Identification of bHLH Family Genes in Rhododendron hainanense Merr. and Verification of Their Role in Heat Tolerance
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth Condition and Evaluation of Heat Resistance of R. hainanense
2.2. Identification of bHLH Members in R. hainanense
2.3. Chromosomal Positioning, Gene Duplication, and Synteny Analysis
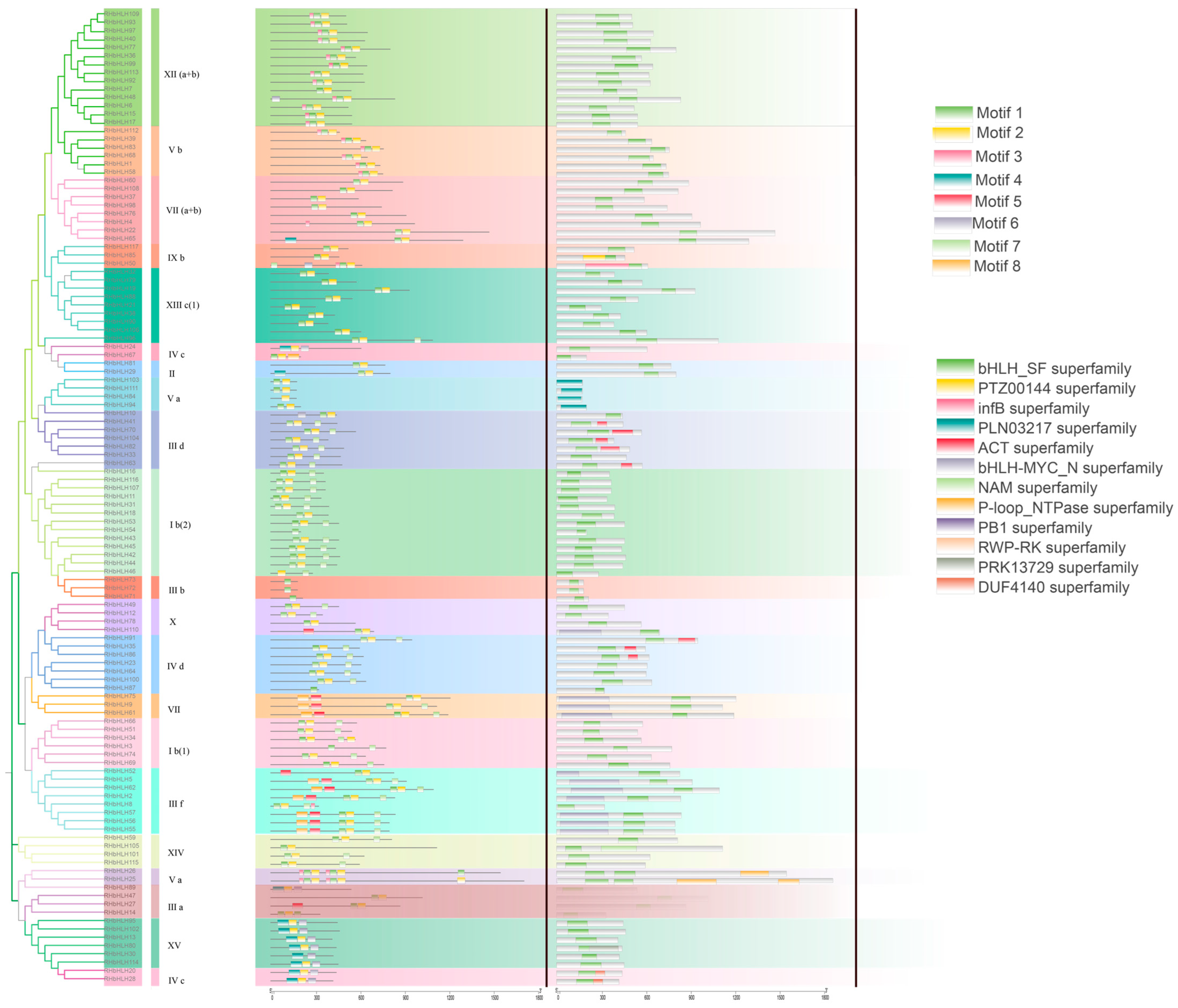
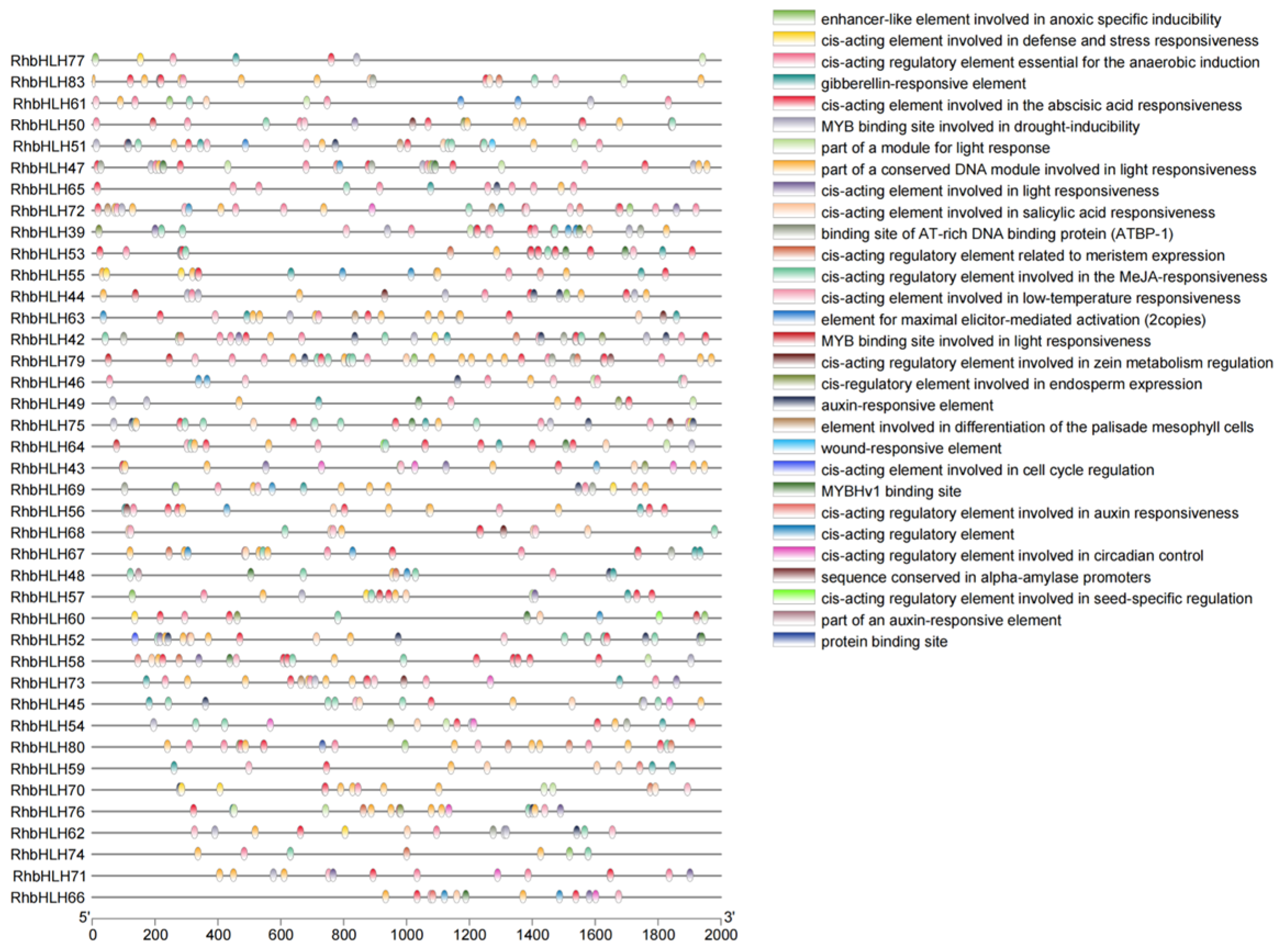
2.4. Analysis of Conserved Motifs, Gene Structure, and Phylogenetic Tree
2.5. Protein Structure Visualization and Protein Interaction Network Analysis
2.6. Expression Patterns and Differential Gene Screening
2.7. Analysis of Differential Metabolites and Association Analysis with Differential Genes
2.8. Real-Time Fluorescence Quantitative Verification
3. Results
3.1. Comparison of High-Temperature Treatment of R. hainanense and R. dauricum
3.1.1. Changes in Hydrogen Peroxide Content
3.1.2. Changes in Proline Content
3.1.3. Changes in MDA Content
3.1.4. Changes in the Activity of the Protective Enzyme SOD
3.2. Identification and Physicochemical Property Analysis of RhbHLH
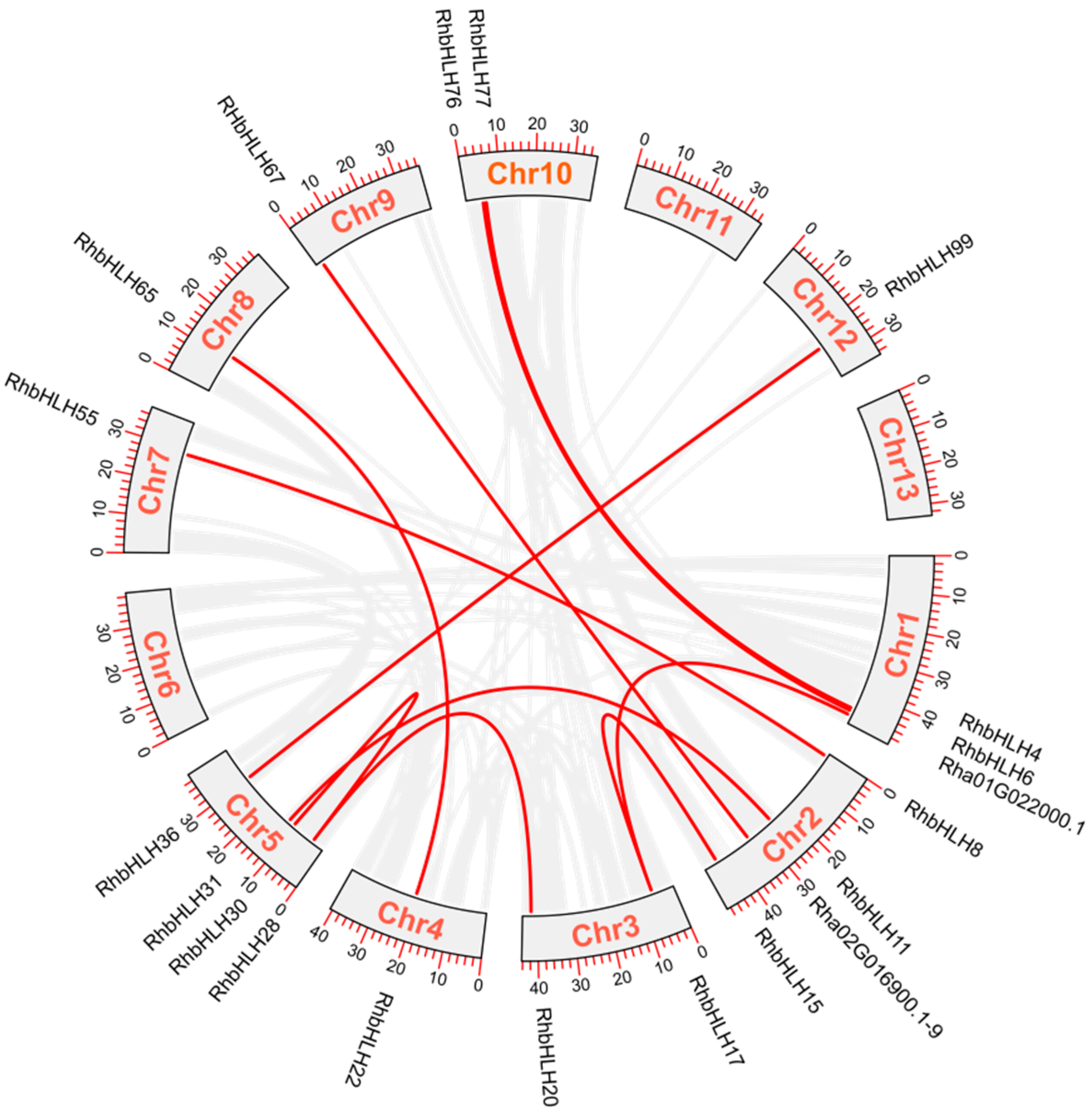
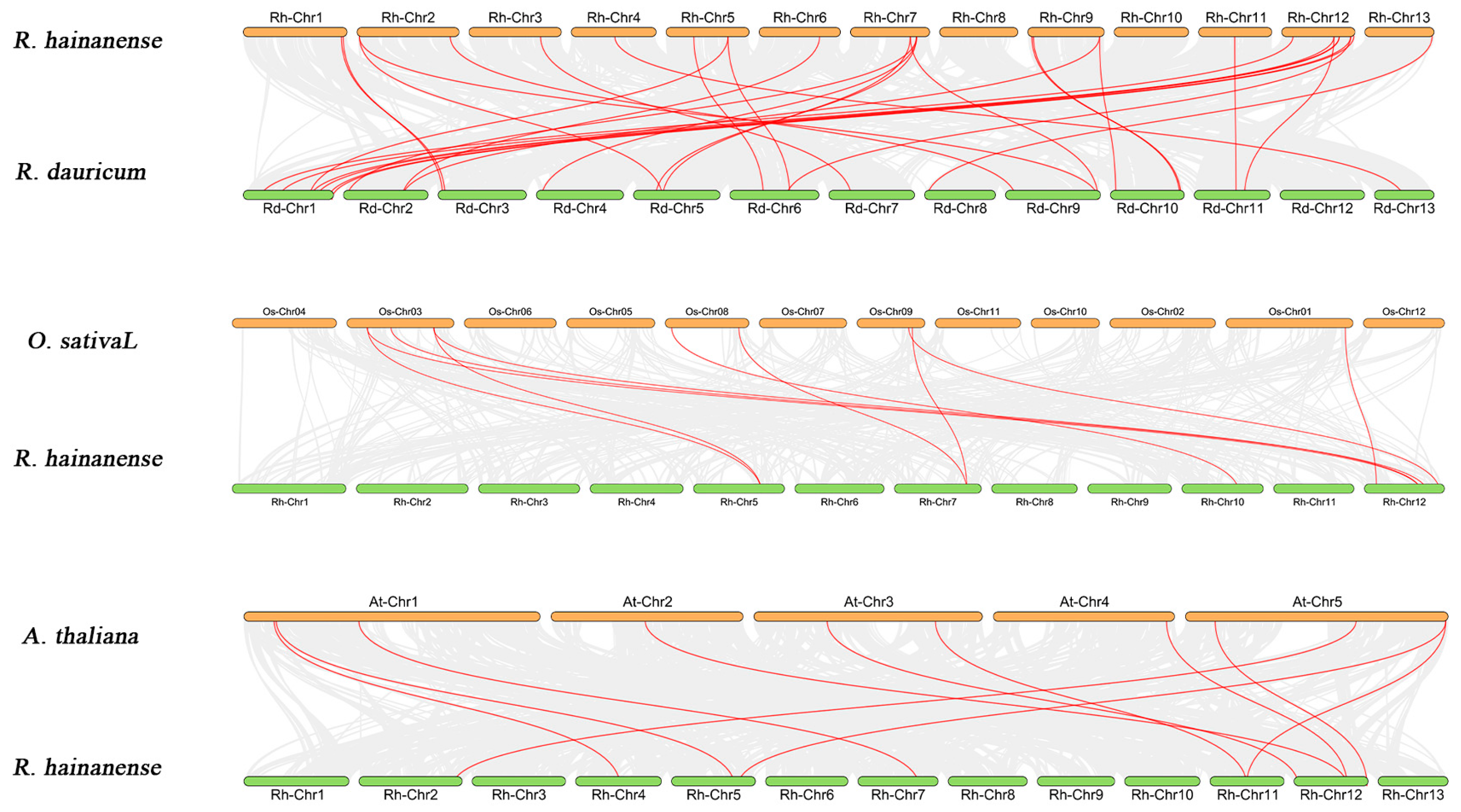
3.3. Chromosomal Location and Its Collinearity Relationship
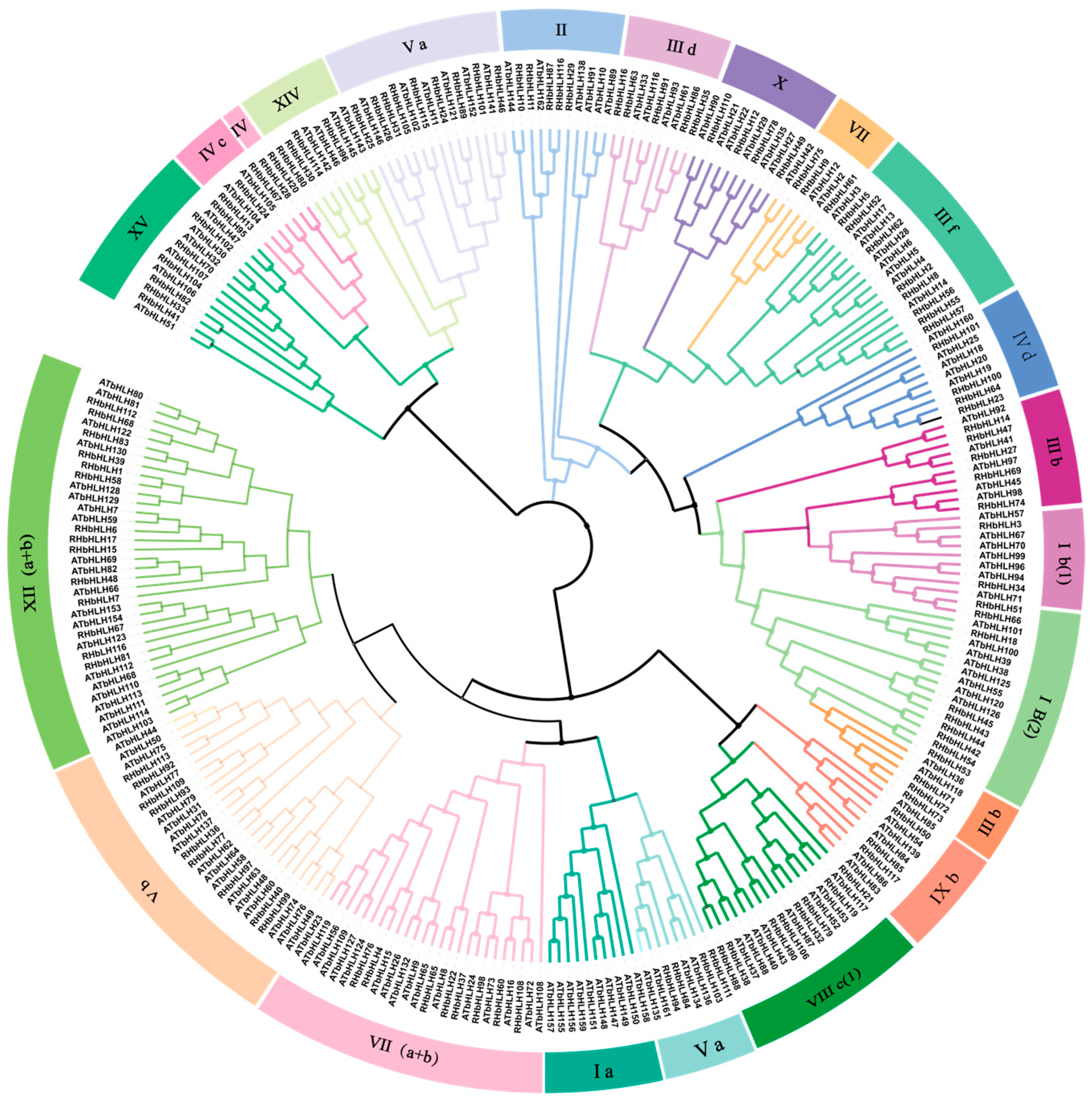
3.4. Sequence Conservatism and Phylogenetic Relationships
3.5. Visualization of bHLH Protein Structure and Interaction Network
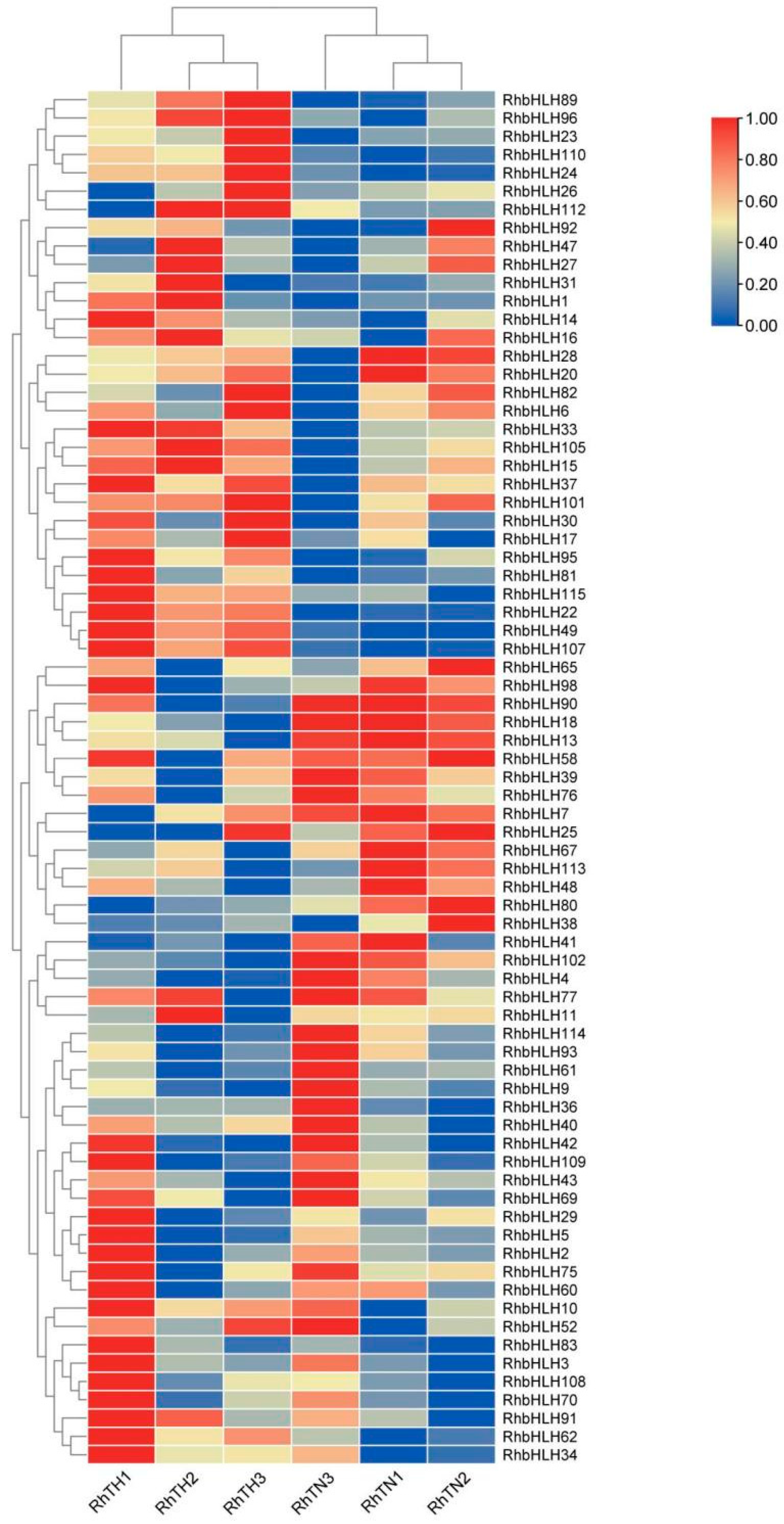
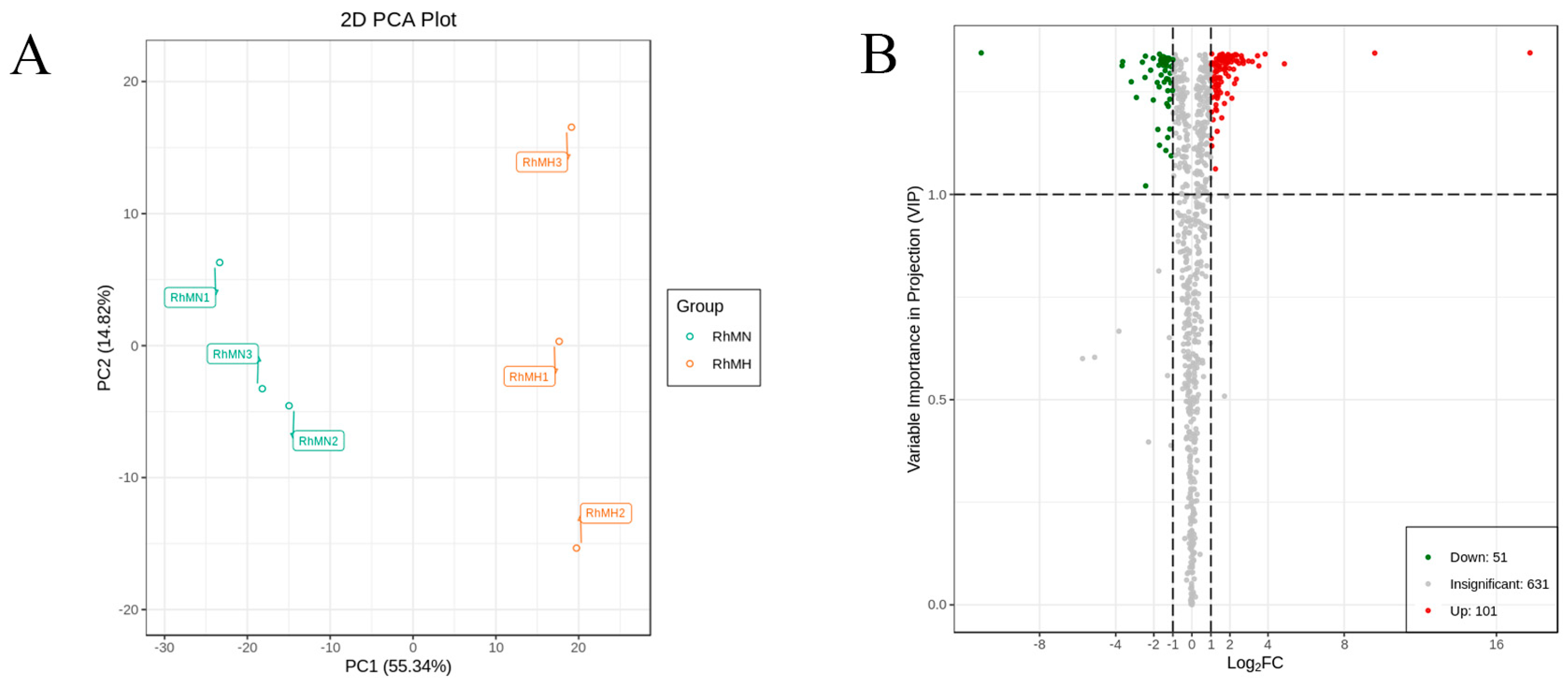
3.6. Expression Patterns and Analysis of Transcriptome Under High-Temperature Stress
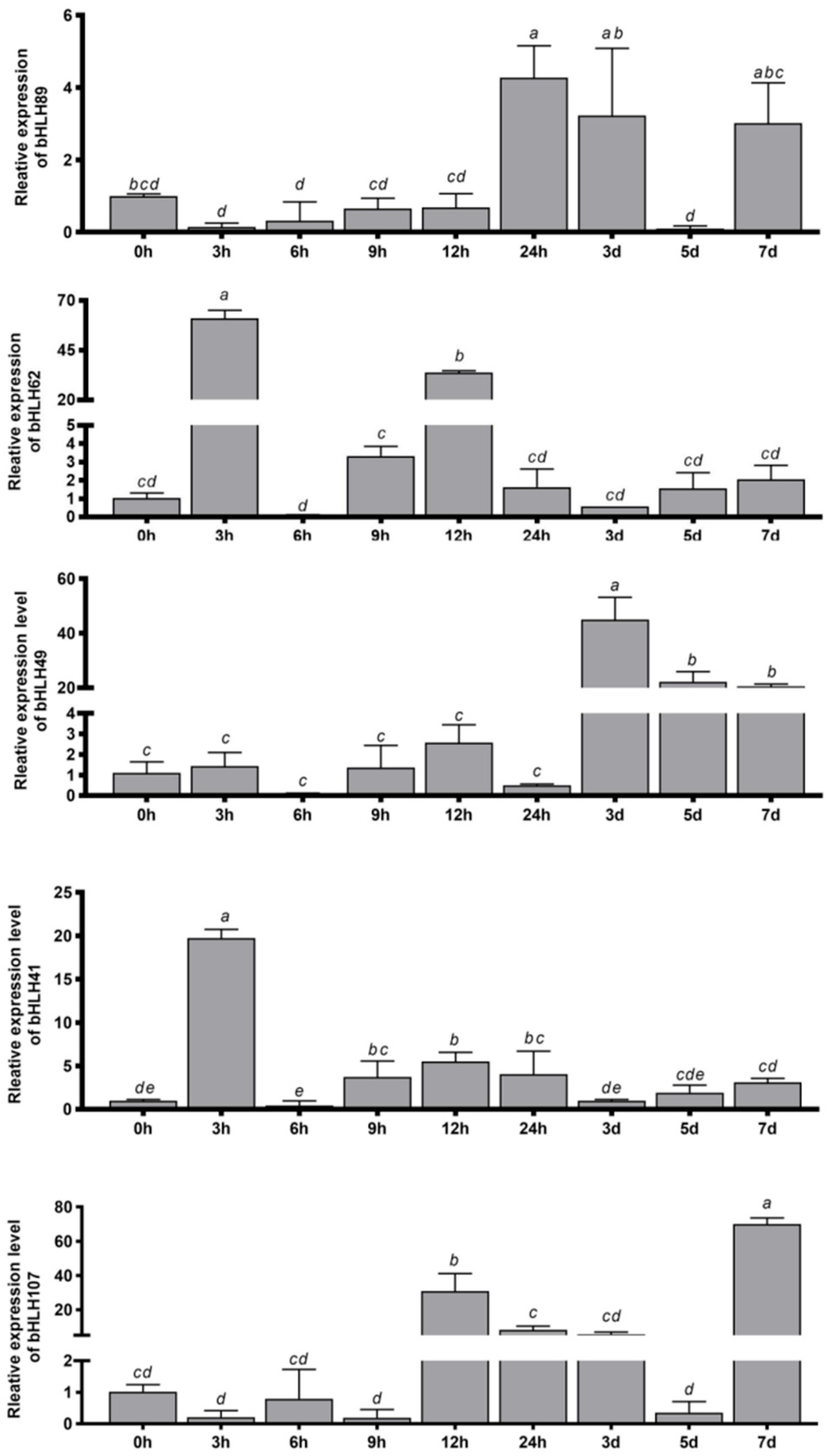
3.7. Real-Time Fluorescence Quantitative Verification and Relative Expression Levels of RhbHLH
3.8. Differential Metabolite Screening
3.9. Analysis of Differential Metabolites and Differential Gene Associations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.C.; Zhao, Y.; Song, X.Q.; Ren, M.X. Difference in spatial distribution patterns and population structures of Rhododendron hainanense between both sides of riparian bends. Chin. J. Plant Ecol 2018, 42, 841–849. [Google Scholar] [CrossRef]
- Zhang, M.L.; Liu, Y.Q. Research on cultivation technology of alpine azalea. Anhui Agric. Sci. 2005, 33, 1008–1009. [Google Scholar]
- Huo, S.J.; Li, Y.F.; Li, R.P.; Chen, R.H.; Xing, H.T.; Wang, J.; Zhao, Y.; Song, X.Q. Genome-wide analysis of the MADS-box gene family in Rhododendron hainanense Merr. and expression analysis under heat and waterlogging stresses. Ind. Crops Prod. 2021, 172, 114007. [Google Scholar] [CrossRef]
- Shi, Y.H.; Li, S.P.; Liang, W.H.; Song, X.Q.; Tan, J.H. Germplasm Resourses of Rhododendron in Hainan. Chin. J. Trop. Crops. 2010, 31, 551–555. [Google Scholar]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef]
- Kolonko, M.; Greb-Markiewicz, B. bHLH-PAS Proteins: Their Structure and Intrinsic Disorder. Int. J. Mol. Sci. 2019, 20, 3653. [Google Scholar] [CrossRef]
- Massari, M.E.; Murre, C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000, 20, 429–440. [Google Scholar] [CrossRef]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Cell. Biol. 2010, 27, 862–874. [Google Scholar] [CrossRef]
- Moore, A.W.; Barbel, S.; Jan, L.Y.; Jan, Y.N. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc. Natl. Acad. Sci. USA 2000, 97, 10436–10441. [Google Scholar] [CrossRef]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop- helix gene family in metazoans: Insights from comparative genomics. BMC Dev. Biol. 2007, 7, 33. [Google Scholar]
- Ledent, V.; Paquet, O.; Vervoort, M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002, 3, research0030. [Google Scholar] [CrossRef] [PubMed]
- Buck, M.; Atchley, W. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 2003, 56, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Brouwer, M. Hypoxia-inducible factor, gsHIF, of the grass shrimp Palaemonetes pugio:molecular characteriza-tion and response to hypoxia. Comp. Biochem. Physiol. B 2007, 147, 11–19. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Komatsu, K.; Maekawa, M.; Ujiie, S.; Satake, Y.; Furutani, I.; Okamoto, H.; Shimamoto, K.; Kyozuka, J. LAX and SPA: Major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA 2003, 100, 11765–11770. [Google Scholar] [CrossRef]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef]
- Ichihashi, Y.; Horiguchi, G.; Gleissberg, S.; Tsukaya, H. The bHLH Transcription Factor SPATULA Controls Final Leaf Size in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 252–261. [Google Scholar] [CrossRef]
- Qi, T.C.; Huang, H.; Song, S.S.; Xie, D.X. Regulation of Jasmonate-Mediated Stamen Development and Seed Production by a bHLH-MYB Complex in Arabidopsis. Plant Cell 2015, 27, 1620–1633. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as Transcriptional Activators in Abscisic Acid Signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis MYB transcription factors interacting with R/B-like bHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Zhang, H.M.; Li, X.L.; Ai, Q.; Yu, D.Q. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis. J. Exp. Bot. 2017, 68, 1743–1755. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, W.; Ohmori, T.; Kageyama, K.; Miyazaki, C.; Saito, A.; Murata, M.; Noda, K.; Maekawa, M. The Purple leaf (Pl) locus of rice; the PlW allele has a complex organizationand includes two genes encoding basic helix-loop-helix proteinsinvolved in anthocyanin biosynthesis. Plant Cell Physiol. 2001, 42, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008, 53, 312–323. [Google Scholar] [CrossRef]
- Koini, M.A.; Alvey, L.; Allen, T.; Tilley, C.A.; Harberd, N.P.; Whitelam, G.C.; Franklin, K.A. High Temperature-Mediated Adaptations in Plant Architecture Require the bHLH Transcription Factor PIF4. Curr. Biol. 2009, 19, 408–413. [Google Scholar] [CrossRef]
- Khan, G.; Nolzen, J.; Schepker, H.; Albach, D.C. Incongruent phylogenies and their implications for the study of diversification, taxonomy, and genome size evolution of Rhododendron. Am. J. Bot. 2021, 108, 1957–1981. [Google Scholar] [CrossRef]
- Lin, W.; Liu, L.; Liang, J.C.; Tang, X.X.; Shi, J.; Zhang, L.; Wu, P.; Lan, S.; Wang, S.; Zhou, Y.; et al. Changes of endophytic microbial community in Rhododendron dauricum roots under heat stress and its correlation with leaf physiological indicators. Front. Microbiol. 2022, 13, 1006686. [Google Scholar] [CrossRef]
- Hodges, D.M.; DeLong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D. Bairoch ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Xie, T.; Chen, C.; Luan, A.; Long, J.; Li, C.; Ding, Y.; He, Y. Genome-wide organization and expression profiling of the R2R3-MYB transcription factor family in pineapple (Ananas comosus). BMC Genom. Data 2017, 18, 503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Chen, H. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Anam, S.; Hilal, B.; Fariduddin, Q. Polyamines and hydrogen peroxide: Allies in plant resilience against abiotic stress. Chemosphere 2024, 366, 143438. [Google Scholar] [CrossRef]
- Ghosh, U.K.; Islam, M.N.; Siddiqui, M.N.; Cao, X.; Khan, M.A.R. Proline, a multifaceted signalling molecule in plant responses to abiotic stress: Understanding the physiological mechanisms. J. Integr. Plant Biol. 2022, 24, 227–239. [Google Scholar] [CrossRef]
- Hnilickova, H.; Kraus, K.; Vachova, P.; Hnilicka, F. Salinity Stress Affects Photosynthesis, Malondialdehyde Formation, and Proline Content in Portulaca oleracea L. Plants 2021, 10, 845. [Google Scholar] [CrossRef]
- Bowler, C.; Van Camp, W.; Van Montagu, M.; Inzé, D.; Asada, K. Superoxide Dismutase in Plants. CRC Crit. Rev. Plant Sci. 1994, 13, 199–218. [Google Scholar] [CrossRef]
- Purugganan, M.D. The evolution of rice: Molecular vignettes on its origins and spread. Archaeol Anthropol Sci. 2010, 2, 61–68. [Google Scholar] [CrossRef]
- Baumgarten, A.; Cannon, S.; Spangler, R.; May, G. Genome-Level Evolution of Resistance Genes in Arabidopsis thaliana. Genetics 2003, 165, 309–319. [Google Scholar] [CrossRef]
- Polezhaeva, M.A.; Pimenova, E.A.; Tikhonova, N.A.; Korchagina, O.S. Plastid DNA diversity and genetic divergence within Rhododendron dauricum s.l. (R. dauricum s.s., R. ledebourii, R. sichotense and R. mucronulatum; Ericaceae). Plant Syst Evol. 2018, 304, 763–774. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Jaime, F.; García, M.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell. 2016, 127, 269–287. [Google Scholar] [CrossRef]
- Alexandre, P.; Andrea, L.M.; Eveland, A.L.; Kaufmann, K.; Nathan, M.S. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49–60. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Cai, Z.; He, F.; Feng, X.; Liang, T.; Wang, H.W.; Ding, S.C.; Tian, X.H. Transcriptomic Analysis Reveals Important Roles of Lignin and Flavonoid Biosynthetic Pathways in Rice Thermotolerance During Reproductive Stage. Front. Genet. 2020, 11, 1664–8021. [Google Scholar] [CrossRef]
- Gabriela, T.O.; Huq, E.; Peter, H.Q. The Arabidopsis Basic/Helix-Loop-Helix Transcription Factor Family. Plant Cell 2003, 15, 1749–1770. [Google Scholar]
- Li, J.; Zhang, Z.; Vang, S.; Yu, J.; Wong, G.K.-S.; Wang, J. Correlation Between Ka/Ks and Ks is Related to Substitution Model and Evolutionary Lineage. J. Mol. Evol. 2009, 68, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Gudesblat, G.; Schneider-Pizoń, J.; Betti, C.; Mayerhofer, J.; Vanhoutte, I.; van Dongen, W.; Boeren, S.; Zhiponova, M.; de Vries, S.; Jonak, C.; et al. SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 2012, 14, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.C.; Yu, S.Y.; Wang, Y.D.; Jinn, T.L. Guard Cell-Specific Pectin METHYLESTERASE53 Is Required for Abscisic Acid-Mediated Stomatal Function and Heat Response in Arabidopsis. Front. Plant Sci. 2022, 13, 836151. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Fan, S.; Zhang, Q.; Wang, Y. Effect of heat stress on the MDA, proline and soluble sugar content in leaf lettuce seedlings. Agric. Sci. 2013, 4, 112–115. [Google Scholar] [CrossRef]
- Jiang, L.; Tian, X.; Li, S.; Fu, Y.; Xu, J.; Wang, G. The AabHLH35 Transcription Factor Identified from Anthurium andraeanum is Involved in Cold and Drought Tolerance. Plants 2019, 8, 216. [Google Scholar] [CrossRef]
- Wang, P.; Wang, H.; Wang, Y.; Ren, F.; Liu, W. Analysis of bHLH genes from foxtail millet (Setaria italica) and their potential relevance to drought stress. PLoS ONE 2018, 13, e0207344. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and Mechanism of Jasmonic Acid in Plant Responses to Abiotic and Biotic Stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Su, Y.; Huang, Y.; Dong, X.; Wang, R.; Tang, M.; Cai, J.; Chen, J.; Zhang, X.; Nie, G. Exogenous methyl jasmonate improves heat tolerance of perennial ryegrass through alteration of osmotic adjustment, antioxidant defense, and expression of jasmonic acid-responsive genes. Front. Plant Sci. 2021, 12, 665. [Google Scholar] [CrossRef]
- Zhang, F.; Yao, J.; Ke, J.; Zhang, L.; Lam, V.Q.; Xin, X.-F.; Zhou, X.E.; Chen, J.; Brunzelle, J.; Griffin, P.R.; et al. Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling. Nature 2015, 525, 269–273. [Google Scholar] [CrossRef]
- Cao, F.; Zheng, Y.H.; Wang, K.T.; Rui, H. Methyl jasmonate reduces chilling injury and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chem. 2009, 115, 1458–1463. [Google Scholar] [CrossRef]
- Baskar, V.; Venkatesh, R.; Ramalingam, S. Flavonoids (Antioxidants Systems) in Higher Plants and Their Response to Stresses. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D., Palma, J., Corpas, F., Eds.; Springer: Cham, Switzerland, 2018; pp. 253–268. [Google Scholar]
- Coberly, L.C.; Rausher, M.D. Analysis of a chalcone synthase mutant in Ipomoea purpurea reveals a novel function for flavonoids: Amelioration of heat stress. Mol. Ecol. 2003, 12, 1113–1124. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Rong, X.; Li, X.; Ma, Y.; Cheng, H.; Sheng, J.; Huang, L.; Jin, S. Transcriptome and Flavonoid Compounds Metabolome Analyses Reveal the Mechanisms of Heat Stress in Rhododendron with Exogenously Applied Calcium. Agronomy. 2024, 14, 1282. [Google Scholar] [CrossRef]
- Woch, N.; Laha, S.; Gudipalli, P. Salicylic acid and jasmonic acid induced enhanced production of total phenolics, flavonoids, and antioxidant metabolism in callus cultures of Givotia moluccana (L.) Sreem. In Vitro Cell. Dev. Biol. Plant 2023, 59, 227–248. [Google Scholar] [CrossRef]
- Cao, J.; Li, M.; Chen, J.; Liu, P.; Li, Z. Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Sci. Rep. 2016, 6, 37674. [Google Scholar] [CrossRef] [PubMed]
- Du, T.T.; Fan, Y.X.; Cao, H.Y.; Song, Z.; Dong, B.; Liu, T.; Yang, W.; Wang, M.; Niu, L.; Yang, Q.; et al. Transcriptome analysis revealed key genes involved in flavonoid metabolism in response to jasmonic acid in pigeon pea (Cajanus cajan (L.) Millsp.). Plant Physiol. Biochem. 2021, 168, 410–422. [Google Scholar] [CrossRef]







| Duplicated Gene Pairs | Non Synonymous (Ka) | Synonymous (Ks) | Ka/Ks |
|---|---|---|---|
| RhbHLH76 & RhbHLH4 | 0.2254 | 0.8422 | 0.2676 |
| RhbHLH17 & RhbHLH6 | 0.2277 | 1.3497 | 0.1687 |
| RhbHLH36 & RhbHLH99 | 0.4423 | 2.0141 | 0.2196 |
| RhbHLH17 & RhbHLH15 | 0.1184 | 0.7172 | 0.1651 |
| RhbHLH55 & RhbHLH8 | 0.3218 | 0.9632 | 0.3341 |
| RhbHLH28 & RhbHLH20 | 0.0990 | 0.6096 | 0.1624 |
| RhbHLH65 & RhbHLH22 | 0.2189 | 0.5992 | 0.3654 |
| RhbHLH30 & RhbHLH28 | 0.2460 | 2.3699 | 0.1038 |
| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| RhTH1 | 50,883,988 | 49,954,064 | 7.49 | 0.03 | 97.81 | 93.81 | 46.8 |
| RhTH2 | 43,558,656 | 42,651,196 | 6.4 | 0.03 | 97.89 | 93.68 | 46.57 |
| RhTH3 | 44,111,830 | 43,263,532 | 6.49 | 0.03 | 97.85 | 93.56 | 46.61 |
| RhTN1 | 41,280,128 | 40,342,882 | 6.05 | 0.03 | 97.93 | 93.84 | 46.89 |
| RhTN2 | 44,580,586 | 43,394,078 | 6.51 | 0.03 | 97.93 | 93.84 | 46.95 |
| RhTN3 | 70,401,558 | 68,999,938 | 10.35 | 0.03 | 97.96 | 93.83 | 47.09 |
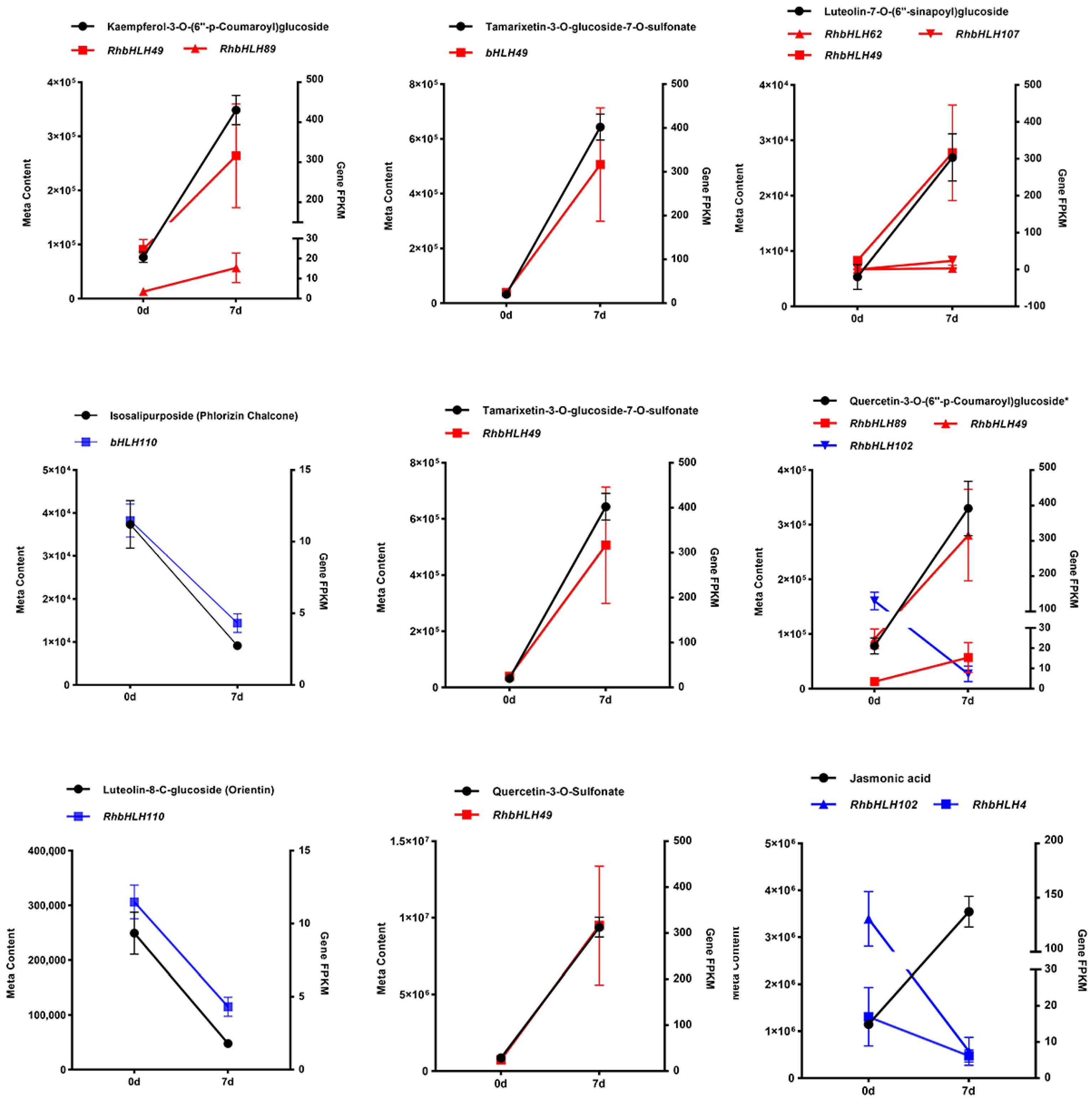
| Compounds | Class | Gene Name | PCC | PCCP |
|---|---|---|---|---|
| 7-O-Methyleriodictyol | Flavanols | RhbHLH49 | 0.967 | 0.001607786 |
| Isosalipurposide (Phlorizin Chalcone) | Chalcones | RhbHLH110 | −0.946 | 0.0043221 |
| Kaempferol-3-O-(6″-p-Coumaroyl)glucoside | Flavonols | RhbHLH49 | 0.968 | 0.001487601 |
| Kaempferol-3-O-(6″-p-Coumaroyl)glucoside | Flavonols | RhbHLH89 | 0.925 | 0.008120533 |
| Luteolin-7-O-(6″-sinapoyl)glucoside | Flavonoid | RhbHLH49 | 0.971 | 0.001227645 |
| Luteolin-7-O-(6″-sinapoyl)glucoside | Flavonoid | RhbHLH62 | 0.944 | 0.004665124 |
| Luteolin-7-O-(6″-sinapoyl)glucoside | Flavonoid | RhbHLH107 | 0.933 | 0.006563481 |
| Luteolin-8-C-glucoside (Orientin) | Flavonoid carbonoside | RhbHLH110 | −0.97 | 0.001349201 |
| Quercetin-3-O-(6″-p-Coumaroyl)glucoside* | Flavonols | RhbHLH89 | 0.946 | 0.004371937 |
| Quercetin-3-O-(6″-p-Coumaroyl)glucoside* | Flavonols | RhbHLH49 | 0.942 | 0.004896722 |
| Quercetin-3-O-(6″-p-Coumaroyl)glucoside* | Flavonols | RhbHLH102 | −0.976 | 0.000866412 |
| Quercetin-3-O-Sulfonate | Flavonols | RhbHLH49 | 0.977 | 0.00078461 |
| Tamarixetin-3-O-glucoside-7-O-sulfonate | Flavonoid | RhbHLH49 | 0.936 | 0.005967489 |
| Jasmonic acid | Organic acids | RhbHLH4 | −0.949 | 0.003875338 |
| Jasmonic acid | Organic acids | RhbHLH102 | −0.936 | 0.006062754 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Z.; Wang, E.; Zhai, M.; Li, Y.; Li, Y.; Shi, J.; Feng, X.; Wang, J.; Song, X.; Shi, Y.; et al. Identification of bHLH Family Genes in Rhododendron hainanense Merr. and Verification of Their Role in Heat Tolerance. Forests 2025, 16, 416. https://doi.org/10.3390/f16030416
Cao Z, Wang E, Zhai M, Li Y, Li Y, Shi J, Feng X, Wang J, Song X, Shi Y, et al. Identification of bHLH Family Genes in Rhododendron hainanense Merr. and Verification of Their Role in Heat Tolerance. Forests. 2025; 16(3):416. https://doi.org/10.3390/f16030416
Chicago/Turabian StyleCao, Zidan, Enbo Wang, Minghui Zhai, Yufei Li, Yifan Li, Jiaxuan Shi, Xueping Feng, Jian Wang, Xiqiang Song, Youhai Shi, and et al. 2025. "Identification of bHLH Family Genes in Rhododendron hainanense Merr. and Verification of Their Role in Heat Tolerance" Forests 16, no. 3: 416. https://doi.org/10.3390/f16030416
APA StyleCao, Z., Wang, E., Zhai, M., Li, Y., Li, Y., Shi, J., Feng, X., Wang, J., Song, X., Shi, Y., & Zhao, Y. (2025). Identification of bHLH Family Genes in Rhododendron hainanense Merr. and Verification of Their Role in Heat Tolerance. Forests, 16(3), 416. https://doi.org/10.3390/f16030416







