Integrating Hyperspectral Images and LiDAR Data Using Vision Transformers for Enhanced Vegetation Classification
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.1.1. The Trento Dataset
2.1.2. The Houston 2013 Dataset
2.2. Methods
2.2.1. Overall Framework
2.2.2. Spectral–Spatial Extractor Module (SSE)
- 1.
- Feature HSI
- 2.
- Feature LiDAR
2.2.3. Multimodal Fusion Integrator (MFI)
2.2.4. Deep Feature Classifier (DFC)
- 1.
- Patch Embedding
- 2.
- Positional Encoding
- 3.
- Transformer Blocks
- 4.
- Classification
3. Experiments and Results
3.1. Experimental Setup
3.2. Implementation Details and Evaluation
3.2.1. Model Implementation and Training
- Optimizer and learning rate. The Adam optimizer was used for training with a learning rate of 0.01. A comparison among 0.001, 0.005, 0.01, and 0.05 indicated that 0.01 provided the best balance between convergence speed and accuracy. Betas and Epsilon were set to default values, supported by prior studies on ViT in RS.
- Loss function: Soft-Target Cross-Entropy was employed to enhance class separability and mitigate class imbalance, which is common in RS datasets.
- Batch size: A batch size of 32 was chosen, as it balances computational efficiency and generalization ability. Tests with 16, 32, and 64 showed that 32 provided stable gradient updates without excessive memory consumption.
- Number of training epochs: The model was trained for 100 epochs, determined through validation loss convergence analysis. Training with 50 epochs was insufficient, while exceeding 100 epochs led to diminishing returns and increased overfitting risk.
- Regularization techniques: To prevent overfitting, L2 regularization (weight decay = 1 × 10−4) and Dropout (0.3) were applied in the transformer layers, following best practices in DL.
3.2.2. Model Evaluation and Metrics
- Overall accuracy (OA): measures the proportion of correctly predicted observations to the total observations, providing a holistic view of model accuracy.
- Average accuracy (AA): represents the average of accuracies computed for each class, offering insights into class-wise performance.
- Kappa coefficient (): quantifies the agreement of prediction with the true labels, corrected by the randomness of agreement, serving as a robust statistic for classification accuracy.
3.3. Experimental Results
3.3.1. Performance on the Trento Dataset
3.3.2. Performance on the Houston 2013 Dataset
4. Discussion
4.1. Performance Comparison on the Trento Dataset
4.2. Performance Comparison on the Houston 2013 Dataset
4.3. Learning Curve Insights
4.4. Model Strengths and Advantages
- The integration of high-resolution HSI and LiDAR data enables PlantViT to effectively combine spectral and spatial features, which are essential for accurate vegetation and land cover classification. With its multimodal fusion capability, the model excels in detecting fine-scale landscape features, making it particularly advantageous for monitoring both urban and natural ecosystems.
- Advanced DL architecture: The incorporation of LightViT, a lightweight Vision Transformer, significantly enhances computational efficiency while preserving high classification accuracy. By selectively focusing on the most relevant features within the dataset, LightViT minimizes the need for overly complex network architectures and excessive computational resources, ensuring scalability for large-scale applications.
- Generalization across datasets: The consistent performance of PlantViT on both the Trento and Houston 2013 datasets highlights its robustness and adaptability across different geographical regions and land cover types. This strong generalization capability makes it well-suited for large-scale RS tasks, including vegetation analysis and land-use mapping.
4.5. Conclusion of Comparative Performance
5. Conclusions
- Cross-environment adaptability. By integrating HSI and LiDAR data, the model effectively enhances its generalization capability across diverse environmental conditions. Utilizing multiple datasets, including the Trento and Houston 2013 datasets, we demonstrated its adaptability across distinct geographical regions and plant communities. This advancement directly addresses a well-documented challenge in RS—ensuring classification accuracy across heterogeneous ecosystems.
- Overcoming CNN limitations in complex environments. Traditional convolutional neural networks (CNNs) often encounter difficulties in extracting fine-grained spatial–spectral features within intricate plant environments. To mitigate this issue, we incorporated involution-based operations, enabling the model to efficiently capture subtle feature variations while reducing the computational burden typically associated with CNN-based approaches. This enhancement is particularly advantageous for distinguishing morphologically similar plant species within highly heterogeneous landscapes.
- Cross-environment attention mechanism. The fusion of involution with LightViT introduces an innovative attention mechanism that dynamically adjusts to varying environmental contexts. This mechanism enables the model to selectively focus on the most relevant plant features, thereby improving classification robustness and accuracy across diverse ecosystems.
- Future work will involve evaluating the model on additional RS datasets to further assess its robustness and applicability to broader ecological and environmental monitoring tasks.
- Investigating the potential of transfer learning across diverse ecosystems could further enhance the model’s adaptability and generalization capabilities.
- Future research should explore the feasibility of deploying this model in real-time large-scale wetland monitoring. Additionally, incorporating supplementary data sources, such as UAV-based RGB imagery and satellite observations, could expand its practical applications and operational scalability.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ma, Y.; Zhao, Y.; Im, J.; Zhao, Y.; Zhen, Z. A Deep-Learning-Based Tree Species Classification for Natural Secondary Forests Using Unmanned Aerial Vehicle Hyperspectral Images and LiDAR. Ecol. Indic. 2024, 159, 111608. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, T.; Skidmore, A.K.; Holzwarth, S.; Heiden, U.; Heurich, M. Mapping Individual Silver Fir Trees Using Hyperspectral and LiDAR Data in a Central European Mixed Forest. Int. J. Appl. Earth Obs. Geoinf. 2021, 98, 102311. [Google Scholar] [CrossRef]
- de Almeida, D.R.A.; Broadbent, E.N.; Ferreira, M.P.; Meli, P.; Zambrano, A.M.A.; Gorgens, E.B.; Resende, A.F.; de Almeida, C.T.; do Amaral, C.H.; Corte, A.P.D.; et al. Monitoring Restored Tropical Forest Diversity and Structure through UAV-Borne Hyperspectral and Lidar Fusion. Remote Sens. Environ. 2021, 264, 112582. [Google Scholar] [CrossRef]
- Mohan, M.; Richardson, G.; Gopan, G.; Aghai, M.M.; Bajaj, S.; Galgamuwa, G.A.P.; Vastaranta, M.; Arachchige, P.S.P.; Amorós, L.; Corte, A.P.D.; et al. UAV-Supported Forest Regeneration: Current Trends, Challenges and Implications. Remote Sens. 2021, 13, 2596. [Google Scholar] [CrossRef]
- Guo, F.; Li, Z.; Meng, Q.; Ren, G.; Wang, L.; Wang, J.; Qin, H.; Zhang, J. Semi-Supervised Cross-Domain Feature Fusion Classification Network for Coastal Wetland Classification with Hyperspectral and LiDAR Data. Int. J. Appl. Earth Obs. Geoinf. 2023, 120, 103354. [Google Scholar] [CrossRef]
- Baena, S.; Moat, J.; Whaley, O.; Boyd, D.S. Identifying Species from the Air: UAVs and the Very High Resolution Challenge for Plant Conservation. PLoS ONE 2017, 12, e0188714. [Google Scholar] [CrossRef]
- Roy, S.K.; Deria, A.; Hong, D.; Rasti, B.; Plaza, A.; Chanussot, J. Multimodal Fusion Transformer for Remote Sensing Image Classification. IEEE Trans. Geosci. Remote Sens. 2023, 61, 5515620. [Google Scholar] [CrossRef]
- Wang, R.; Ma, L.; He, G.; Johnson, B.A.; Yan, Z.; Chang, M.; Liang, Y. Transformers for Remote Sensing: A Systematic Review and Analysis. Sensors 2024, 24, 3495. [Google Scholar] [CrossRef]
- Asner, G.P.; Ustin, S.L.; Townsend, P.; Martin, R.E. Forest Biophysical and Biochemical Properties from Hyperspectral and LiDAR Remote Sensing. In Remote Sensing Handbook, Volume IV; CRC Press: Boca Raton, FL, USA, 2024; ISBN 978-1-003-54117-2. [Google Scholar]
- Adegun, A.A.; Viriri, S.; Tapamo, J.-R. Review of Deep Learning Methods for Remote Sensing Satellite Images Classification: Experimental Survey and Comparative Analysis. J. Big Data 2023, 10, 93. [Google Scholar] [CrossRef]
- Dao, P.D.; Axiotis, A.; He, Y. Mapping Native and Invasive Grassland Species and Characterizing Topography-Driven Species Dynamics Using High Spatial Resolution Hyperspectral Imagery. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102542. [Google Scholar] [CrossRef]
- Li, Q.; Wong, F.K.K.; Fung, T. Mapping Multi-Layered Mangroves from Multispectral, Hyperspectral, and LiDAR Data. Remote Sens. Environ. 2021, 258, 112403. [Google Scholar] [CrossRef]
- Yang, C. Remote Sensing and Precision Agriculture Technologies for Crop Disease Detection and Management with a Practical Application Example. Engineering 2020, 6, 528–532. [Google Scholar] [CrossRef]
- Esmaeili, M.; Abbasi-Moghadam, D.; Sharifi, A.; Tariq, A.; Li, Q. ResMorCNN Model: Hyperspectral Images Classification Using Residual-Injection Morphological Features and 3DCNN Layers. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2024, 17, 219–243. [Google Scholar] [CrossRef]
- Norton, C.L.; Hartfield, K.; Collins, C.D.H.; van Leeuwen, W.J.D.; Metz, L.J. Multi-Temporal LiDAR and Hyperspectral Data Fusion for Classification of Semi-Arid Woody Cover Species. Remote Sens. 2022, 14, 2896. [Google Scholar] [CrossRef]
- Dang, Y.; Zhang, X.; Zhao, H.; Liu, B. DCTransformer: A Channel Attention Combined Discrete Cosine Transform to Extract Spatial–Spectral Feature for Hyperspectral Image Classification. Appl. Sci. 2024, 14, 1701. [Google Scholar] [CrossRef]
- Kuras, A.; Brell, M.; Rizzi, J.; Burud, I. Hyperspectral and Lidar Data Applied to the Urban Land Cover Machine Learning and Neural-Network-Based Classification: A Review. Remote Sens. 2021, 13, 3393. [Google Scholar] [CrossRef]
- Zhang, M.; Li, W.; Zhang, Y.; Tao, R.; Du, Q. Hyperspectral and LiDAR Data Classification Based on Structural Optimization Transmission. IEEE Trans. Cybern. 2023, 53, 3153–3164. [Google Scholar] [CrossRef]
- Kashefi, R.; Barekatain, L.; Sabokrou, M.; Aghaeipoor, F. Explainability of Vision Transformers: A Comprehensive Review and New Perspectives. arXiv 2023, arXiv:2311.06786. [Google Scholar]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-Based Learning Applied to Document Recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Li, J.; Hong, D.; Gao, L.; Yao, J.; Zheng, K.; Zhang, B.; Chanussot, J. Deep Learning in Multimodal Remote Sensing Data Fusion: A Comprehensive Review. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102926. [Google Scholar] [CrossRef]
- Shen, X.; Deng, X.; Geng, J.; Jiang, W. SFE-FN: A Shuffle Feature Enhancement-Based Fusion Network for Hyperspectral and LiDAR Classification. IEEE Geosci. Remote Sens. Lett. 2023, 20, 5501605. [Google Scholar] [CrossRef]
- Shi, Y.; Skidmore, A.K.; Wang, T.; Holzwarth, S.; Heiden, U.; Pinnel, N.; Zhu, X.; Heurich, M. Tree Species Classification Using Plant Functional Traits from LiDAR and Hyperspectral Data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 207–219. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, L. A Review on Unmanned Aerial Vehicle Remote Sensing: Platforms, Sensors, Data Processing Methods, and Applications. Drones 2023, 7, 398. [Google Scholar] [CrossRef]
- Ding, J.; Ren, X.; Luo, R. An Adaptive Learning Method for Solving the Extreme Learning Rate Problem of Transformer. In Proceedings of the Natural Language Processing and Chinese Computing; Liu, F., Duan, N., Xu, Q., Hong, Y., Eds.; Springer: Cham, Switzerland, 2023; pp. 361–372. [Google Scholar]
- Tusa, E.; Laybros, A.; Monnet, J.-M.; Dalla Mura, M.; Barré, J.-B.; Vincent, G.; Dalponte, M.; Féret, J.-B.; Chanussot, J. Chapter 2.11—Fusion of Hyperspectral Imaging and LiDAR for Forest Monitoring. In Data Handling in Science and Technology; Amigo, J.M., Ed.; Hyperspectral Imaging; Elsevier: Amsterdam, The Netherlands, 2019; Volume 32, pp. 281–303. [Google Scholar]
- Huang, T.; Huang, L.; You, S.; Wang, F.; Qian, C.; Xu, C. LightViT: Towards Light-Weight Convolution-Free Vision Transformers. arXiv 2022, arXiv:2207.05557. [Google Scholar]
- Wäldchen, J.; Rzanny, M.; Seeland, M.; Mäder, P. Automated Plant Species Identification—Trends and Future Directions. PLoS Comput. Biol. 2018, 14, e1005993. [Google Scholar] [CrossRef]
- Ding, M.; Xiao, B.; Codella, N.; Luo, P.; Wang, J.; Yuan, L. DaViT: Dual Attention Vision Transformers. In Proceedings of the Computer Vision—ECCV 2022, Tel Aviv, Israel, 23–27 October 2022; Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T., Eds.; Springer: Cham, Switzerland, 2022; pp. 74–92. [Google Scholar]
- Li, D.; Hu, J.; Wang, C.; Li, X.; She, Q.; Zhu, L.; Zhang, T.; Chen, Q. Involution: Inverting the Inherence of Convolution for Visual Recognition. arXiv 2021, arXiv:2103.06255. [Google Scholar]
- Available online: https://github.com/shuxquan/PlantViT/tree/main/trento (accessed on 30 March 2025).
- Available online: https://github.com/shuxquan/PlantViT/tree/main/Houston%202013 (accessed on 30 March 2025).
- Hong, D.; Gao, L.; Hang, R.; Zhang, B.; Chanussot, J. Deep Encoder–Decoder Networks for Classification of Hyperspectral and LiDAR Data. IEEE Geosci. Remote Sens. Lett. 2022, 19, 5500205. [Google Scholar] [CrossRef]
- Wang, J.; Tan, X. Mutually Beneficial Transformer for Multimodal Data Fusion. IEEE Trans. Circuits Syst. Video Technol. 2023, 33, 7466–7479. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Shi, Y.; Lai, J.; Tan, X. AM3Net: Adaptive Mutual-Learning-Based Multimodal Data Fusion Network. IEEE Trans. Circuits Syst. Video Technol. 2022, 32, 5411–5426. [Google Scholar] [CrossRef]
- Wang, W.; Liu, L.; Zhang, T.; Shen, J.; Wang, J.; Li, J. Hyper-ES2T: Efficient Spatial–Spectral Transformer for the Classification of Hyperspectral Remote Sensing Images. Int. J. Appl. Earth Obs. Geoinf. 2022, 113, 103005. [Google Scholar] [CrossRef]
- Song, T.; Zeng, Z.; Gao, C.; Chen, H.; Li, J. Joint Classification of Hyperspectral and LiDAR Data Using Height Information Guided Hierarchical Fusion-and-Separation Network. IEEE Trans. Geosci. Remote Sens. 2024, 62, 5505315. [Google Scholar] [CrossRef]
- Xu, X.; Li, W.; Ran, Q.; Du, Q.; Gao, L.; Zhang, B. Multisource Remote Sensing Data Classification Based on Convolutional Neural Network. IEEE Trans. Geosci. Remote Sens. 2018, 56, 937–949. [Google Scholar] [CrossRef]
- Ma, Y.; Lan, Y.; Xie, Y.; Yu, L.; Chen, C.; Wu, Y.; Dai, X. A Spatial–Spectral Transformer for Hyperspectral Image Classification Based on Global Dependencies of Multi-Scale Features. Remote Sens. 2024, 16, 404. [Google Scholar] [CrossRef]
- Roy, S.K.; Deria, A.; Shah, C.; Haut, J.M.; Du, Q.; Plaza, A. Spectral–Spatial Morphological Attention Transformer for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2023, 61, 5503615. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, J.; Feng, Y.; Wang, L. MS2CANet: Multiscale Spatial–Spectral Cross-Modal Attention Network for Hyperspectral Image and LiDAR Classification. IEEE Geosci. Remote Sens. Lett. 2024, 21, 5501505. [Google Scholar] [CrossRef]
- Shen, X.; Zhu, H.; Meng, C. Refined Feature Fusion-Based Transformer Network for Hyperspectral and LiDAR Classification. In Proceedings of the 2023 International Conference on Image Processing, Computer Vision and Machine Learning (ICICML), Chengdu, China, 3–5 November 2023; pp. 789–793. [Google Scholar]
- Liu, H.; Li, W.; Xia, X.-G.; Zhang, M.; Tao, R. Multiarea Target Attention for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2023, 61, 5524916. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Z.; Li, A.; Jiang, H. Cross-Stage Fusion Network Based Multi-Modal Hyperspectral Image Classification. In Proceedings of the 6GN for Future Wireless Networks, Shanghai, China, 7–8 October 2023; Li, A., Shi, Y., Xi, L., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2023; pp. 77–88. [Google Scholar]
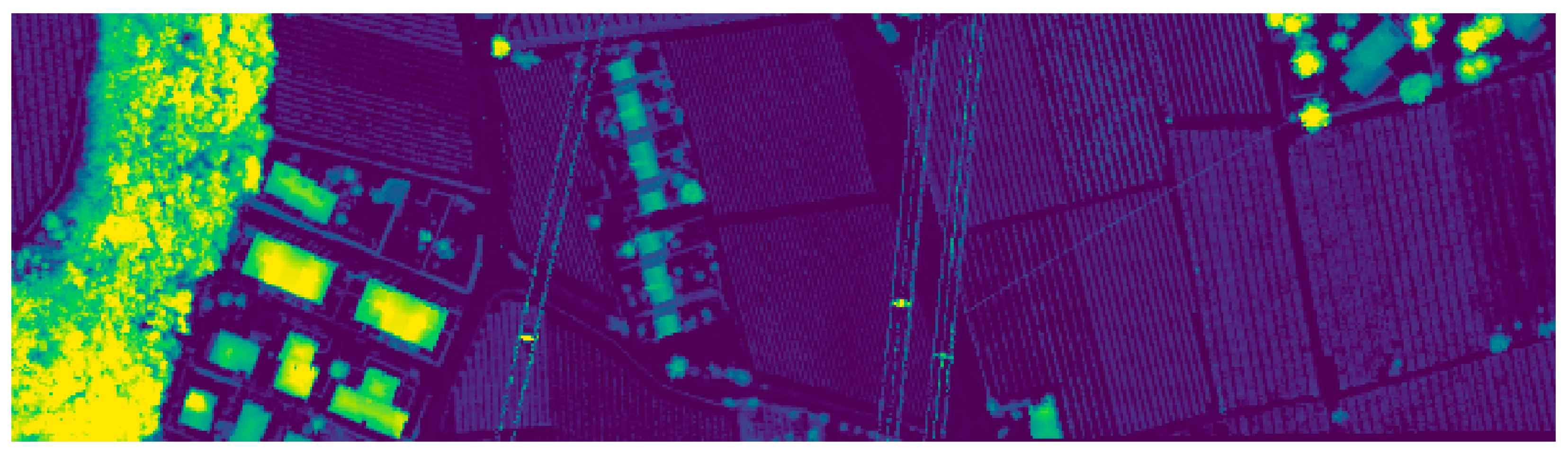

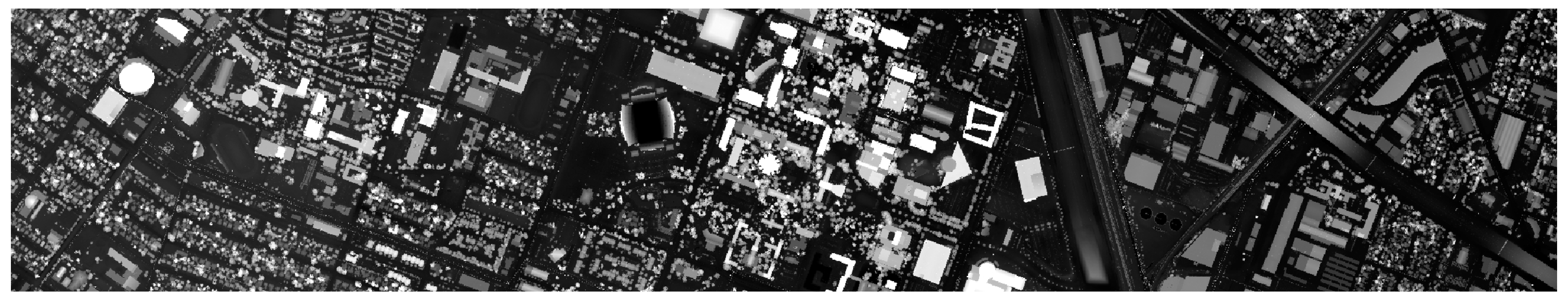

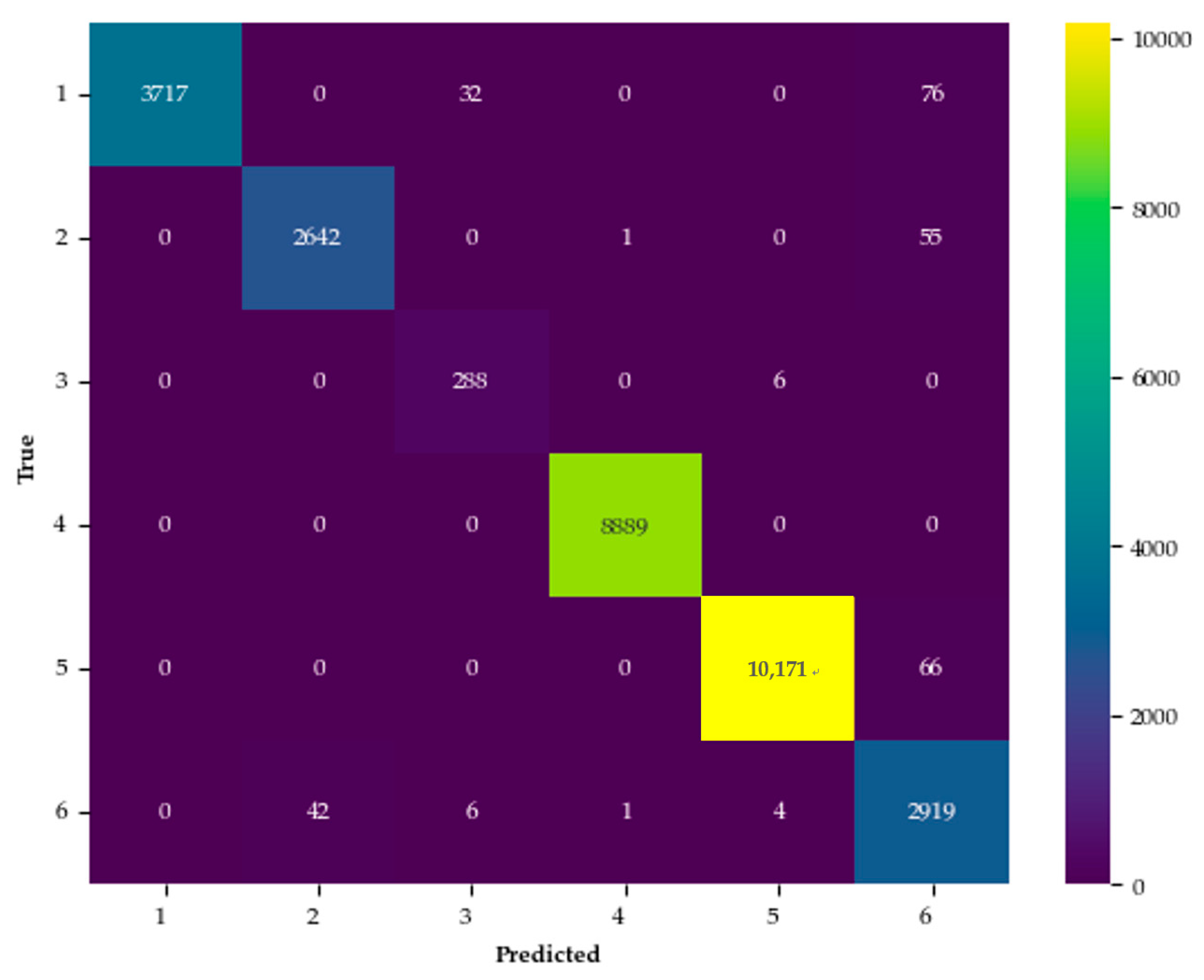

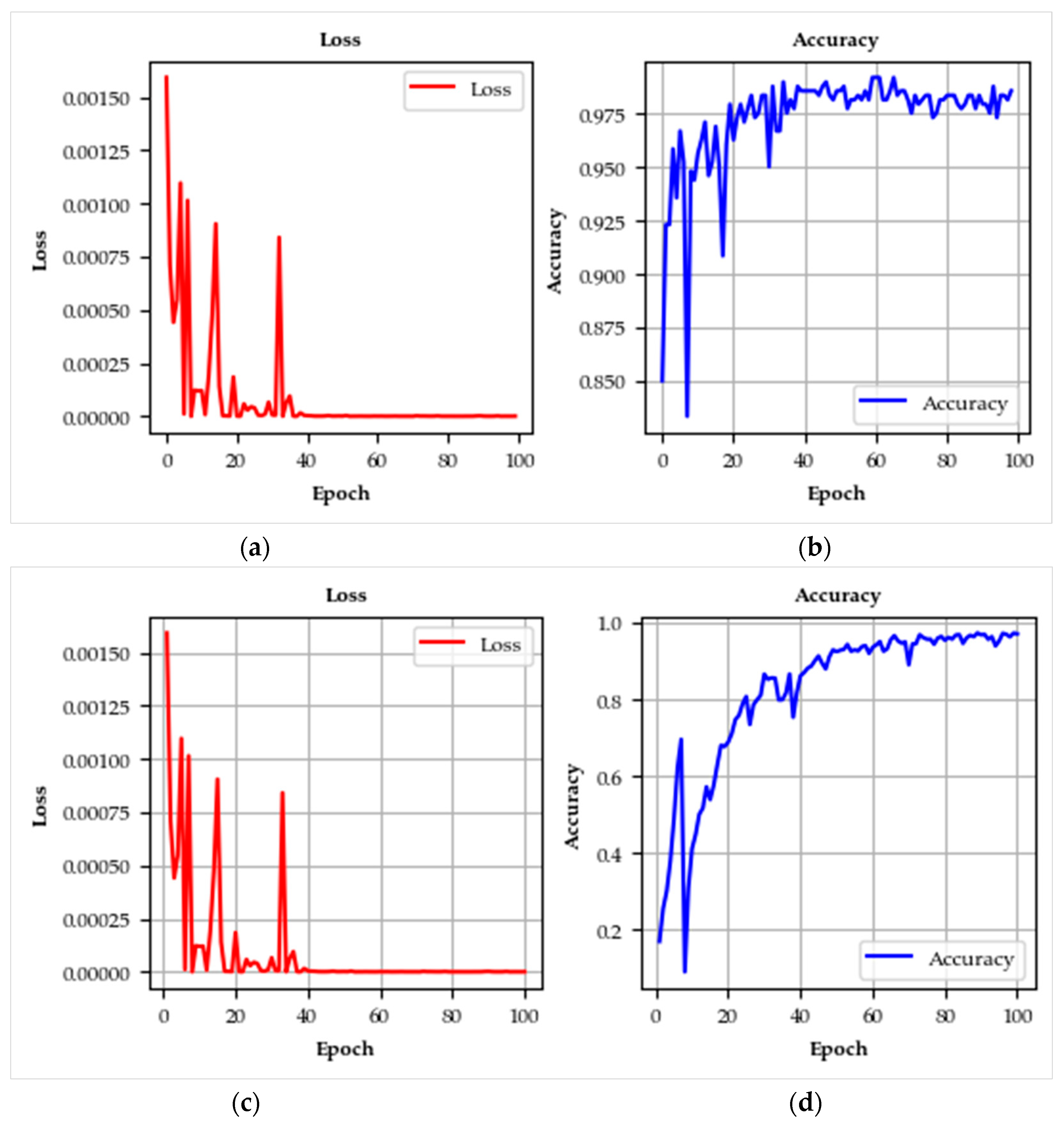
| No. | Class Name | Training | Test | Samples |
|---|---|---|---|---|
| 1 | Apple Trees | 129 | 3905 | 4034 |
| 2 | Buildings | 125 | 2778 | 2903 |
| 3 | Ground | 105 | 374 | 479 |
| 4 | Wood | 154 | 8969 | 9123 |
| 5 | Vineyard | 184 | 10,317 | 10,501 |
| 6 | Roads | 122 | 3052 | 3174 |
| Total | 819 | 29,395 | 30,214 |
| No. | Class Name | Training | Test | Samples |
|---|---|---|---|---|
| 1 | Healthy Grass | 198 | 1053 | 1251 |
| 2 | Stressed Grass | 190 | 1064 | 1254 |
| 3 | Synthetic Grass | 192 | 505 | 697 |
| 4 | Tree | 188 | 1056 | 1244 |
| 5 | Soil | 186 | 1056 | 1242 |
| 6 | Water | 182 | 143 | 325 |
| 7 | Residential | 196 | 1072 | 1268 |
| 8 | Commercial | 191 | 1053 | 1244 |
| 9 | Road | 193 | 1059 | 1252 |
| 10 | Highway | 191 | 1036 | 1227 |
| 11 | Railway | 181 | 1054 | 1235 |
| 12 | Parking Lot 1 | 192 | 1041 | 1233 |
| 13 | Parking Lot 2 | 184 | 285 | 469 |
| 14 | Tennis Court | 181 | 247 | 428 |
| 15 | Running Track | 187 | 473 | 660 |
| Total | 2832 | 12,197 | 15,029 |
| No. | Class Name | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| 1 | Apple trees | 97.18 | 99.18 | 97.33 | 98.25 |
| 2 | Buildings | 97.92 | 98.23 | 97.92 | 98.07 |
| 3 | Ground | 97.96 | 95.68 | 97.96 | 96.81 |
| 4 | Wood | 100.00 | 99.99 | 99.99 | 99.99 |
| 5 | Vineyard | 99.36 | 99.35 | 99.35 | 99.35 |
| 6 | Roads | 98.22 | 96.82 | 97.46 | 97.14 |
| OA | 99.00 | ||||
| AA | 98.44 | ||||
| 98.66 | |||||
| No. | Class Name | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|---|
| 1 | Healthy Grass | 97.15 | 97.00 | 98.00 | 98.00 |
| 2 | Stressed Grass | 97.46 | 97.00 | 98.00 | 98.00 |
| 3 | Synthetic Grass | 99.80 | 99.00 | 99.00 | 99.00 |
| 4 | Trees | 95.55 | 100.00 | 98.00 | 97.00 |
| 5 | Soil | 100.00 | 100.00 | 100.00 | 100.00 |
| 6 | Water | 97.20 | 97.00 | 98.00 | 98.00 |
| 7 | Residential | 92.82 | 97.00 | 95.00 | 94.00 |
| 8 | Commercial | 96.39 | 96.00 | 96.00 | 95.00 |
| 9 | Road | 97.92 | 98.00 | 97.00 | 97.00 |
| 10 | Highway | 100.00 | 100.00 | 100.00 | 100.00 |
| 11 | Railway | 96.39 | 97.00 | 96.00 | 95.00 |
| 12 | Parking Lot 1 | 99.62 | 100.00 | 99.00 | 99.00 |
| 13 | Parking Lot 2 | 98.60 | 100.00 | 99.00 | 99.00 |
| 14 | Tennis Court | 100.00 | 100.00 | 100.00 | 100.00 |
| 15 | Running Track | 94.93 | 95.00 | 96.00 | 94.00 |
| OA | 97.41 | ||||
| AA | 97.59 | ||||
| 97.19 | |||||
| No. | Class Name | PlantViT | EndNet [33] | DCT [16] | MBF [34] | AM3Net [35] | HES2T [36] | BTRF [37] | TTCNN [38] |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Apple Trees | 97.18 | 88.19 | 99.13 | 98.71 | 97.12 | 99.46 | 95.92 | 98.07 |
| 2 | Buildings | 97.92 | 98.49 | 99.44 | 98.87 | 96.73 | 91.65 | 98.04 | 95.21 |
| 3 | Ground | 97.96 | 95.19 | 99.94 | 97.79 | 98.96 | 87.43 | 80.33 | 93.32 |
| 4 | Wood | 100.00 | 99.30 | 92.76 | 99.89 | 99.12 | 98.33 | 99.83 | 99.93 |
| 5 | Vineyard | 99.36 | 91.96 | 86.37 | 99.25 | 98.22 | 97.65 | 99.29 | 98.78 |
| 6 | Roads | 98.22 | 90.14 | 92.78 | 92.38 | 93.76 | 68.94 | 97.66 | 89.98 |
| OA | 99.00 | 94.17 | 97.45 | 98.62 | 97.75 | 94.42 | 98.66 | 97.92 | |
| AA | 98.44 | 93.88 | 95.07 | 98.14 | 97.32 | 90.58 | 95.30 | 96.19 | |
| 98.66 | 92.22 | 96.58 | 97.81 | 97.00 | 92.54 | 98.20 | 96.81 |
| No. | Class Name | PlantViT | MSST [39] | mFormer [40] | S2CA [41] | RFFT [42] | MATA [43] | CSF [44] | SFE-FN [22] |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Healthy Grass | 97.15 | 89.12 | 82.37 | 84.62 | 87.96 | 91.64 | 82.72 | 95.06 |
| 2 | Stressed Grass | 97.46 | 92.85 | 85.03 | 95.68 | 95.90 | 83.83 | 82.24 | 96.52 |
| 3 | Synthetic Grass | 99.80 | 95.43 | 98.09 | 100.00 | 97.41 | 100.00 | 100.00 | 100.00 |
| 4 | Trees | 95.55 | 91.13 | 95.23 | 99.62 | 97.74 | 99.24 | 92.52 | 100.00 |
| 5 | Soil | 100.00 | 100.00 | 99.15 | 100.00 | 95.32 | 99.34 | 98.67 | 99.81 |
| 6 | Water | 97.20 | 86.31 | 98.14 | 100.00 | 97.84 | 100.00 | 95.10 | 97.20 |
| 7 | Residential | 92.82 | 72.35 | 89.61 | 87.03 | 92.57 | 85.91 | 80.97 | 93.00 |
| 8 | Commercial | 96.39 | 76.87 | 72.33 | 93.02 | 95.42 | 94.59 | 81.67 | 88.98 |
| 9 | Road | 97.92 | 79.40 | 88.86 | 89.71 | 91.74 | 91.78 | 86.87 | 87.44 |
| 10 | Highway | 100.00 | 99.57 | 61.97 | 71.43 | 99.10 | 91.12 | 79.63 | 98.17 |
| 11 | Railway | 96.39 | 98.06 | 96.24 | 98.96 | 96.86 | 97.91 | 83.30 | 93.55 |
| 12 | Parking Lot 1 | 99.62 | 98.09 | 94.46 | 98.94 | 94.56 | 95.39 | 88.95 | 96.35 |
| 13 | Parking Lot 2 | 98.60 | 80.35 | 87.02 | 85.67 | 95.30 | 92.98 | 87.37 | 81.40 |
| 14 | Tennis Court | 100.00 | 100.00 | 99.73 | 91.90 | 93.22 | 100.00 | 100.00 | 100.00 |
| 15 | Running Track | 94.93 | 100.00 | 96.69 | 100.00 | 97.57 | 100.00 | 99.52 | 100.00 |
| OA | 97.41 | 90.29 | 87.85 | 92.52 | 95.01 | 93.83 | 89.42 | 95.11 | |
| AA | 97.59 | 90.63 | 89.66 | 93.11 | 95.23 | 94.92 | 89.24 | 95.17 | |
| 97.19 | 89.50 | 86.81 | 92.52 | 94.60 | 93.31 | 88.51 | 94.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, X.; Ma, L.; Chang, F. Integrating Hyperspectral Images and LiDAR Data Using Vision Transformers for Enhanced Vegetation Classification. Forests 2025, 16, 620. https://doi.org/10.3390/f16040620
Shu X, Ma L, Chang F. Integrating Hyperspectral Images and LiDAR Data Using Vision Transformers for Enhanced Vegetation Classification. Forests. 2025; 16(4):620. https://doi.org/10.3390/f16040620
Chicago/Turabian StyleShu, Xingquan, Limin Ma, and Fengqin Chang. 2025. "Integrating Hyperspectral Images and LiDAR Data Using Vision Transformers for Enhanced Vegetation Classification" Forests 16, no. 4: 620. https://doi.org/10.3390/f16040620
APA StyleShu, X., Ma, L., & Chang, F. (2025). Integrating Hyperspectral Images and LiDAR Data Using Vision Transformers for Enhanced Vegetation Classification. Forests, 16(4), 620. https://doi.org/10.3390/f16040620







