1. Introduction
Image segmentation is the process of dividing the pixel points of the original image into different categories of sub-regions according to attributes such as color, texture, and intensity value, and then completing the process of extracting the target in the image [
1]. Standard segmentation methods include the thresholding method [
2,
3,
4,
5], region technique [
6], watershed algorithm [
7], and deep learning [
8,
9,
10]. The region-based segmentation method has a severe segmentation deficiency for aggregated particle adhesion [
11]. The watershed algorithm has the disadvantage of over-segmentation and reflected light interfering with the image [
12]. Deep learning is widely used in forest canopy image segmentation owing to its high segmentation accuracy, robustness, adaptability, and ability to handle large-scale data. U-Net, Mask R-CNN and Transformers are classical segmentation modules in deep learning. However, segmentation methods based on deep learning also suffer from high training cost. They are long and time-consuming, have limitations in being sensitive to noise, and are not suitable for segmenting small-sample images with little feature information and high randomness. Compared to the deep learning method, the threshold segmentation method utilizes the information in the image histogram to find the threshold value of the image. It distinguishes different target and background regions by setting multiple threshold values [
13]. Since it relies on simple mathematical operations, it outperforms complex deep learning models in terms of processing speed. The key to threshold segmentation is selecting an appropriate segmentation method to determine the segmentation threshold. Threshold selection methods based on information entropy have attracted considerable attention owing to their clear mathematical concepts. Otsu entropy [
14], Renyi entropy [
15], and Kapur entropy [
16] are typical information entropy methods. The Otsu method selects segmentation thresholds to maximize the variance between the target and background classes. It is suitable for the case where the area occupied by the target and the background in the image are close, and the Otsu segmentation method will be invalid when the area occupied by the two is disparate [
17]. The essence of Renyi entropy is to use the subordination function to convert the image histogram information into a fuzzy domain and then obtain the optimal threshold value to segment the image based on the principle of maximum entropy [
18]. The adjustable parameter
is an essential parameter in Renyi entropy, and the value of
determines the form of Renyi entropy. Although different segmentation effects can be obtained by adjusting
, it is a challenge to select the optimal parameter reasonably. Improper selection may lead to over-segmentation or under-segmentation phenomenon. The segmentation principle of Kapur entropy is that the total entropy value of the target class and the background class is the maximum. Because Kapur entropy is based on the grey level intra-class probability when calculating the entropy value, it is not sensitive to the size of the divided target and background, can better retain small targets in the image [
19], and is widely used in color image segmentation.
Forest canopy images have various complex backgrounds, and the gray levels vary widely, requiring multiple thresholds for accurate segmentation. Traditional image segmentation methods use enumeration, and when performing threshold segmentation, each combination must be individually tested to select the optimal threshold. The computational complexity increases exponentially with an increase in the number of thresholds. To overcome these limitations, scholars view finding the optimal thresholds in multithreshold segmentation as an optimization problem with constraints. Heuristic optimization algorithms search for optimal thresholds to reduce the computational complexity. Intelligent optimization algorithms, such as the Golden Jackal Optimization Algorithm [
20], Elephant Herding Optimization Algorithm [
21], Sand Cat Optimization Algorithm [
22], and Dung Beetle Optimization Algorithm [
23] have been successfully applied to image segmentation. Studies have shown that the performance of intelligent algorithms significantly affects the segmentation accuracy and efficiency of the images. Therefore, improving intelligent algorithms to enhance their performance and image segmentation accuracy and efficiency has become the focus of research. Sukanta Nama proposes a quasi-reflected slime mold (QRSMA) method combined with a quasi-reflective learning mechanism. The results of the CEC2020 benchmark function tests showed that QRSMA outperformed the statistical, convergence, and diversity measures of the slime mold algorithm. It also outperformed other existing methods for segmenting COVID-19 X-ray images [
24]. Jiquan Wang et al. proposed a Whale Optimization Algorithm combining mutation and de-similarity to address the limitations of the whale optimization Algorithm easily falling into local optimal solutions. Multilevel threshold segmentation was performed on ten grayscale images using Kapur and Otsu as the objective function. The experimental results showed that CRWOA outperformed the five compared algorithms in terms of convergence and segmentation quality [
25]. Essam H. Houssein et al. proposed a chimpanzee optimization algorithm (IChOA) based on the strategy of contrastive learning and Levy Flight improvement. IChOA was tested on a dataset from the database of infrared images using Otsu and Kapur methods. The experimental results show that the IChOA is robust for segmenting various positive and negative cases [
26]. Laith Abualigah et al. proposed a novel hybrid algorithm (MPASSA) to determine the optimal thresholds in multi-threshold segmentation by borrowing the properties of the Marine Predator and Salp Swarm Algorithm. Multiple benchmark images were used to verify the performance of the MPASSA. The experimental results show that MPASSA avoids the problem of falling into the local optima. It also has a strong solution capability for image-segmentation problems [
27].
To address the problems of poor segmentation accuracy of forest canopy images due to complex structure and uneven illumination, Wu et al. proposed a segmentation method based on differential evolutionary whale optimization algorithm [
28]; Tong et al. proposed a segmentation method based on improved northern goshawk algorithm by using multi-strategy fusion [
29]; Wang et al. proposed a segmentation method based on improved tuna swarm optimization [
30]. The experimental results show that compared with the original algorithm, the improved algorithm obtains higher segmentation accuracy in segmenting forest canopy images. It proves the effectiveness and necessity of the improved intelligent algorithm in multi-threshold segmentation of forest canopy images. In recent years, scholars have been enthusiastic about innovative algorithms, and many intelligent algorithms with excellent performance have appeared, bringing new vitality to the research on image multi-threshold segmentation. The BMO is a novel bionic evolutionary optimization algorithm proposed by Sulaiman et al. (2018) that simulates the mating behavior of barnacles in nature to solve the optimization problem [
31]. Since its proposal, it has received extensive attention from scholars. In [
32], quasi-inverse dyadic learning and logistic chaotic mapping were used to solve the problem of the premature convergence of the BMO algorithm. Literature [
33] used random wandering was used to replace the random logarithm
in the remote insemination iteration formula. The parameter
p was replaced with a logistic chaotic mapping operator to improve the problem of premature convergence of the BMO algorithm. Literature [
34] improved the penis coefficient
and the remote insemination iterative formula using a logistic model to achieve adaptive transformation of parameters as well as to avoid falling into local optima. In [
35], cubic chaos initialization was used to improve the diversity of the initial population, the parameter
p was replaced with a hyperbolic sinusoidal modulation factor to improve the convergence speed of the algorithm, and Gaussian and Cosey variants were introduced to solve the shortcomings of the BMO, which is prone to fall into local optimother. The literature [
36] integrates barnacle larvae settlement and adhesion behavior to increase population diversity and improve the global exploration performance of the algorithm. It also uses positive and negative decline casting strategies to improve the local mining ability.
The BMO algorithm exhibited an excellent performance and fewer control parameters. However, these parameters have a more significant impact on the performance of the BMO algorithm. Many improvements have also focused on the importance of the parameters of the BMO algorithm, and the direction of its improvement is almost always centered around the parameters. This is explicitly embodied in the following aspects: (1) The fixed penis coefficient makes the exploration and exploitation process unbalanced, although the literature [
34] realized the adaptive conversion of
, the range of variation of
was small, and there was not much difference with the fixed penis coefficient. (2) The Hardy-Weinberg law coefficient
p plays a decisive role in the exploitation ability of the BMO algorithm, and fixed coefficients cannot maintain the diversity of the populations, which weakens the exploitation ability of the BMO algorithm. Literature [
33] utilized Logistic Chaos coefficients instead of the parameter
p. Although Logistic Chaos coefficients have strong ergodicity, their iteration sequences are unevenly distributed, which is not conducive to increasing population diversity. Reference [
35] designed a nonlinearly decreasing parameter
p in the interval
, which aims to accelerate the convergence speed of the algorithm, but
p tends to 0 in the late iteration, which also results in the loss of population diversity. In addition, to improve the ability of the BMO algorithm to jump out of the local optimal solution, ref. [
35] added a double perturbation, which effectively enhances the ability of the algorithm to jump out of the local optimal solution but also slows down the convergence speed of the algorithm.
This paper proposes a PEBMO algorithm and applies it to the multi-threshold segmentation of forest canopy images. Specifically, the main contributions of this study are (a) to address the limitations of the BMO algorithm, a wide range of nonlinear increasing penis coefficients, dynamically decreasing reproduction coefficients, and Chebyshev chaotic perturbation strategies are employed to improve the balance between exploration and exploitation, the exploitation ability, and the resistance to premature maturation of the BMO algorithm; (b) the PEBMO algorithm is combined with Kapur entropy to determine the optimal segmentation threshold to improve the segmentation efficiency and accuracy of forest canopy images; and (c) to investigate the performance of PEBMO and PEBMO-based segmentation methods on the 2017 test function and forest canopy image problems, respectively. (d) We compared PEBMO with five BMO variant algorithms. PEBMO performed better than the other BMO-variant algorithms. The forest canopy image segmentation results show that PEBMO-based segmentation methods outperform the artificial bee colony optimization algorithm (ABC) [
37], chimp optimization algorithm (ChOA) [
38], cuckoo algorithm (CS) [
39], flower pollination algorithm (FPA) [
40], mayfly algorithm (MA) [
41], improved mayfly algorithm (IMA) [
42], and BMO.
2. Materials and Methods
2.1. Materials
The canopy images used in the experiment were obtained from the Liangshui Experimental Forestry Farm of Northeast Forestry University. They were acquired randomly by a Panasonic DMC-LX5 camera (Panasonic Company, Kadoma City, Osaka, Japan) fisheye lens Samyang AE 8/3.5 Aspherical IF MC Fisheye (Samyang Optics, Changwon, Gyeongsangnam-do, Republic of Korea). The Liangshui Experimental Forest Farm is located in the southern section of the Xiaoxinganling Mountain Range (eastern slope of the Dali Belt Range branch) in the eastern mountains of northeast China. The geographic coordinates are 128°–128° E, 47°–47° N. This location is on the eastern edge of the Eurasia continent, which is deeply influenced by the oceanic climate and has prominent temperate continental monsoon climate characteristics. In winter, under the control of the metamorphic continental air masses, the climate is cold, dry, and windy. In summer, under the influence of subtropical metamorphic oceanic air masses, precipitation is concentrated (of which June–August accounts for more than 60% of annual precipitation), and temperature is high. The climate in spring and fall is variable and spring is windy, with little precipitation, and prone to drought. In the fall, the temperature drops sharply, and early frosts occur more often. The soils in the region are divided into four soil classes (drenching soil class, Semi-hydro pedogenic class, pedogenic class, and organic soil class), four soil classes (dark brown loam, meadow soil, swamp soil, and peat soil), and 14 subclasses. Because the altitude of this area is not high, there is no apparent high mountain, and the vertical distribution of soil is not apparent, only a territorial distribution pattern. Red pine, spruce forests and birch forests dominate in the forest field, accounting for 63.7%, 11.4% and 6.4% of the forested area respectively. The forest cover rate is 98%.
2.2. Basic BMO
Initialization: In the BMO algorithm, barnacles are candidate solutions and the following matrix represents their population vectors:
N is the number of control variables (problem dimensions) and
n is the population size or the number of barnacles. The upper-and lower-bound constraints for each control variable are as follows:
where
and
denote the upper and lower bounds of the
ith variable, respectively. First, the initial population
X is evaluated, then the fitness value of each barnacle is ranked, and the barnacle with the best current fitness value is ranked at the top of
X.
Selection: Randomly selecting the father and mother to be mated from the barnacle population, the formulae for selection are shown in (4) and (5):
where
denotes the father to be mated and
denotes the mother to be mated.
Reproduction: The value
is essential to determine the exploration and exploitation processes. The value
in the basic BMO algorithm is set to 7. If
, the barnacle algorithm reproduces the offspring according to the Hardy-Weinberg theorem, which is the exploitation phase of the BMO algorithm. The barnacle mating iteration formula is given by (6).
where
p and
q are the percentages of characteristics of the father and mother in the next generation, respectively. Where
p is a random number uniformly distributed in
and
, and are the variables of father and mother selected by Equations (4) and (5), respectively.
If
, the offspring are produced by remote insemination, and this process can be considered as an exploration phase with the iterative formula shown in (7):
where
is a random number within
, which can be seen from Equation (7) that the mother determines the generation of offspring during remote insemination.
2.3. PEBMO
2.3.1. Large Range of Non-Linear Increasing Penis Coefficients
The penis coefficient
directly determines the transition between the exploration and exploitation of BMO algorithms. In the basic BMO algorithm,
is fixed value 7. A fixed penis coefficient defeats the original purpose of the metaheuristic algorithm of dynamic balance between exploration and exploitation. Therefore, it is necessary to design a dynamic adaptive penis coefficient to improve the flexibility between exploration and exploitation. The improved
parameter is given by Equation (8).
where
t is the current number of iterations, and
T is the maximum number of iterations.
As can be seen from Equation (8), consists of three parts: is the range adjustment part that determines the overall range of . was initially designed to utilize the nonlinearity of to enhance the balance between algorithm exploration and exploitation, and the nonlinearity of the exponential function meets this requirement. The designed increases nonlinearly in the interval [7, 26.95], which makes the value smaller at the beginning of the iteration and gradually increases at the end of the iteration. determines the overall trend of and is the core part of . However, gradually reached its maximum value at the end of the iteration and remained there until the end. The that no more changes increases the probability of the algorithm falling into a locally optimal solution. Therefore, it is necessary to make change slightly at the end to enhance the diversity of the population. However, a significant increase is detrimental to the fast convergence of the algorithm. Therefore, a nonlinearly increasing in the interval [0, 1.5506] compensates for these shortcomings.
A
parameter ranging in the interval [7, 28] and increasing nonlinearly with the number of iterations was designed. At the beginning of the iteration, the
value was small, and most of the barnacle particles were outside the range of the
value to fully explore the solution space. As the number of iterations increased, the
value increased nonlinearly, and the algorithm gradually transitioned from the exploration phase to the exploitation phase. At the end of the iteration, the
value reached approximately 28, at which time most particles were in the exploitation stage. However, a small number of barnacle particles still perform global exploration to find possible global optimal solutions. This design not only makes the exploration and exploitation process of the BMO algorithm well balanced and speeds up the algorithm’s convergence but also increases the adaptivity of the algorithm. The proposed optimization mechanism is illustrated in
Figure 1.
2.3.2. Dynamically Decreasing Reproduction Factor
In the exploitation phase, the parameter p significantly affects the exploitation capability of the BMO algorithm. In the basic BMO algorithm, p is a fixed value of 0.6 for each iteration, and the fixed parameter decreases the diversity of offspring propagated by Equation (6), resulting in a weak local exploitation capability of the algorithm and a slow convergence rate.
The dynamic decreasing coefficients in the white-crowned chicken optimization algorithm [
43] provide a faster convergence rate and stronger exploitation capability. Inspired by the white-crowned chicken optimization algorithm, this study uses the dynamic parameter instead of
p in the exploitation phase of the white-crowned chicken optimization algorithm. The iterative formula is as follows:
where
t is the number of current iterations;
T is the maximum number of iterations;
is a random number in [−1, 1]; and
R is a random number in the interval (0, 1). It can be seen from
Figure 2a that
fluctuates uniformly in the interval (−1, 1). The introduction of
increases the variability of
. From
Figure 2b, the value of
decreases gradually from the (−2, 2) interval to the (−1, 1) interval. Even at the end of the iteration,
fluctuates in the (−1, 1) interval, which fully retains the diversity of the population and improves the exploitation ability of the algorithm.
decreases linearly with the number of iterations in the (1, 2) interval, and it can be seen from the comparison of
Figure 2a,b that the addition of
not only improves the range of fluctuation of
but also accelerates the convergence speed of the algorithm. The improved iteration formula is as follows:
2.3.3. Optimal Neighbourhood Chebyshev Chaotic Perturbations
When the algorithm is stalled, there are two cases: (1) the algorithm finds the global optimal solution, and the search for the optimal solution is complete; (2) the algorithm is stuck in the local optimal solution. It is not sufficiently comprehensive to perturb the algorithm solely because it stagnates. Some scholars add perturbation at the optimal solution, which increases the population diversity near the optimal solution and prevents the algorithm from maturing prematurely at the optimal solution; however, it also affects the convergence speed. Therefore, in this study, we added Chebyshev chaotic perturbation to the mother generation of barnacle particles at
. The iterative formula of Chebyshev chaotic mapping is shown below:
where
k is the order.
When
, the perturbation equation is as follows:
Figure 3 shows ten common chaotic mappings. The figure shows that the Chebyshev chaotic perturbation has good randomness with a perturbation range of [−1, 1], which is more significant than other chaotic perturbations and produces a wider global distribution of particles with more comprehensive information. Adding the perturbation at
= 0.5 can maximize the retention of possible optimal solutions in the late iteration and increase the algorithm’s ability to jump out of the local optimal solution.
2.3.4. Convergence Rate Analysis
The PEBMO algorithm is designed to achieve improved performance by improving the algorithm parameters. The large range of nonlinear increasing penis coefficients, dynamic decreasing reproduction coefficients, and optimal neighborhood Chebyshev chaotic perturbations all have some effect on the convergence speed. It is analyzed explicitly as follows:
In intelligent algorithm optimization, the balance between exploration and exploitation has a significant impact on the convergence speed of the algorithm. If the algorithm is too biased towards exploration (global search), it may waste a lot of time in irrelevant regions, resulting in slow convergence. If the algorithm is too biased toward exploitation (local search), it may fall into the local optimal solution prematurely and fail to find the global optimum. The is a parameter that regulates the balance between the exploration and exploitation of the BMO algorithm, and it is a fixed value 7. A fixed search range may result in the algorithm not being able to cover the global space at the initial stage or not being able to optimize it at a later stage. Therefore, in this study, a large range of non-linear increasing penis coefficients is designed to better balance the BMO algorithm exploration and exploitation process. At the beginning of the iteration, the value is large, and the vast majority of barnacle particles are in the exploration stage to explore the solution space. As the number of iterations increases, the value decreases, and most of the particles are in the exploration stage, finely exploiting the searched region. The large range of non-linear increasing penis coefficients enables the algorithm to find the best trade-off between global search and local optimization, thus finding the optimal solution more efficiently and significantly accelerating the convergence speed.
The parameter p of the BMO algorithm in the exploitation phase directly determines the behavior of the algorithm in the local search and optimization process. If the exploitation intensity is too great, it may lead to a decrease in population diversity, and the algorithm falls into a local optimum; if the exploitation intensity is too low, it cannot fully utilize the regional information, and the convergence speed is slow. In the BMO algorithm, p is a random number between (0, 1). Although the random number can increase the population diversity in the exploitation phase, over-reliance on random exploration will lead to unclear search direction and slow convergence. The dynamic decreasing reproduction coefficient varies in a large range in the early period of iteration, giving the algorithm a strong ability to fully exploit the explored region in search of a possible optimal solution. As the number of iterations increases, the range of p decreases, avoiding over-exploitation and speeding up convergence. The design of the dynamically decreasing reproduction coefficient better balances the exploitation intensity and thus accelerates the convergence rate.
In order to improve the ability of the algorithm to jump out of the local optimal solution, adding perturbations to the optimal solution to increase the diversity of the population is an effective strategy for improvement. However, it also brings the defect of slow convergence speed of the algorithm. Therefore, in this study, the ability of the algorithm to jump out of the local optimal solution is increased by adding the Chebyshev chaotic perturbation at = 0.5. The limitation of excessive perturbation at the optimal solution, which leads to non-convergence of the algorithm, is avoided.
2.4. PEBMO Algorithm Design
In this study, the exploration and exploitation process of the BMO algorithm is made more homogeneous by the design of a large range of penis coefficients; the introduction of the dynamic decreasing reproduction coefficients increases the diversity of the populations and improves the exploitation capability of the algorithm; and the Chebyshev chaotic perturbation at
= 0.5 increases the ability of the algorithm to jump out of the local optimal solution. The flowchart of PEBMO is shown in
Figure 4.
The basic steps of the PEBMO algorithm are as follows:
1. Randomly generate an initial population with a population size of N. Initialize the position of each barnacle particle using Equation (1) and set the parameters required for the execution of the algorithm: N, n, T, .
2. Calculate the fitness value of each barnacle particle and rank them, The optimal solution at the top ( = the current optimal solution).
3. Calculate value according to Equation (8); Calculate value according to Equation (12); Calculate value according to Equation (14).
4. Select dad according to Equation (4) and Mum according to Equation (5).
5. If the penis length of the chosen father is less than , the offspring are generated according to Equation (13); otherwise, the offspring are generated according to Equation (7).
6. If the penis length of the selected parent is 0.5, offspring are generated according to Equation (15).
7. The fitness value of each barnacle particle is calculated.
8. Sorting and update .
9. Determine whether the iteration termination condition is met; if so, exit the loop; otherwise, proceed to Step 2.
2.5. Forest Canopy Image Segmentation Based on the PEBMO Algorithm
2.5.1. Multi-Level Threshold Image Segmentation
Threshold segmentation can be classified into second-and multilevel thresholds according to the number of thresholds [
44]. Second-level segmentation divides an image into two parts, target and background, by a single threshold and is suitable for segmenting simple images. Multilevel threshold segmentation divides an image into regions with different gray levels using various thresholds, which is ideal for segmenting complex multi-target images. Canopy images contain more information and single-threshold segmentation is insufficient to complete the segmentation task. For multilevel threshold segmentation, suppose
n thresholds are
, and the grey level mapping is
where
is
gray level of the segmented image;
. Equation (16) shows that the key step of threshold segmentation is to select the optimal threshold related to the segmentation quality. Kapur entropy was used as the objective function in this study.
2.5.2. Kapur Entropy
Kapur entropy is a more commonly used information-entropy-based threshold selection method that is widely used in the multi-threshold segmentation of complex images. The principle of multi-threshold image segmentation based on Kapur entropy is as follows:
Assuming that the image gray level is
L and the gray range is
, the probability of a pixel in this gray level can be expressed as
where
represents the sum of the number of pixels with gray level
i and
N represents the total number of pixels in the image. The Kapur entropy objective function for multi-thresholding is defined as:
Among them,
is the entropy of different categories after segmentation,
is the probability of pixel points occurrence in each category, and
is the probability of pixel points with grey value
j.
A judgment is made according to Equation (23) to select the optimal combination of thresholds.
The maximized combination is the optimal threshold sought to solve the problem of the best combination of thresholds. Therefore, using an intelligent optimization algorithm to select the optimal threshold can effectively solve the problems of low efficiency and accuracy of traditional methods. In this study, PEBMO was used to select an optimal threshold.
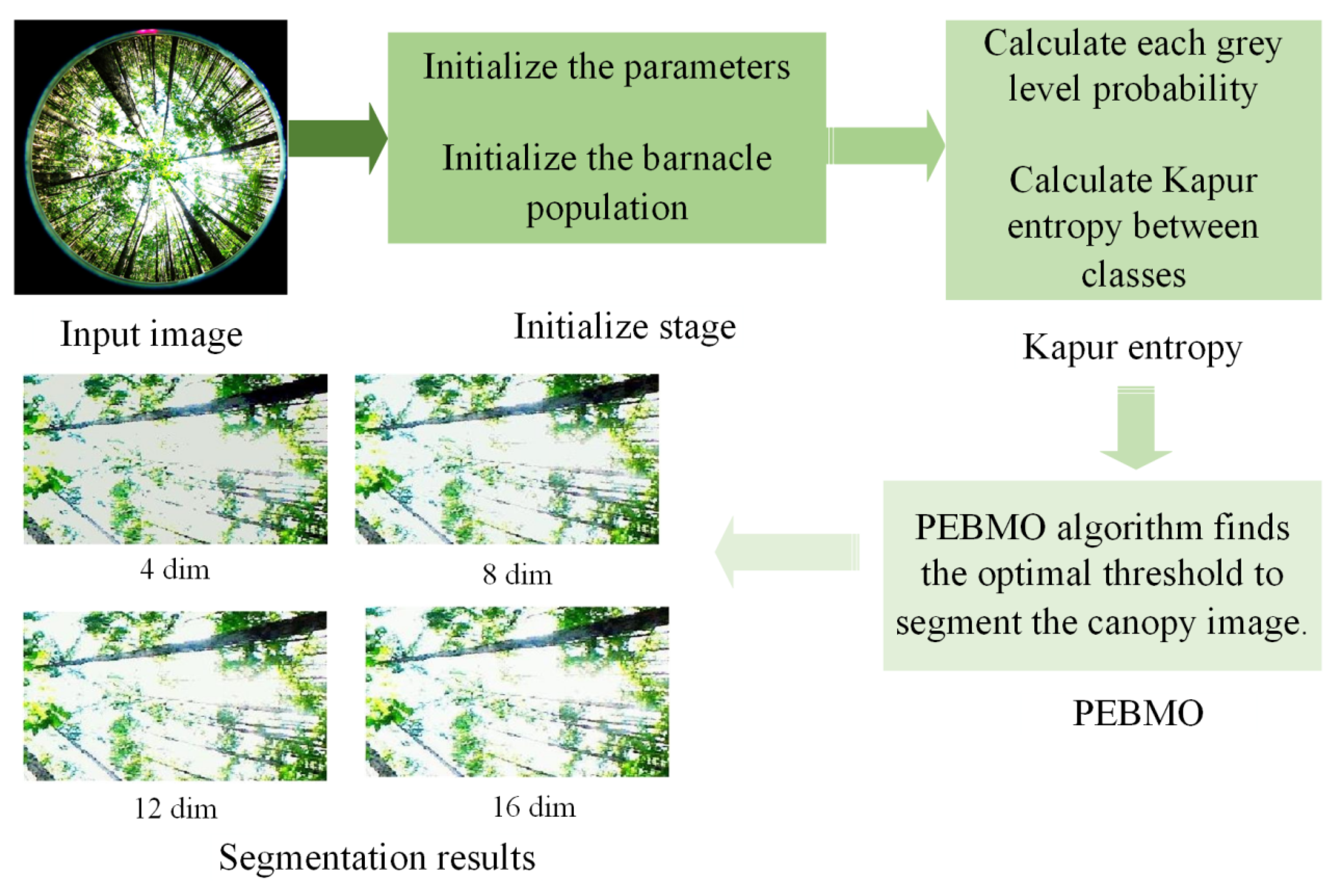
2.5.3. Implementation of the PEBMO-Based Segmentation Algorithm
In order to improve the segmentation performance of the canopy image, Kapur entropy is the fitness function, and the PEBMO algorithm searches for the optimal threshold to segment the canopy image accurately. Figrue
Figure 5 shows the flowchart of the proposed multi-threshold segmentation of the canopy image based on the PEBMO algorithm. The framework of the proposed algorithm is as follows:
1. Input the image to be segmented.
2. Generate the population and parameters of PEBMO.
3. Calculate each grey level’s probability (22).
4. Calculate the fitness value of each search agent by using (19) for Kapur entropy.
5. PEBMO algorithm finds the optimal threshold to segment the canopy image.
6. Output the segmentation result.
4. Discussion
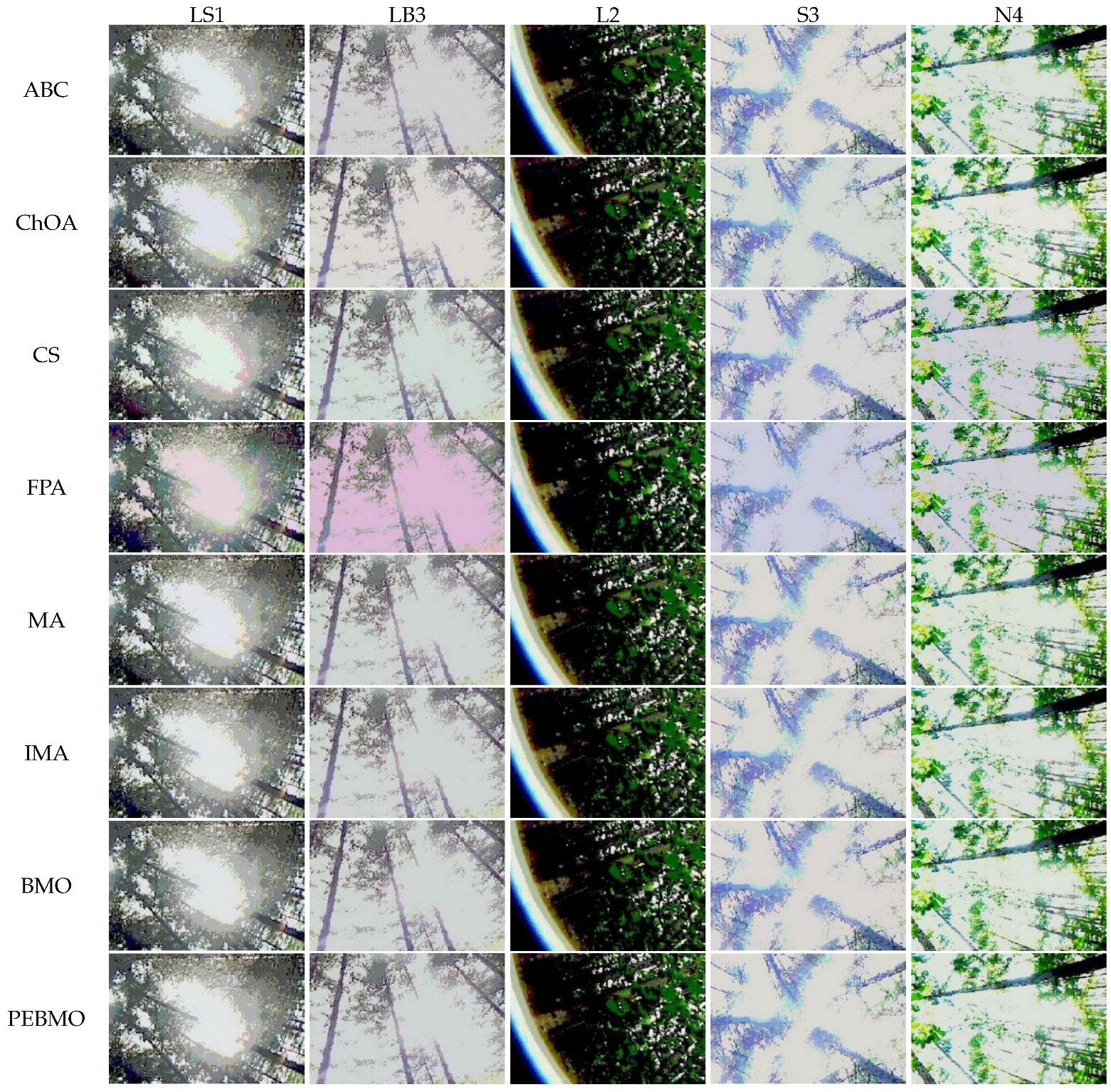
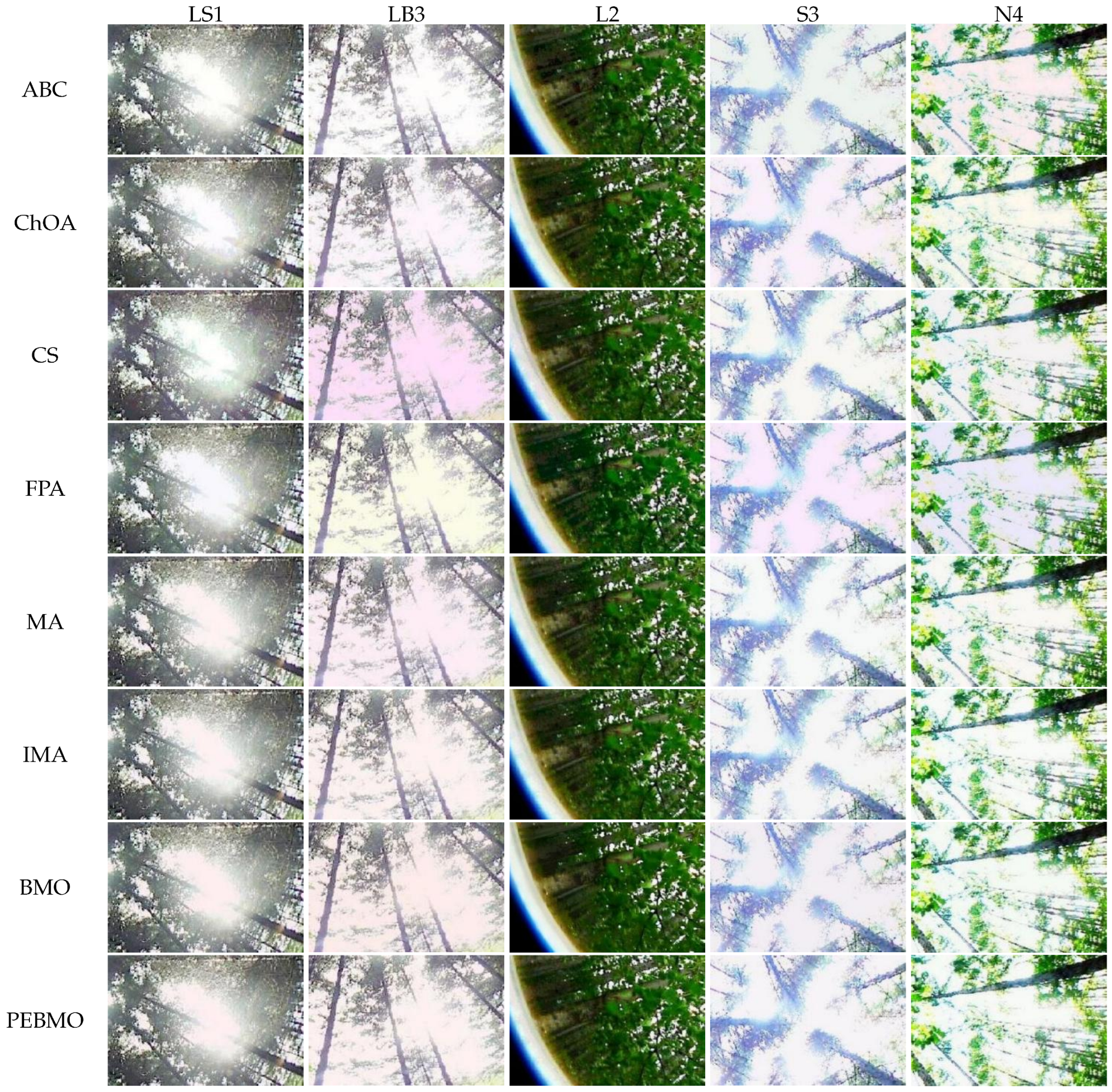
The results of the subjective and objective evaluations of each algorithm on the four types of forest canopy images show that the proposed PEBMO algorithm has obvious advantages over other intelligent algorithms and improved intelligent algorithms. The effectiveness of the improved PEBMO algorithm is demonstrated. However, there is no perfect algorithm to solve these problems. A canopy image has a significant gap in the complexity of the target, light intensity, and image clarity, which is a significant challenge to the accuracy and stability of image segmentation.
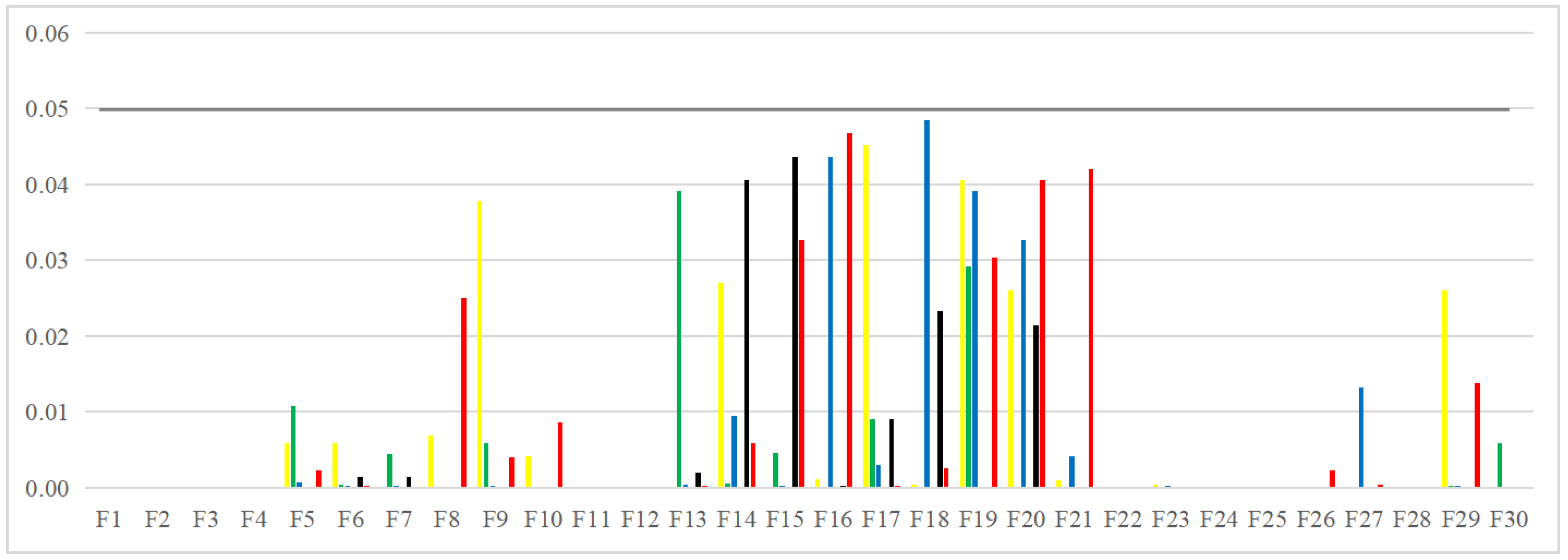
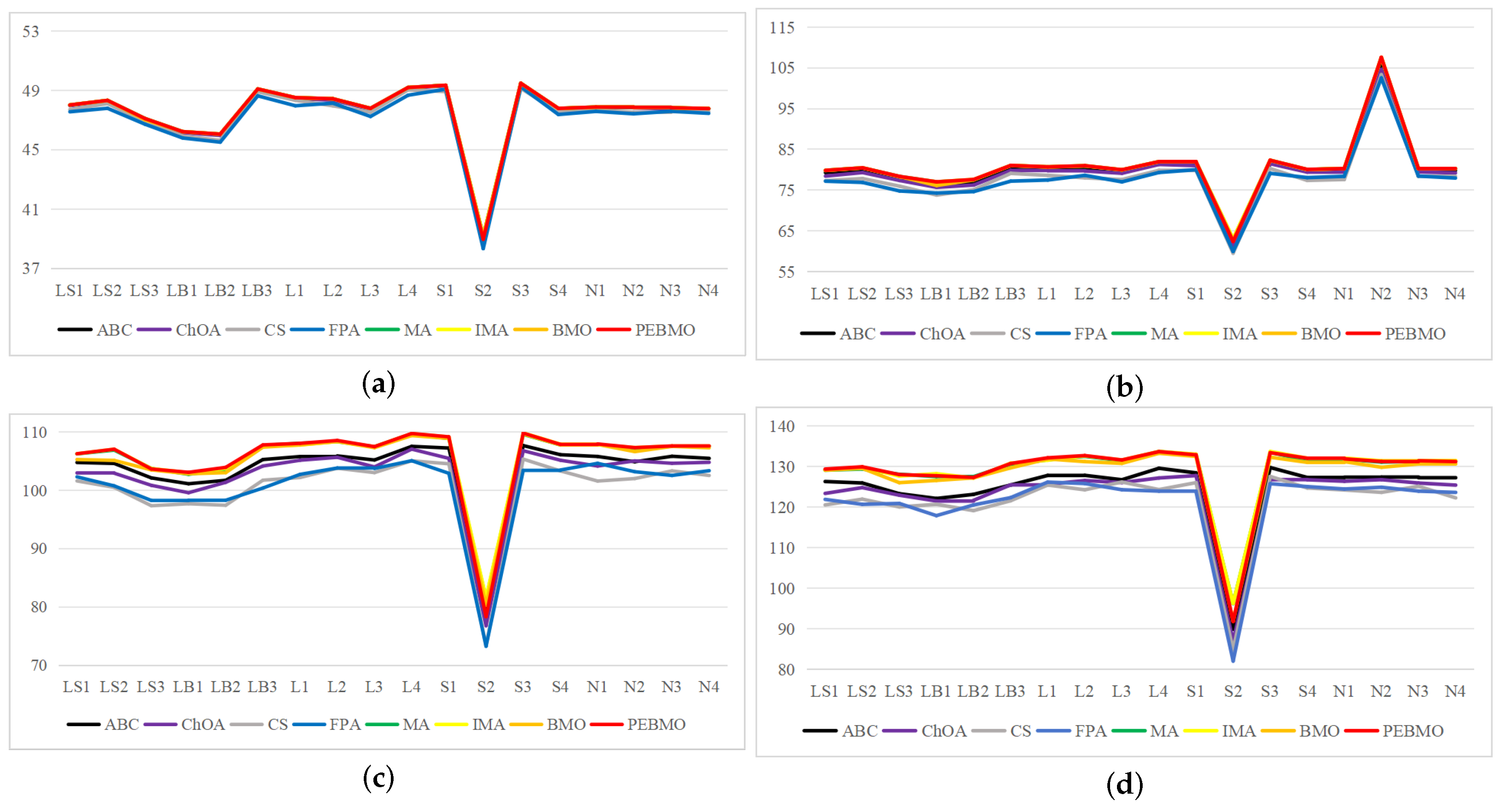
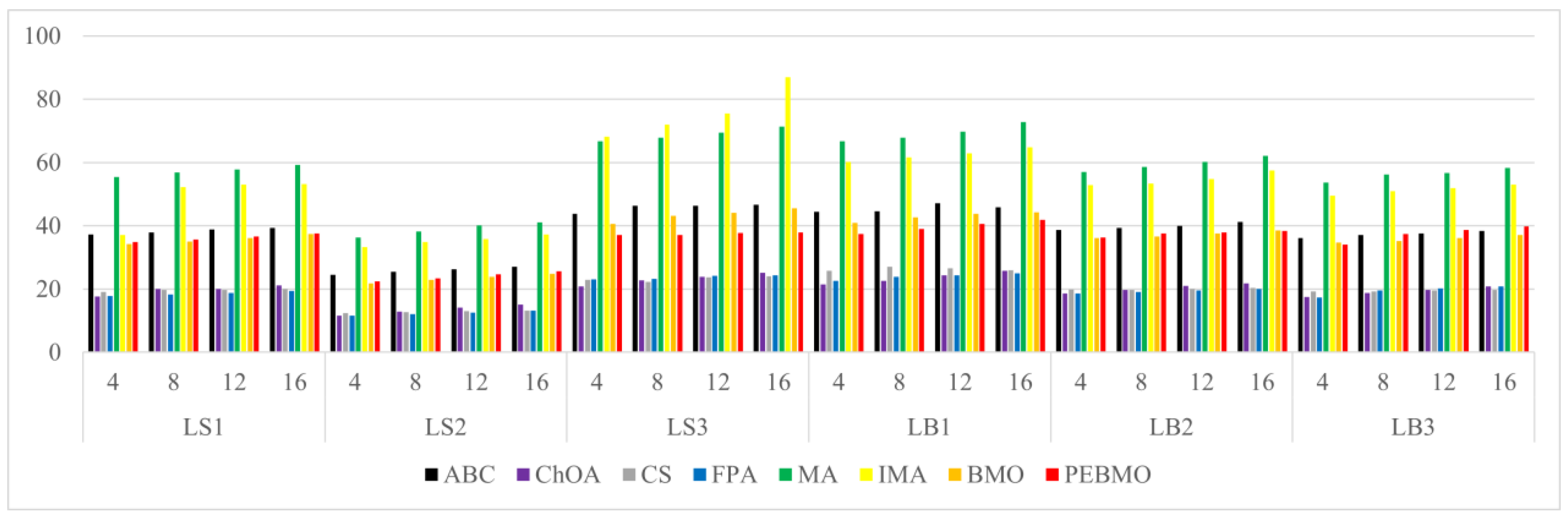
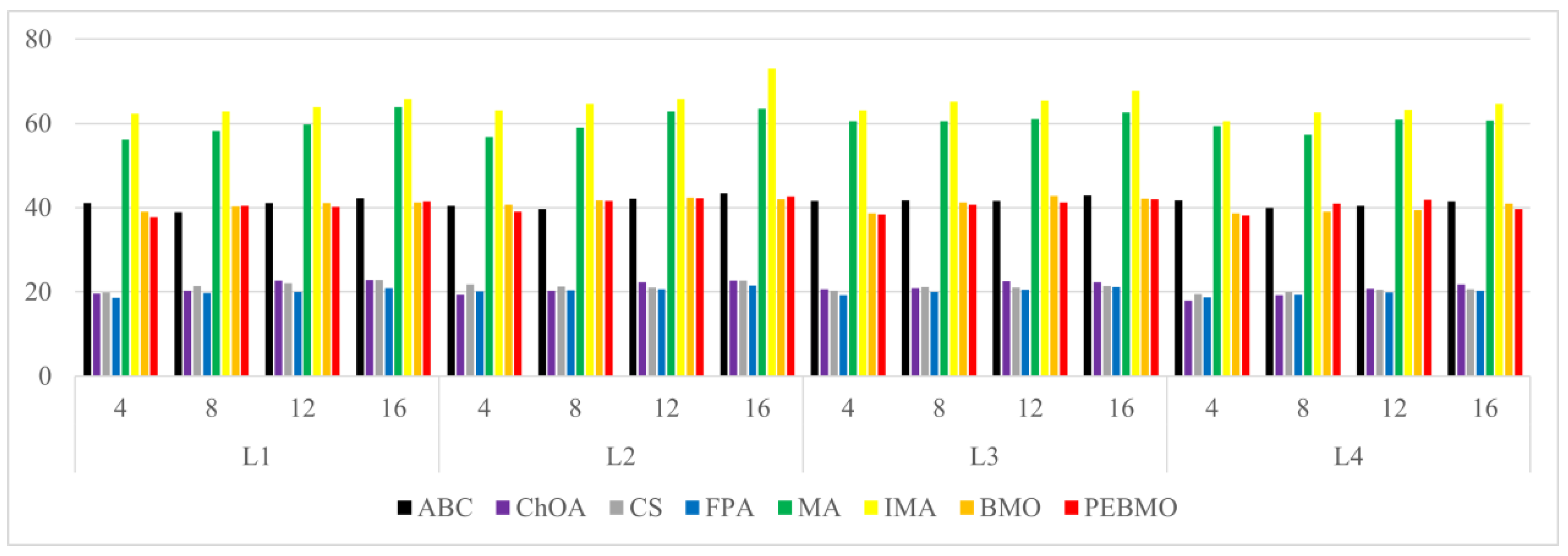
Image segmentation accuracy: 17 equal sets of optimal fitness values emerged in acquiring fitness values, of which 94.12% were distributed when the number of thresholds was 4. This stems from the fact that none of the algorithms can accurately segment the canopy image when the number of thresholds is not big enough, and the difference between the adaptative values obtained is insignificant. As the number of thresholds increases, the differences between the algorithms are gradually noticeable. The PEBMO and IMA algorithms have apparent advantages, 40.28% and 30.56% better than the other algorithms, respectively; 90.9% and 81.36% better than BMO and MA algorithms. It shows that it is necessary to improve the intelligent algorithms to improve the segmentation accuracy of canopy images. Although the PEBMO algorithm 56.36% outperforms IMA. However, the IMA algorithm outperforms PEBMO by 69.23% and 66.67% in segmenting strong and normal light canopy images. This indicates that the PEBMO algorithm has higher segmentation accuracy in segmenting non-uniform and low-illuminated canopy images. At the same time, IMA excels in the segmentation of strong and normal light canopy images. The other algorithms perform poorly in segmentation accuracy, and only MA obtains two optimal adaptation values.
Image segmentation stability: the PEBMO algorithm has better stability in segmenting the forest canopy image, obtaining 55.56% of the optimal standard deviation value. The IMA algorithm is 79.17% better than the MA algorithm, although it only obtains 23.61% of the optimal standard deviation value. It shows that the IMA algorithm’s improvement strategy effectively improves the MA algorithm’s segmentation stability algorithm. ABC, ChOA, CS, FPA, MA, and BMO algorithms perform in general and fluctuate more in segmenting non-uniform and strongly illuminated canopy images. This is due to the difficulty of segmenting non-uniform and strongly illuminated canopy images. Uneven illumination and details hidden under strong light can easily cause poorly performing algorithms to fall into local optima and fail in segmenting such images. Hence, there is less stability in segmenting such images.
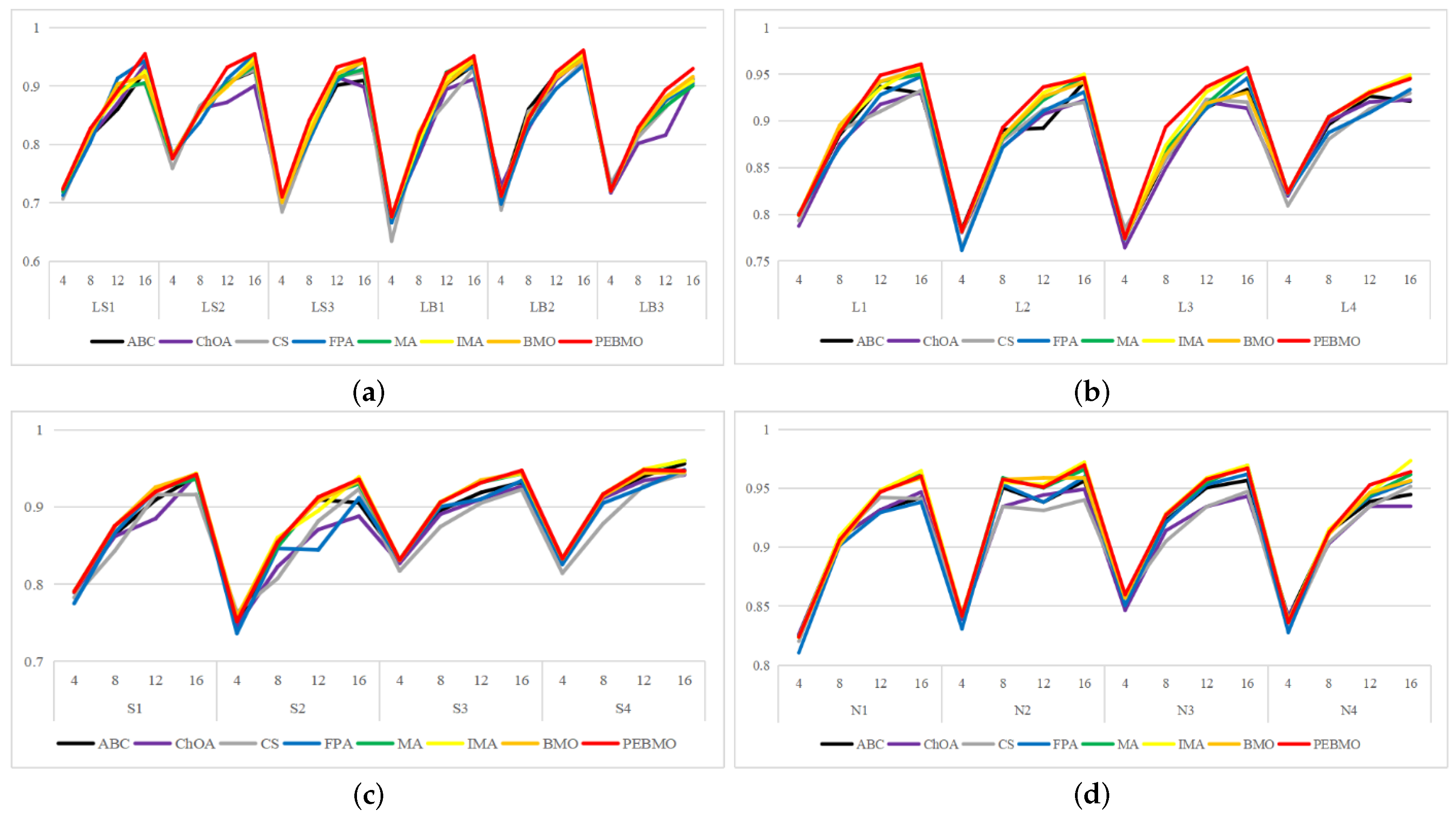
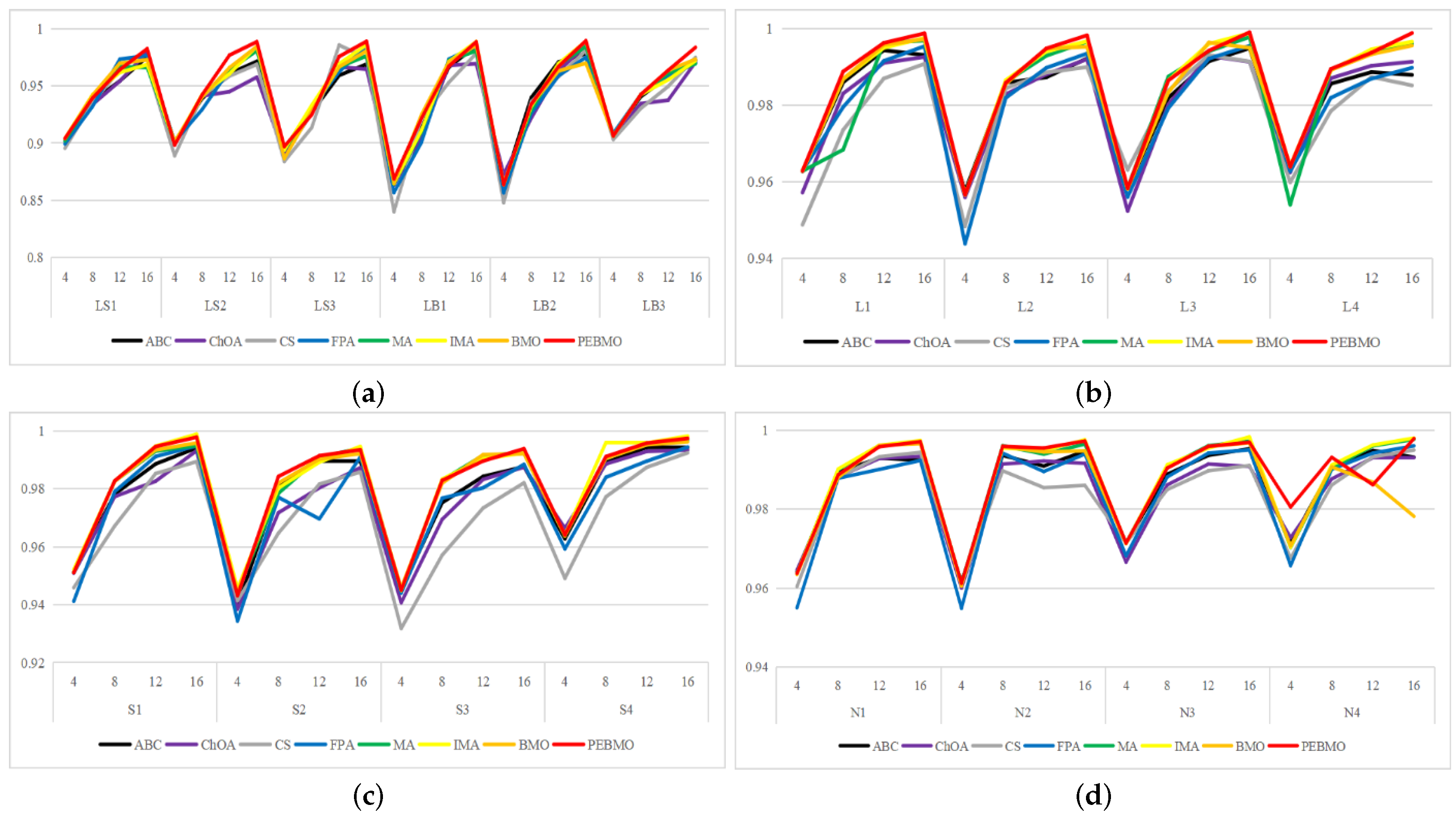
Feature similarity analysis: The optimal SSIM values obtained by ABC, ChOA, CS, FPA, MA, IMA, BMO, and PEBMO are 5, 3, 4, 3, 4, 19, 7, and 27, respectively. Of which are 1, 1, 2, 2, 1, 1, 1, 3, and 13 on the non-uniform canopy images; on low-illumination canopy images are 1, 0, 1, 1, 1, 1, 3, 1, 8; on strong light canopy images are 2, 0, 1, 0, 1, 6, 2, 4; and on normal illumination canopy images are 1, 2, 0, 0, 1, 1, 9, 1, 2. Regarding the optimal SSIM values distribution, the overall IMA and PEBMO algorithms have a clear advantage, obtaining 63.89% of the optimal SSIM values. IMA’s strengths are centered on strong and normally illuminated canopy images, while the PEBMO algorithm outperforms the comparison algorithm on non-uniform and low-illumination canopy images. The performance of the other algorithms is rather mediocre.
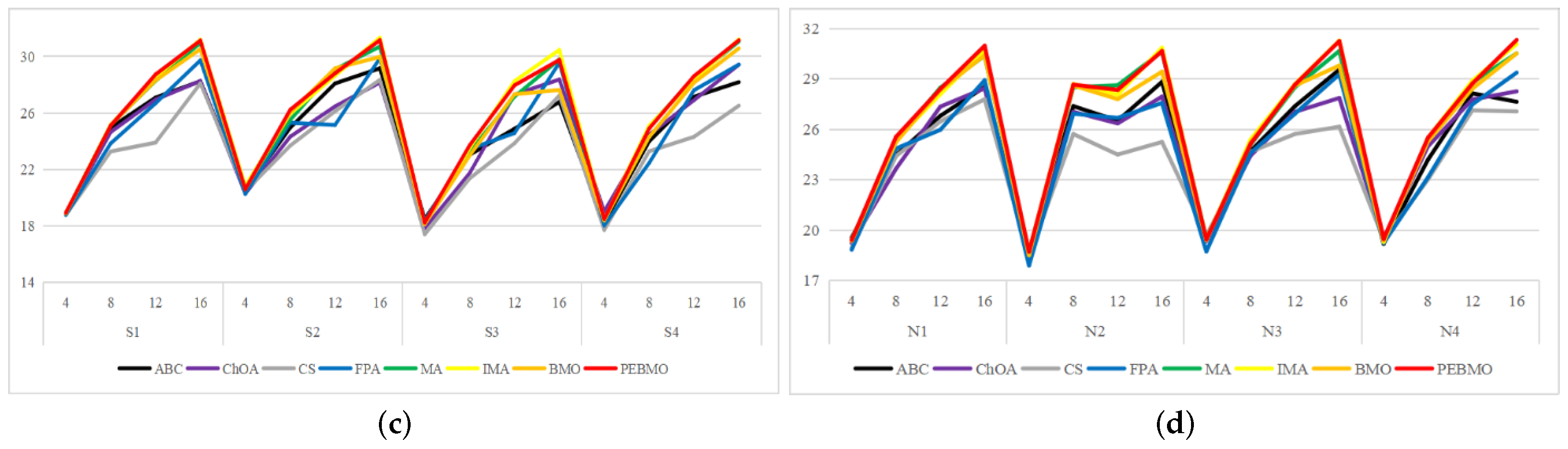
Degree of image distortion: the ABC, ChOA, CS, PFA, MA, IMA, BMO, and PEBMO algorithms obtained 4, 2, 4, 3, 5, 19, 6, and 29 optimal PSNR values, respectively. The IMA and PEBMO algorithms outperformed the others, indicating that their segmented canopy images were less distorted and of better quality. The performance of the algorithms on non-uniform canopy images is more balanced. The PEBMO algorithm obtains 10 out of 24 optimal values on non-uniform canopy images, which is slightly better than the comparison algorithms. The PEBMO algorithm has a clear advantage in low-illumination canopy images, obtaining 62.5% of the optimal PSNR values. In contrast, the MA and CS algorithms are slightly inferior, obtaining two optimal values, respectively. In the strong and normal illumination images, the IMA algorithm obtained 14 optimal values, while the PEBMO algorithm obtained 8, indicating that the IMA algorithm segmented images with better quality.
Feature similarity analysis: 6.94%, 2.78%, 2.78%, 5.56%, 9.72%, 15.28%, 30.56%, and 33.33% of the optimal FSIM values were obtained for ABC, ChOA, CS, FPA, MA, BMO, IMA and PEBMO, respectively. MA and BMO algorithms outperform ABC, ChOA, CS, and FPA, which indicates that they also have good optimization-seeking performance. IMA and PEBMO improve the FSIM values of both MA and BMO by 73.61%. Therefore, it is essential to increase the feature similarity between the segmentated canopy image and the original one by improving the performance of the intelligent algorithms.
Image segmentation efficiency: by analyzing the computation time data of each algorithm, it can be concluded that all algorithms take no more than 5s when the number of thresholds increases from 4 to 16. The limitation that the computational complexity increases exponentially with increasing the number of thresholds in multi-threshold segmentation is overcome. The FPA, CS, and ChOA algorithms have apparent advantages. The MA and IMA algorithms have the lowest segmentation efficiency, which is inferior to the others. The gap between the BMO and PEBMO algorithms is not large, which suggests that improving the PEBMO algorithm does not increase the segmentation efficiency of the BMO algorithm. The segmentation efficiency of ABC is between that of PEBMO and MA.
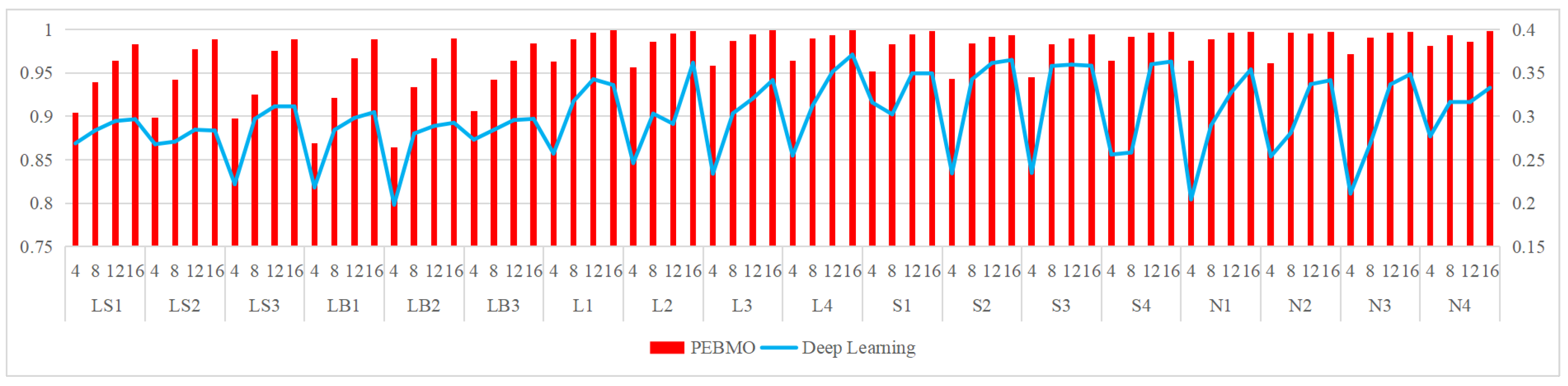
Comparison with deep learning segmentation methods: in comparison with deep learning methods, the PEBMO algorithm outperforms deep learning methods in terms of fitness value, standard deviation value, FSIM, and computation time, while deep learning outperforms the PEBMO algorithm in terms of SSIM and PSNR. Both algorithms have advantages and disadvantages. Deep learning has a clear advantage in segmentation quality. However, it also has the limitations of poor stability and high data dependency. The PEBMO algorithm, on the contrary, is stable, computationally efficient, and easy to implement. Still, it also suffers from significant performance degradation in low-dimensional or complex cases and high distortion.
In further research, overall, the PBEMO algorithm is better than the IMA algorithm, but its advantage is mainly concentrated in non-uniform and low-illumination canopy images. The IMA algorithm is 53.126% better than the PEBMO algorithm in segmenting normal illumination and strong illumination images. Therefore, further improvement of the BMO algorithm is necessary to improve its segmentation accuracy on high-brightness canopy images. In addition, this study provides a comprehensive comparison with the deep learning segmentation method, in which both algorithms have strengths and weaknesses. Deep learning has apparent advantages in segmentation quality. However, it also has the limitations of poor stability and excessive dependence on data, while the PEBMO algorithm is the opposite, with good stability, high computational efficiency, and ease of implementation. Therefore, different segmentation methods can be used based on these requirements.