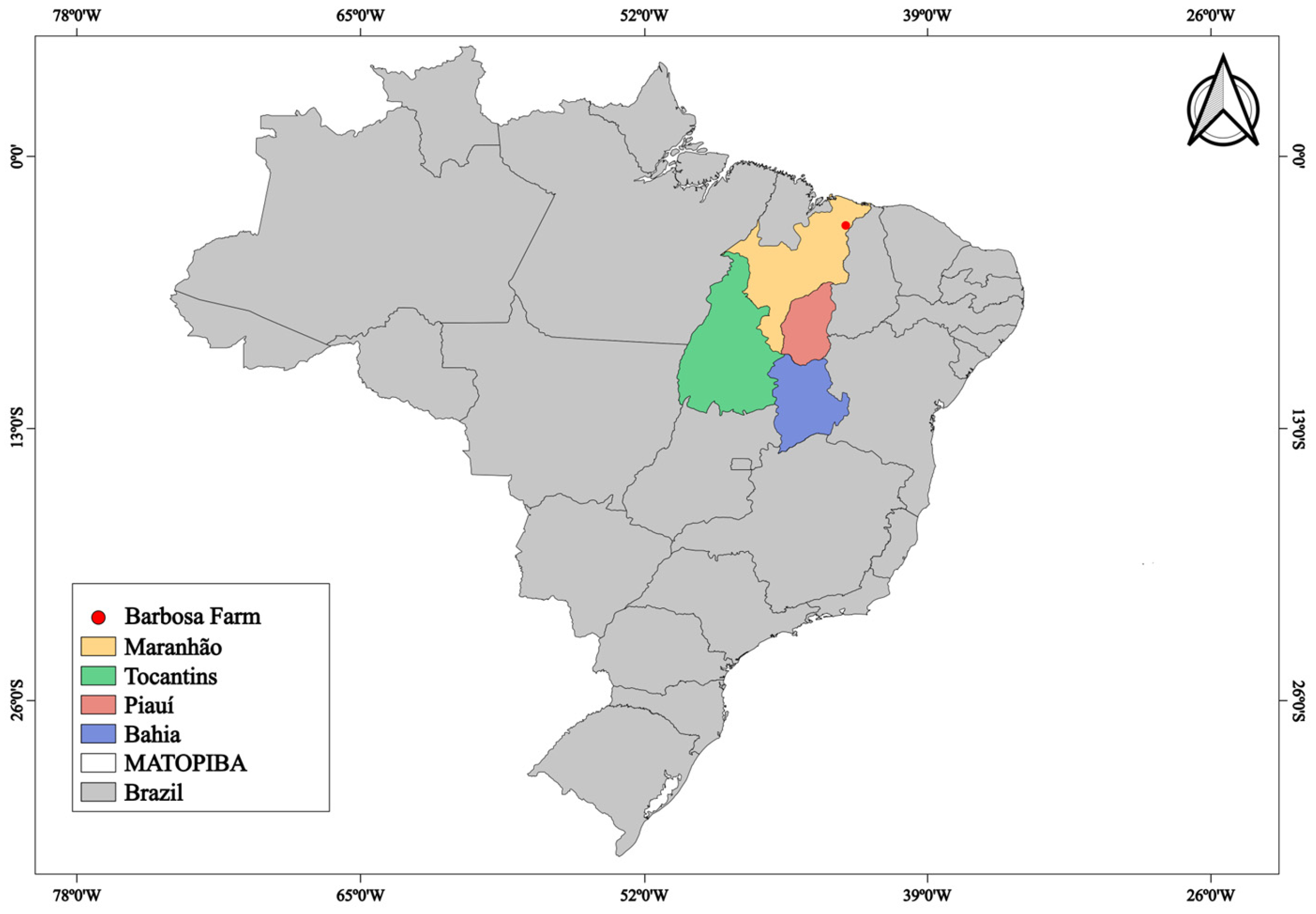
2.1. Soil Sampling
2.2. Determination of Soil Physical–Chemical Parameters
2.3. DNA Extraction
2.4. Library Preparation and Sequencing
2.5. Metataxonomic Data
2.6. Microbiome and Statistical Analyses
2.6.1. Alpha and Beta Diversity
2.6.2. Taxonomic Composition
2.6.3. Differentially Abundant Taxa
3. Results
3.1. Sequencing and Data Processing
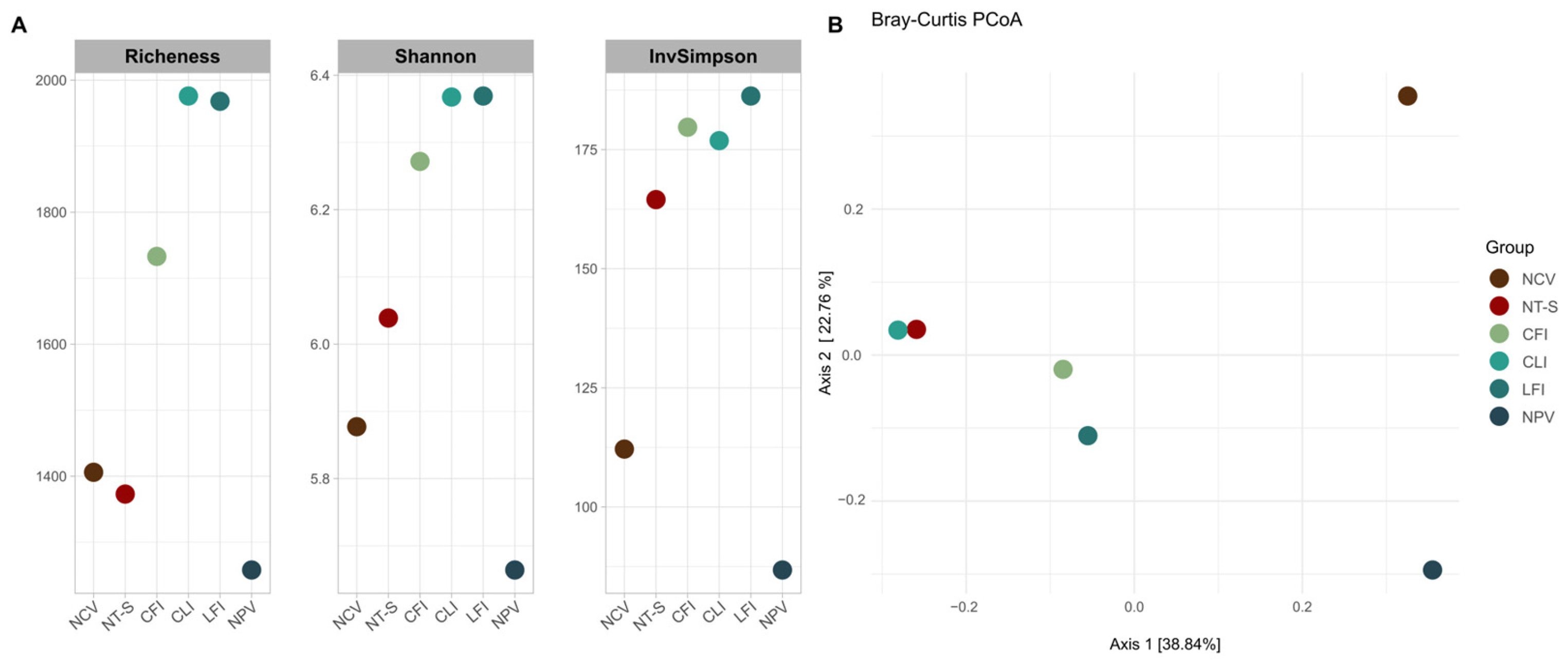
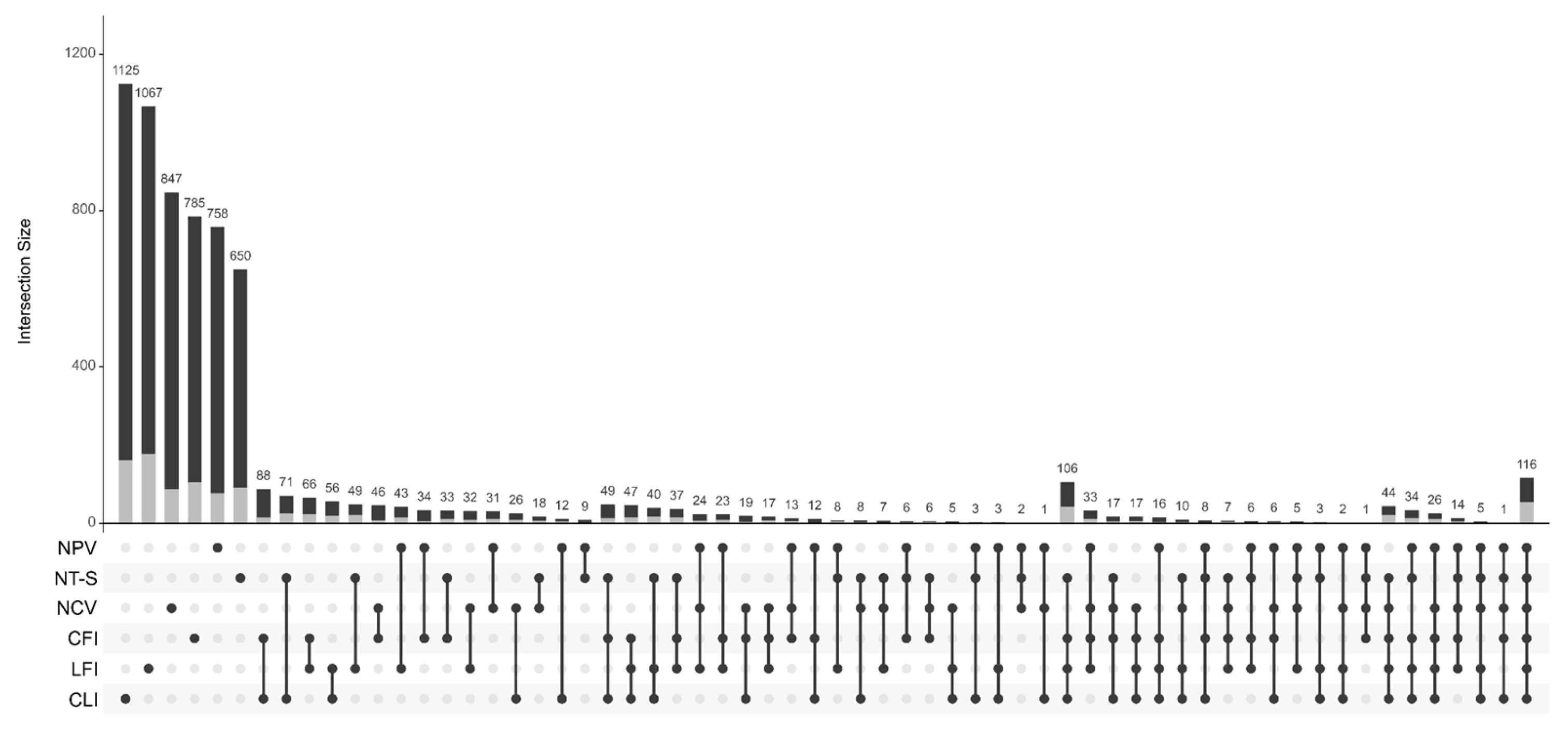
3.2. Structure and Diversity of Bacterial Communities
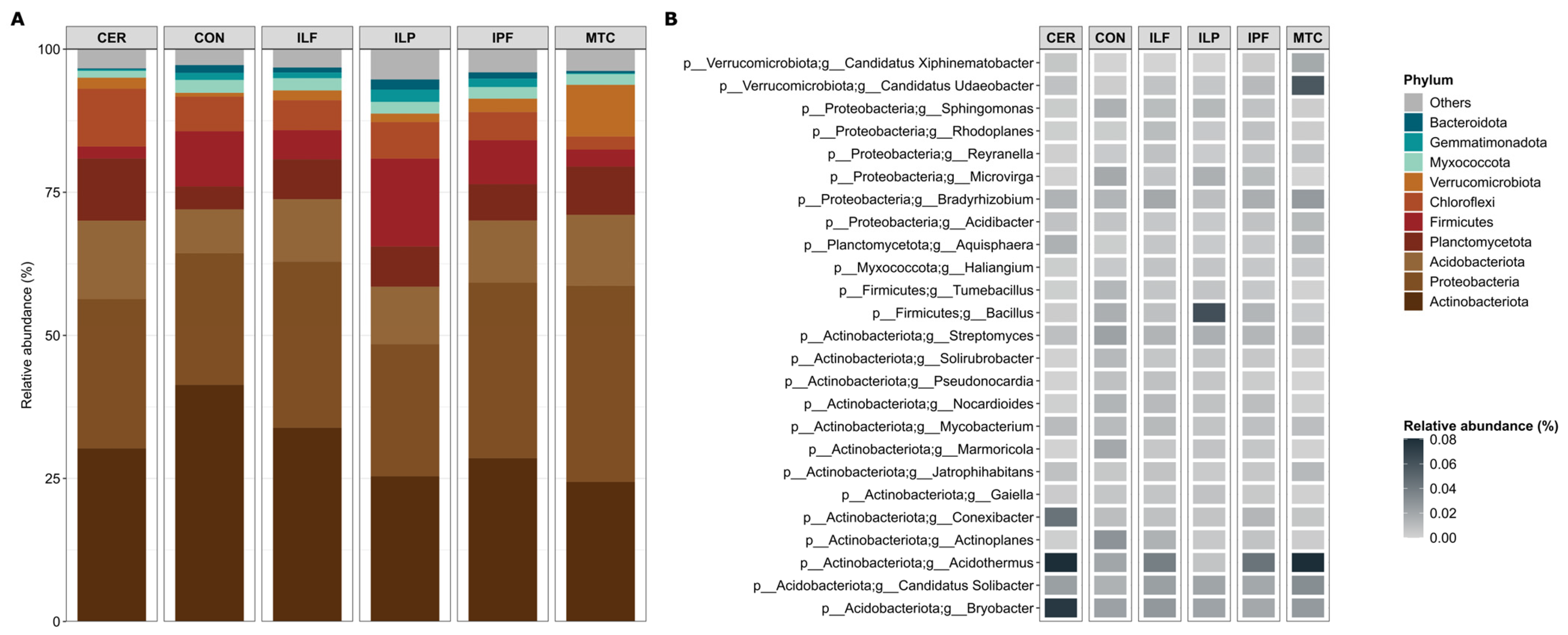
3.3. Taxonomic Composition of Bacterial Communities Associated with Different Management Systems
3.4. Differential Abundance of Taxa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lima, M.; Silva Junior, C.A.D.; Rausch, L.; Gibbs, H.K.; Johann, J.A. Demystifying Sustainable Soy in Brazil. Land Use Policy 2019, 82, 349–352. [Google Scholar] [CrossRef]
- Pires, G.F.; Abrahão, G.M.; Brumatti, L.M.; Oliveira, L.J.C.; Costa, M.H.; Liddicoat, S.; Kato, E.; Ladle, R.J. Increased Climate Risk in Brazilian Double Cropping Agriculture Systems: Implications for Land Use in Northern Brazil. Agric. For. Meteorol. 2016, 228–229, 286–298. [Google Scholar] [CrossRef]
- de Almeida, R.E.M.; de Souza, H.A.; Evangelista, B.A.; Uhlmann, A.; Ramos, M.R.; Sagrilo, E.; dos Santos Dias, T.S.; de Sousa Paz Oliveira, L.R.; Costa, N.R. Challenges to Managing Soil Health in the Newest Agricultural Frontier in Brazil. In ASA, CSSA, and SSSA Books; Mendes, I.C., Cherubin, M.R., Eds.; Wiley: Hoboken, NJ, USA, 2024; pp. 327–374. ISBN 978-0-89118-743-1. [Google Scholar]
- Lustosa Filho, J.; Souza, H.; Almeida, R.; Leite, L. Conservação e Manejo Da Fertilidade Do Solo No Cerrado Do Matopiba. In Cerrado: Capital Natural e Serviços Ambientais; Paco Editorial: Jundiaí, Brazil, 2021; pp. 75–97. [Google Scholar]
- Souza, H.A.; Sagrilo, E.; Oliveira Junior, J.O.L.; Leite, L.F.C.; Vogado, R.F.; Santos, S.F.C.B.; Clark, M.V.G.; Barbosa, L.R.; Brito, L.C.R. Integração Lavoura-Pecuária-Floresta Com Componente Soja No Maranhão: Resultados de Pesquisa Em Unidades de Referência Tecnológica. In Soja Sustentável No Leste Maranhense: Realidade e Perspectivas; da Silva Cruz, A.P., Sander, N.L., de Oliveira, A.B., Ferreira, I.G.M., Eds.; CRV: Curitiba, Brazil, 2023; Volume 1, pp. 37–54. [Google Scholar]
- Teixeira Neto, M.L.; Carvalho, G.M.C.; de Araujo Neto, R.B.; de Azevedo, D.M.P.; da Frota, M.N.L.; das Chagas Monteiro, F.; de Souza, H.A.; de Alcantara, R.M.C.M.; de Andrade Junior, A.S.; Cardoso, M.J.; et al. Integração Lavoura-Pecuária-Floresta (ILPF) Nos Cerrados Do Piauí e Do Maranhão: Estratégia de Produção Para Quatro Safras Ao Ano Só Com Chuvas. In Solos Sustentáveis para a Agricultura No Nordeste; de Souza, H.A., Leite, L.F.C., Medeiros, J.C., Eds.; Embrapa: Brasília, Brazil, 2021. [Google Scholar]
- Silva, A.A.; Lacerda, J.J.D.J.; de Araújo-Neto, R.B.; Sagrilo, E.; Lustosa-Filho, J.F.; de Andrade, H.A.; de Souza, H.A. Integrated Agroforestry System Affects the Dynamics of Inorganic Phosphorus Fractions in the Savanna of Brazilian Northeast. Can. J. Soil Sci. 2024, 104, 181–190. [Google Scholar] [CrossRef]
- Vogado, R.F.; de Souza, H.A.; Sagrilo, E.; de Brito, L.D.C.R.; Matias, S.S.R.; Neto, M.L.T.; de Oliveira Junior, J.O.L.; de Andrade, H.A.F.; Leite, L.F.C. Soil Organic Carbon Stocks and Fractions Under Integrated Systems and Pasture in the Cerrado of Northeast Brazil. Catena 2024, 243, 108196. [Google Scholar] [CrossRef]
- Costa, R.M.; Araujo, E.M.B.; Silva, D.E.O.; Rocha, S.M.B.; Bonifacio, A.; Sousa, R.S.; de Araujo Pereira, A.P.; de Medeiros, E.V.; Sagrilo, E.; de Oliveira Junior, J.O.L.; et al. Seasonal Responses of Soil Microbial Biomass C and Enzymatic Activity Comparing No-Tillage and Integrated Crop-Livestock Systems. Eur. J. Soil Biol. 2024, 121, 103628. [Google Scholar] [CrossRef]
- Bernardino, F.D.S.; Garcia, R. Sistemas Silvipastoris. Pesqui. Florest. Bras. 2009, 60, 77–87. [Google Scholar]
- Macedo, R.; Vale, A.; Venturin, N. Eucalipto Em Sistemas Agroflorestais: Lavras; Editora da UFLA: Lavras, Brazil, 2010. [Google Scholar]
- Salton, J.C.; Mielniczuk, J.; Bayer, C.; Fabrício, A.C.; Macedo, M.C.M.; Broch, D.L. Teor e Dinâmica Do Carbono No Solo Em Sistemas de Integração Lavoura-Pecuária. Pesqui. Agropecuária Bras. 2011, 46, 1349–1356. [Google Scholar] [CrossRef]
- Carvalho, J.L.N.; Avanzi, J.C.; Silva, M.L.N.; de Mello, C.R.; Cerri, C.E.P. Potencial de Sequestro de Carbono Em Diferentes Biomas Do Brasil. Rev. Bras. Ciência Solo 2010, 34, 277–290. [Google Scholar] [CrossRef]
- Salton, J.C.; Mercante, F.M.; Tomazi, M.; Zanatta, J.A.; Concenço, G.; Silva, W.M.; Retore, M. Integrated Crop-Livestock System in Tropical Brazil: Toward a Sustainable Production System. Agric. Ecosyst. Environ. 2014, 190, 70–79. [Google Scholar] [CrossRef]
- Leite, L.F.C.; de Freitas, R.D.C.A.; Sagrilo, E.; Galvão, S.R.D.S. Decomposição e Liberação de Nutrientes de Resíduos Vegetais Depositados Sobre Latossolo Amarelo No Cerrado Maranhense. Rev. Ciência Agronômica 2010, 41, 29–35. [Google Scholar]
- Costa, S.; Souza, E.; Anghinoni, I.; Carvalho, P.; Martins, A.; Kunrath, T.; Cecagno, D.; Balerini, F. Impact of an Integrated No-till Crop–Livestock System on Phosphorus Distribution, Availability and Stock. Agric. Ecosyst. Environ. 2014, 190, 43–51. [Google Scholar] [CrossRef]
- Marchão, R.L.; Lavelle, P.; Celini, L.; Balbino, L.C.; Vilela, L.; Becquer, T. Soil Macrofauna under Integrated Crop-Livestock Systems in a Brazilian Cerrado Ferralsol. Pesqui. Agropecuária Bras. 2009, 44, 1011–1020. [Google Scholar] [CrossRef]
- Malewski, T.; Borowik, P.; Golińska, P.; Okorski, A.; Olejarski, I.; Oszako, T. Organic Inputs Positively Alter the Bacteriome of Post-Agricultural Soils. Forests 2023, 14, 1711. [Google Scholar] [CrossRef]
- Raimi, A.R.; Ezeokoli, O.T.; Adeleke, R.A. Soil Nutrient Management Influences Diversity, Community Association and Functional Structure of Rhizosphere Bacteriome Under Vegetable Crop Production. Front. Microbiol. 2023, 14, 1229873. [Google Scholar] [CrossRef]
- Passos, M.L.V.; Zambrzycki, G.C.; Pereira, R.S. Balanço Hídrico Climatológico e Classificação Climática Para o Município de Balsas-MA. Sci. Agrar. 2017, 18, 83–89. [Google Scholar] [CrossRef]
- Dantas, J.S.; Marques Júnior, J.; Martins Filho, M.V.; Resende, J.M.D.A.; Camargo, L.A.; Barbosa, R.S. Gênese de Solos Coesos Do Leste Maranhense: Relação Solo-Paisagem. Rev. Bras. Ciênc. Solo 2014, 38, 1039–1050. [Google Scholar] [CrossRef]
- de Sousa, D.M.G.; Lobato, E. Cerrado: Correção Do Solo e Adubação, 2nd ed.; Embrapa Informação Tecnológica; Embrapa Cerrados: Brasília, DF, Brazil, 2004. [Google Scholar]
- Furley, P.A. The Nature and Diversity of Neotropical Savanna Vegetation with Particular Reference to the Brazilian Cerrados. Glob. Ecol. Biogeogr. 1999, 8, 223–241. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Babraham, UK, 2010. [Google Scholar]
- Didion, J.P.; Martin, M.; Collins, F.S. Atropos: Specific, Sensitive, and Speedy Trimming of Sequencing Reads. PeerJ 2017, 5, e3720. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. Using DECIPHER V2.0 to Analyze Big Biological Sequence Data in R. R J. 2016, 8, 352–359. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. Microeco: An R Package for Data Mining in Microbial Community Ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Oksanen, J. Vegan: Community Ecology Package. 2010. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 2 March 2025).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R Package for the Visualization of Intersecting Sets and Their Properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data Quality Aware Analysis of Differential Expression in RNA-Seq with NOISeq R/Bioc Package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Tarazona, S.; García-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential Expression in RNA-Seq: A Matter of Depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Kassambara, A. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. 2023. Available online: https://cran.r-project.org/web/packages/rstatix/index.html (accessed on 2 March 2025).
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The Interplay Between Microbial Communities and Soil Properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Muneer, M.A.; Hou, W.; Li, J.; Huang, X.; Ur Rehman Kayani, M.; Cai, Y.; Yang, W.; Wu, L.; Ji, B.; Zheng, C. Soil pH: A Key Edaphic Factor Regulating Distribution and Functions of Bacterial Community Along Vertical Soil Profiles in Red Soil of Pomelo Orchard. BMC Microbiol. 2022, 22, 38. [Google Scholar] [CrossRef] [PubMed]
- de Brito, L.D.C.R.; Souza, H.A.D.; Araújo Neto, R.B.D.; Azevedo, D.M.P.D.; Sagrilo, E.; Vogado, R.F.; Carvalho, S.P.; Ferreira, A.C.D.M.; Cavigelli, M.A. Improved Soil Fertility, Plant Nutrition and Grain Yield of Soybean and Millet Following Maize Intercropped with Forage Grasses and Crotalaria in the Brazilian Savanna. Crop Pasture Sci. 2023, 74, 438–448. [Google Scholar] [CrossRef]
- de, C.R.; de Brito, L.; de Souza, H.A.; Deon, D.S.; de Souza, I.M.; dos Santos, S.F.D.C.B.; Sobral, A.H.S. Greenhouse Gas Emissions and Chemical and Physical Soil Attributes of Off-Season Agricultural Production Systems in The Savannah of Maranhão State, Brazil. Eng. Agrícola 2023, 43, e20220181. [Google Scholar]
- dos Santos, S.F.D.C.B.; de Souza, H.A.; de Araújo Neto, R.B.; Sagrilo, E.; Ferreira, A.C.M.; Carvalho, S.P.; de Brito, L.D.C.R.; Leite, L.F.C. Soil Microbiological Attributes and Soybean Grain Yield in Succession to Corn Intercropped with Forage in the Maranhão Eastern Cerrado. Int. J. Plant Prod. 2021, 15, 669–677. [Google Scholar] [CrossRef]
- Silva, A.A.; Lacerda, J.J.D.J.; Carvalho, S.P.; Ferreira, R.D.S.; Brito, R.R.D.; Vogado, R.F.; Araújo Neto, R.B.D.; Sagrilo, E.; Cavigelli, M.A.; Souza, H.A.D. Chemical and Biological Attributes of Soil and Soybean (Glycine max) Yield in Integrated Systems in the Cerrado of North-East Brazil. Soil Res. 2024, 62, SR23120. [Google Scholar] [CrossRef]
- Souza, I.M.D.; Sagrilo, E.; de Oliveira Júnior, J.O.L.; Araújo, M.D.M.; Muniz, L.C.; Costa, J.B.; Pompeu, R.C.F.F.; de Sousa, D.C.; de Andrade, H.A.F.; de Oliveira Neto, E.D.; et al. Soil Chemical Quality in Integrated Production Systems with the Presence of Native and Exotic Tree Components in the Brazilian Eastern Amazon. Forests 2024, 15, 1078. [Google Scholar] [CrossRef]
- Damian, J.M.; Firmano, R.F.; Cherubin, M.R.; Pavinato, P.S.; De Marchi Soares, T.; Paustian, K.; Cerri, C.E.P. Changes in Soil Phosphorus Pool Induced by Pastureland Intensification and Diversification in Brazil. Sci. Total Environ. 2020, 703, 135463. [Google Scholar] [CrossRef]
- Xun, W.; Yan, R.; Ren, Y.; Jin, D.; Xiong, W.; Zhang, G.; Cui, Z.; Xin, X.; Zhang, R. Grazing-Induced Microbiome Alterations Drive Soil Organic Carbon Turnover and Productivity in Meadow Steppe. Microbiome 2018, 6, 170. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, D.; Delgado-Baquerizo, M.; Liu, S.; Wang, B.; Wu, J.; Hu, S.; Bai, Y. Long-Term Regional Evidence of the Effects of Livestock Grazing on Soil Microbial Community Structure and Functions in Surface and Deep Soil Layers. Soil Biol. Biochem. 2022, 168, 108629. [Google Scholar] [CrossRef]
- Selari, P.J.R.G.; Olchanheski, L.R.; Ferreira, A.J.; Paim, T.D.P.; Calgaro Junior, G.; Claudio, F.L.; Alves, E.M.; Santos, D.D.C.; Araújo, W.L.; Silva, F.G. Short-Term Effect in Soil Microbial Community of Two Strategies of Recovering Degraded Area in Brazilian Savanna: A Pilot Case Study. Front. Microbiol. 2021, 12, 661410. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Tsai, S.M.; Navarrete, A.A.; De Hollander, M.; Van Veen, J.A.; Kuramae, E.E. Soil-Borne Microbiome: Linking Diversity to Function. Microb. Ecol. 2015, 70, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Merloti, L.F.; Mendes, L.W.; Pedrinho, A.; De Souza, L.F.; Ferrari, B.M.; Tsai, S.M. Forest-to-Agriculture Conversion in Amazon Drives Soil Microbial Communities and N-Cycle. Soil Biol. Biochem. 2019, 137, 107567. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.-P.; Clément, C.; Ouhdouch, Y.; Van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef]
- Madegwa, Y.M.; Uchida, Y. Liming Improves the Stability of Soil Microbial Community Structures Against the Application of Digestate Made from Dairy Wastes. J. Environ. Manag. 2021, 297, 113356. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes Benefaction Role in Soil and Plant Health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Nemergut, D.R.; Schmidt, S.K.; Townsend, A.R. Increases in Soil Respiration Following Labile Carbon Additions Linked to Rapid Shifts in Soil Microbial Community Composition. Biogeochemistry 2007, 82, 229–240. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7, 1446. [Google Scholar] [CrossRef]
- Reid, R.P.; Oehlert, A.M.; Suosaari, E.P.; Demergasso, C.; Chong, G.; Escudero, L.V.; Piggot, A.M.; Lascu, I.; Palma, A.T. Electrical Conductivity as a Driver of Biological and Geological Spatial Heterogeneity in the Puquios, Salar de Llamara, Atacama Desert, Chile. Sci. Rep. 2021, 11, 12769. [Google Scholar] [CrossRef]
- McDonald, M.D.; Lewis, K.L.; Blazier, J.C.; Gentry, T.J. Semi-Arid Soil Bacterial Communities Are Refined by Altered Plant Selection Pressure Under Conservation Management Practices. Appl. Soil Ecol. 2024, 194, 105191. [Google Scholar] [CrossRef]
- Chinta, Y.D.; Araki, H. Responses of Bulk and Rhizosphere Soil Microbiomes to Different Cover Crop Inputs and Their Connection and Contribution to Soil Fertility and Plant Growth. Pedobiologia 2023, 101, 150907. [Google Scholar] [CrossRef]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D.J. Bacillus Species in Soil as a Natural Resource for Plant Health and Nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Talia, P.; Sede, S.M.; Campos, E.; Rorig, M.; Principi, D.; Tosto, D.; Hopp, H.E.; Grasso, D.; Cataldi, A. Biodiversity Characterization of Cellulolytic Bacteria Present on Native Chaco Soil by Comparison of Ribosomal RNA Genes. Res. Microbiol. 2012, 163, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Liu, K.L.; Zhao, X.Q.; Gao, G.-F.; Wu, Y.H.; Shen, R.F. Microbial Keystone Taxa Drive Crop Productivity Through Shifting Aboveground-Belowground Mineral Element Flows. Sci. Total Environ. 2022, 811, 152342. [Google Scholar] [CrossRef]
- de Sousa, C.E.S.; Amaral Júnior, F.P.; Cardoso, A.D.S.; Ruggieri, A.C.; Van Cleef, F.D.O.S.; de Pádua, F.T.; Almeida, J.C.D.C. Effects of Integrating Legumes or Trees on Soil C Stock and Organic Matter Dynamics in Tropical Grasslands. Appl. Soil Ecol. 2024, 202, 105560. [Google Scholar] [CrossRef]
- Moretti, M.S.; Becker, B.; Kiffer, W.P.; da Penha, L.O.; Callisto, M. Eucalyptus Leaves Are Preferred to Cerrado Native Species but Do Not Constitute a Better Food Resource to Stream Shredders. J. Arid Environ. 2020, 181, 104221. [Google Scholar] [CrossRef]
- Huo, C.; Zhang, J.; Yang, X.; Li, X.; Su, Y.; Chen, Z. Dry Season Irrigation Promotes Nutrient Cycling by Reorganizing Eucalyptus Rhizosphere Microbiome. Sci. Total Environ. 2024, 954, 176307. [Google Scholar] [CrossRef]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a Free-Living Nitrogen-Fixing Bacterium Closely Associated with Grasses: Genetic, Biochemical and Ecological Aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef]
- Gomes, L.; Simões, S.; Dalla Nora, E.; de Sousa-Neto, E.; Forti, M.; Ometto, J. Agricultural Expansion in the Brazilian Cerrado: Increased Soil and Nutrient Losses and Decreased Agricultural Productivity. Land 2019, 8, 12. [Google Scholar] [CrossRef]
- Oliveira, D.M.S.; Santos, R.S.; Chizzotti, F.H.M.; Bretas, I.L.; Franco, A.L.C.; Lima, R.P.; Freitas, D.A.F.; Cherubin, M.R.; Cerri, C.E.P. Crop, Livestock, and Forestry Integration to Reconcile Soil Health, Food Production, and Climate Change Mitigation in the Brazilian Cerrado: A Review. Geoderma Reg. 2024, 37, e00796. [Google Scholar] [CrossRef]


| Description | History |
|---|---|
| Livestock–forestry integration (LFI) | Native forest clearance took place in 2016, followed by conventional soil preparation with the application of 5 t ha−1 of calcitic limestone, which was incorporated using a 28” plough, then scarified and leveled. The next year, the area was converted to rice cultivation (Oryza sativa) with fertilization rates of 250 kg ha−1 of N, 500 kg ha−1 of P2O5, and 300 kg ha−1 of K2O. In 2018, eucalyptus (Eucalyptus globulus) was planted in an east–west orientation in triple rows spaced 4 m apart, with 3 m between plants, creating a 30 m gap between tree rows over a length of 160 m. In the spaces between the eucalyptus rows, a mixed cropping of maize (Zea mays) and Tamani grass (Megathyrsus maximus—BRS Tamani hybrid) was grown. No additional soil preparation was conducted from 2019 onward. In 2020, the maize and Tamani grass intercropping was replanted, and after the maize harvest, cattle were introduced at a stocking rate of 2.5 AU ha−1, with Tamani grass maintained through 2022. Maize fertilization followed the technical recommendations [22]. |
| No-till soybean (NT-S) | This area has been under no-till soybean cultivation (Glycine max) on millet straw (Pennisetum glaucum L.) for 17 years. In 2003, native Cerrado vegetation was cleared, and mechanized agriculture began, involving conventional soil preparation with intensive tillage, application of correctives, and 2 t ha−1 of calcitic limestone. In 2004, conservation soil management practices were introduced, reducing tillage and planting soybeans. By 2005, a no-till system (NT-S) was established and continues today, with a soybean/millet rotation. Soybean seeds are inoculated with Bradyrhizobium japonicum before planting. For the 2022 season, soybean management included (i) desiccation of millet straw with 2 L ha−1 of glyphosate and 1 L ha−1 of 2,4-D amine; (ii) planting fertilization with 150 kg ha−1 of monoammonium phosphate (MAP) and 170 kg ha−1 of potassium chloride (KCl), plus 38 kg ha−1 of MIB 77 (containing 3% S, 1.8% B, 0.8% Cu, 0.1% Mo, 2% Mn, and 9% Zn); and (iii) an additional 100 kg ha−1 of ammonium sulfate applied 10 days post-emergence, following technical guidelines [22]. Micronutrients were applied as foliar sprays during crop growth, with pest, disease, and weed control performed chemically as needed. After each soybean harvest, millet seeds were broadcasted at a rate of 20 kg ha−1 of seed without additional fertilization. |
| Crop–forestry integration (CFI) | The area was cleared in 2004, and upland rice was planted in the following year. From 2006 to 2010, soybeans were cultivated in a monoculture system. Between 2011 and 2016, the land was managed under a crop–livestock integration (CLI) system, with intercropped maize and brachiaria and a soybean/millet rotation for five years. In 2017, a crop–forest integration (CFI) system was introduced, adding three rows of eucalyptus trees spaced 3 m × 4 m within rows and 30 m between rows, where annual crops were cultivated. At the end of 2016, the entire area received 3 t ha−1 of dolomitic limestone (effective calcium carbonate equivalent—ECCE—of 88%) before planting eucalyptus and annual crops, followed by plowing and harrowing for incorporation. In early February 2017, maize was intercropped with forage grasses in the eucalyptus rows, fertilized with 260 kg ha−1 of NPK 13-33-08 in the planting furrow and two subsequent topdressings: first with 280 kg ha−1 of NPK 08-00-36 at the 2–4 leaf stage, and then 150 kg ha−1 of polymerized urea at the 4–6 leaf stage. From 2018 to 2021, soybeans were cultivated in the eucalyptus rows without soil disturbance, with millet seeds (ADR300) broadcast as a cover crop in the off-season after soybean harvest, using 20 kg ha−1 of seed without additional fertilization. Soybean base fertilization followed technical recommendations [22] and matched the amounts used in the no-till area. At the end of 2021, 3 t ha−1 of dolomitic limestone (ECCE of 88%) was again broadcast across the area. In 2022, pigeon pea (Cajanus cajan cv. Mandarin) was planted in the eucalyptus rows, also without soil disturbance. |
| Crop–livestock integration (CLI) | The CLI system involves intercropping maize with brachiaria (Urochloa brizantha cv. Marandu), followed by grazing cattle during the off-season at a stocking rate of 2.5 AU ha−1. This is followed by four years of alternating soybean and millet cultivation, completing a five-year cycle. This approach is applied across the farm as a rotational system in soybean-growing areas. The evaluated area has a similar management history to the soybean no-till system (NT-S) until 2011. In 2012, the soil was plowed and harrowed, with 3.8 t ha−1 of calcitic limestone applied. In 2017, the CLI system (maize + brachiaria) was adopted, with cattle grazing during the off-season at 2.5 AU ha−1. From 2018 to 2020, the area was managed with a no-till system (soybean and millet). In 2021, subsoiling to 0.30 m was carried out, and maize intercropped with brachiaria was reintroduced. When cattle entered, the dry mass of brachiaria was 7780 kg ha−1, and when they left, it was 3550 kg ha−1. In subsequent years, the area continued under no-till management with soybean and millet. Fertilization for soybeans and maize followed technical guidelines [22], with soybean fertilization similar to the NT-S area and maize fertilization comparable to the CFI area. |
| Native Cerrado vegetation (NCV) | Native forest of Cerrado vegetation (area with Cerrado phytophysiognomy stricto sensu [23], with sporadic fires (almost annual) during the dry season. |
| Native Babassu palm vegetation (NPV) | Native Babassu forest vegetation, with a predominance of the Babassu palm tree (Attaleaspeciosa). |
| Parameters | NCV | NT-S | CFI | CLI | LFI | NPV |
|---|---|---|---|---|---|---|
| pH (CaCl2) | 4.44 | 5.34 | 4.9 | 5.43 | 5.39 | 4.68 |
| pH (H2O) | 5.40 | 6.00 | 5.80 | 6.40 | 6.20 | 5.50 |
| Organic matter (dag Kg−1) | 3.32 | 3.78 | 3.09 | 2.28 | 6.29 | 7.06 |
| Potential soil acidity: H + Al (cmolc dm−3) | 6.68 | 3.48 | 4.41 | 2.75 | 6.29 | 9.56 |
| Sum of bases (cmolc dm−3) | 1.06 | 2.79 | 2.04 | 2.04 | 4.60 | 3.76 |
| Cation exchange capacity (cmolc dm−3) | 7.74 | 6.27 | 6.46 | 4.79 | 10.89 | 13.32 |
| Base saturation (V%) | 14 | 44 | 32 | 43 | 42 | 28 |
| Aluminum Saturation (m%) | 22 | 0 | 2 | 0 | 2 | 3 |
| Al3+ (cmolc dm−3) | 0.30 | 0 | 0.05 | 0 | 0 | 0.10 |
| B (mg dm−3) | 0.28 | 0.40 | 0.61 | 0.33 | 0.29 | 0.47 |
| Ca2+ (cmolc dm−3) | 0.54 | 1.86 | 1.32 | 1.10 | 3.24 | 2.72 |
| Cu2+ (mg dm−3) | 0.06 | 0.07 | 0.07 | 0.08 | 0.05 | 0.05 |
| Fe2+ (mg dm−3) | 100.11 | 118.15 | 64.05 | 48.16 | 27.32 | 63.27 |
| K+ (cmolc dm−3) | 0.03 | 0.08 | 0.06 | 0.39 | 0.18 | 0.06 |
| Mg2+ (cmolc dm−3) | 0.49 | 0.85 | 0.67 | 0.55 | 1.18 | 0.98 |
| Mn2+ (mg dm−3) | 0.20 | 0.60 | 0.48 | 0.42 | 1.30 | 1.43 |
| P (mg dm−3) | 5.06 | 24.04 | 28.46 | 32.8 | 34.09 | 4.26 |
| S-SO42− (mg dm−3) | 5.94 | 7.37 | 6.32 | 6.22 | 7.46 | 7.37 |
| Zn2+ (mg dm−3) | 0.32 | 1.77 | 2.27 | 2.52 | 1.11 | 0.52 |
| Clay (%) | 15.99 | 16.19 | 12.71 | 11.79 | 17.94 | 17.22 |
| Silt (%) | 9.07 | 8.88 | 10.64 | 8.64 | 12.3 | 13.64 |
| Total sand (%) | 74.94 | 74.93 | 76.64 | 79.57 | 69.77 | 69.14 |
| Coarse sand (%) | 34.12 | 38.91 | 30.22 | 30.71 | 30.54 | 27.62 |
| Fine sand (%) | 40.82 | 36.02 | 46.42 | 48.86 | 39.23 | 41.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Funnicelli, M.I.G.; Lima, N.S.M.; Sartini, C.C.F.; de Macedo Lemos, E.G.; de Araújo Neto, R.B.; de Souza, H.A.; de Oliveira Junior, J.O.L.; Sagrilo, E.; Blanco, F.F.; de Freitas Andrade, H.A.; et al. Revealing the Bacteriome in Crop–Livestock–Forest Integration Systems in the Cerrado of MATOPIBA, Brazil. Forests 2025, 16, 626. https://doi.org/10.3390/f16040626
Funnicelli MIG, Lima NSM, Sartini CCF, de Macedo Lemos EG, de Araújo Neto RB, de Souza HA, de Oliveira Junior JOL, Sagrilo E, Blanco FF, de Freitas Andrade HA, et al. Revealing the Bacteriome in Crop–Livestock–Forest Integration Systems in the Cerrado of MATOPIBA, Brazil. Forests. 2025; 16(4):626. https://doi.org/10.3390/f16040626
Chicago/Turabian StyleFunnicelli, Michelli Inácio Gonçalves, Natália Sarmanho Monteiro Lima, Camila Cesário Fernandes Sartini, Eliana Gertrudes de Macedo Lemos, Raimundo Bezerra de Araújo Neto, Henrique Antunes de Souza, José Oscar Lustosa de Oliveira Junior, Edvaldo Sagrilo, Flavio Favaro Blanco, Hosana Aguiar de Freitas Andrade, and et al. 2025. "Revealing the Bacteriome in Crop–Livestock–Forest Integration Systems in the Cerrado of MATOPIBA, Brazil" Forests 16, no. 4: 626. https://doi.org/10.3390/f16040626
APA StyleFunnicelli, M. I. G., Lima, N. S. M., Sartini, C. C. F., de Macedo Lemos, E. G., de Araújo Neto, R. B., de Souza, H. A., de Oliveira Junior, J. O. L., Sagrilo, E., Blanco, F. F., de Freitas Andrade, H. A., de Sousa, D. C., Silva, M. L. d. N., Leite, L. F. C., Costa Lima, P. S. d., & Pinheiro, D. G. (2025). Revealing the Bacteriome in Crop–Livestock–Forest Integration Systems in the Cerrado of MATOPIBA, Brazil. Forests, 16(4), 626. https://doi.org/10.3390/f16040626








