Physiological and Molecular Insights into the Development of Single and Double Flowers in Syringa vulgaris L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
- (i)
- Double flowers of the S. vulgaris cultivars (“Fengnian”) (held by Harbin land-scaping research Institute):
- (ii)
- Single flowers of S. vulgaris (held by Harbin landscaping research Institute):
2.2. Morphological Identification
2.3. Physiological and Biochemical Analyses
- (i)
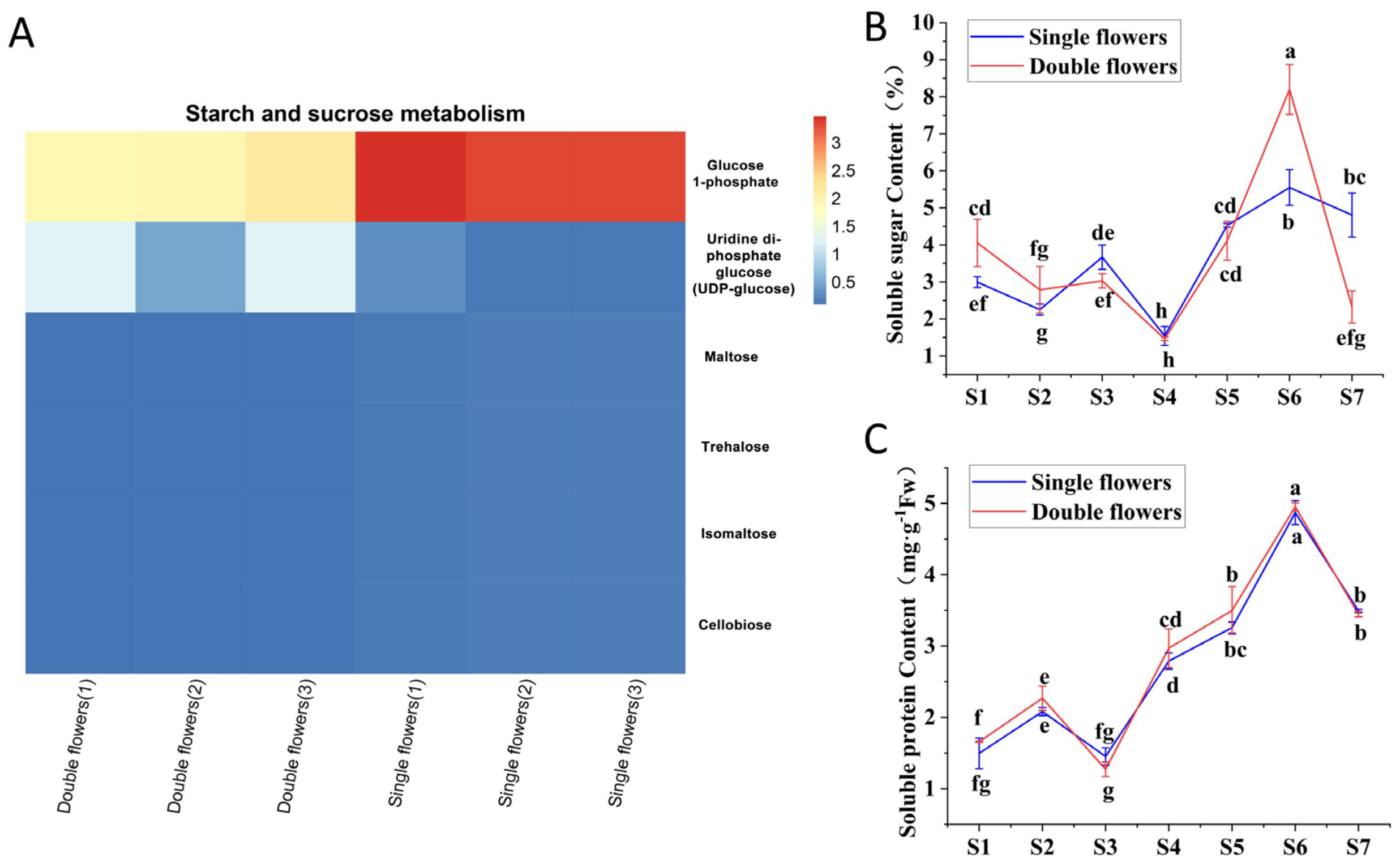
- Soluble sugar content (0.1 g of flower buds per run): This content was determined using the anthrone-sulfuric acid colorimetric method [44].
- (ii)
- Soluble protein content (0.1 g of flower buds per run): This content was measured via the Coomassie brilliant blue colorimetric method [45].
- (iii)
- Antioxidant enzyme activities and MDA content: The peroxidase (POD), superoxide dismutase (SOD), and catalase (CAT) activities and the malondialdehyde (MDA) content were assessed using oxidative stress detection kits. POD, SOD, and CAT activity and MDA content were measured with 0.1 g of material, using an oxidative stress series kit (microplate method) (Suzhou Grace Biotechnology Co., Ltd., Suzhou, China) [46] according to the manufacturer’s instructions.
- (iv)
- Endogenous hormone levels (0.1 g of flower buds per run): The levels of IAA, abscisic acid (ABA), GA3, 1-aminocyclopropane-1-carboxylic acid (ACC), salicylic acid (SA), jasmonic acid (JA), cis-ZR (cZR), and trans-ZR (tZR) were quantified via ultrahigh-performance liquid chromatography coupled with tandem mass spectrometry (UHPLC-MRM-MS/MS) [47]; the concentration ranges of these hormones were as follows [48]:
- IAA concentration range: 10–100 ng/g;
- GA3 concentration range: 5–50 ng/g;
- ABA concentration range: 1–20 ng/g;
- ACC concentration range: 5–100 ng/g;
- SA concentration range: 500–4000 ng/g;
- JA concentration range: 0–100 ng/g;
- cZR concentration range: 2–30 ng/g;
- tZR concentration range: 2–30 ng/g.
2.4. Data Analysis
2.5. Transcriptome Analysis
2.6. Metabolomics Analysis
2.7. Quantitative Real-Time PCR (qRT-PCR) Validation
3. Results
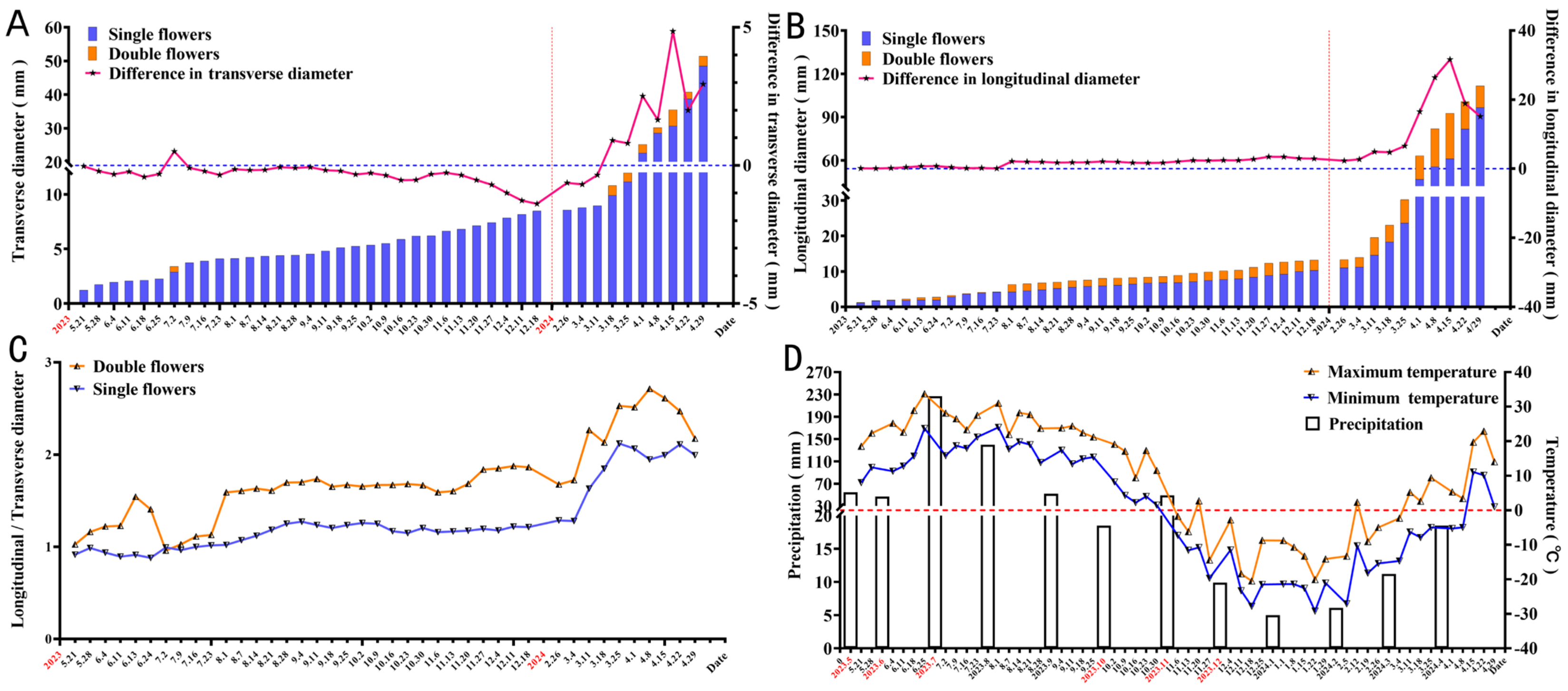
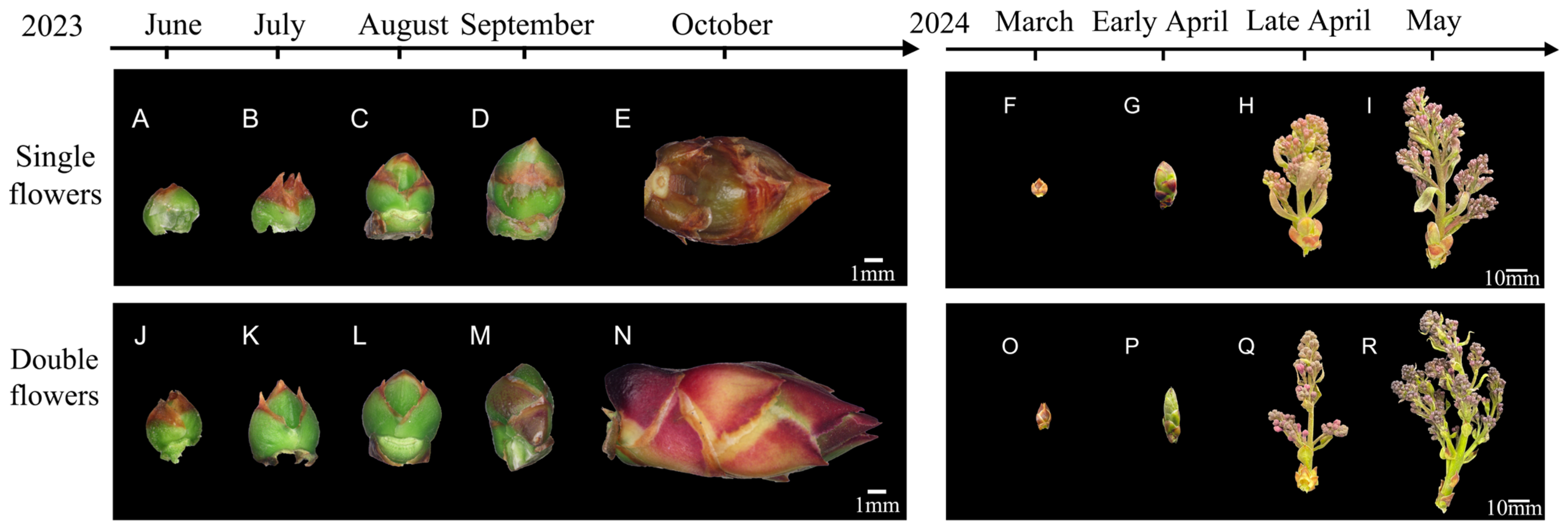
3.1. Morphological Differences Between Single and Double Flower Buds
3.1.1. Flower Bud Morphological Changes
3.1.2. Dynamics of Inflorescence Development
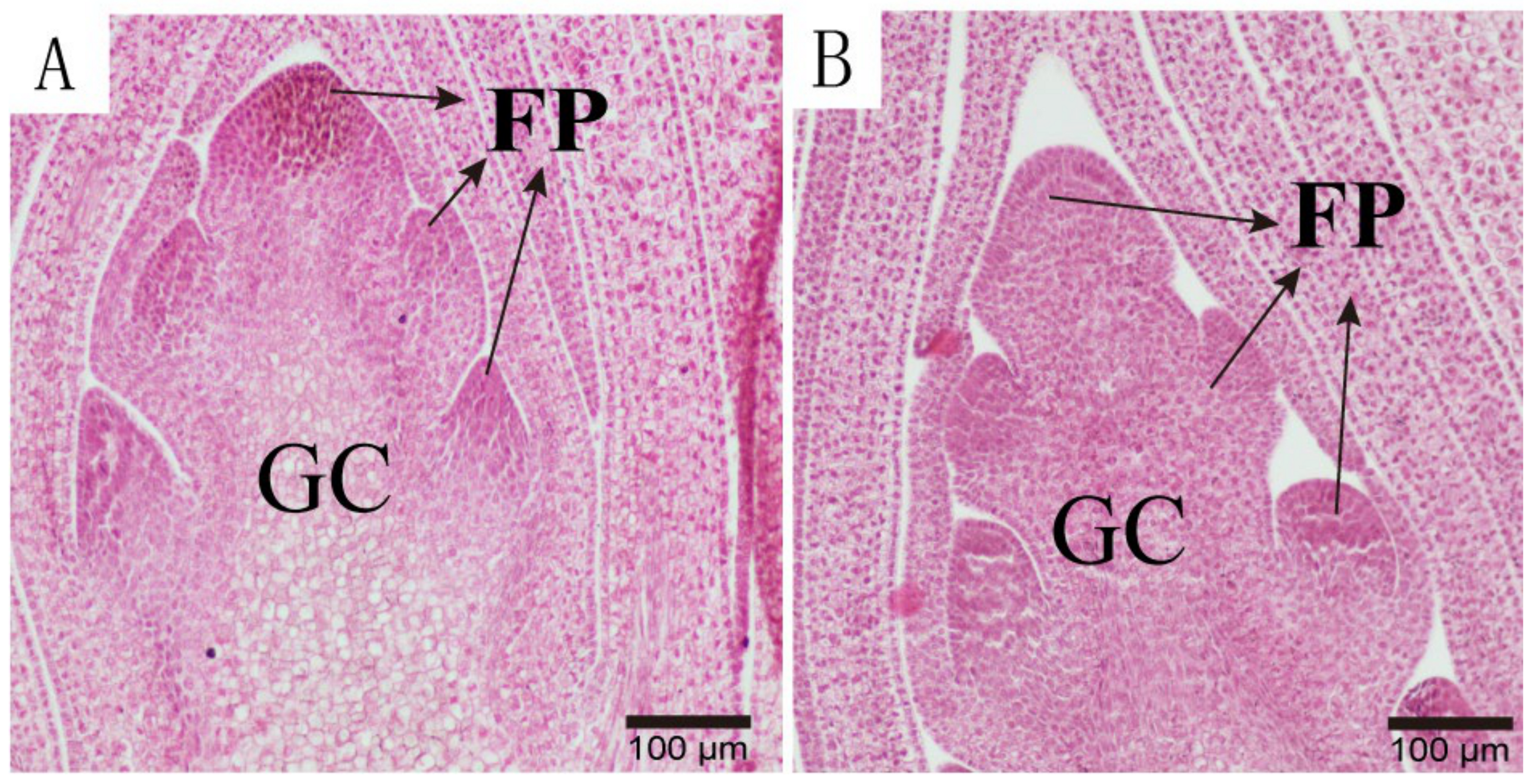
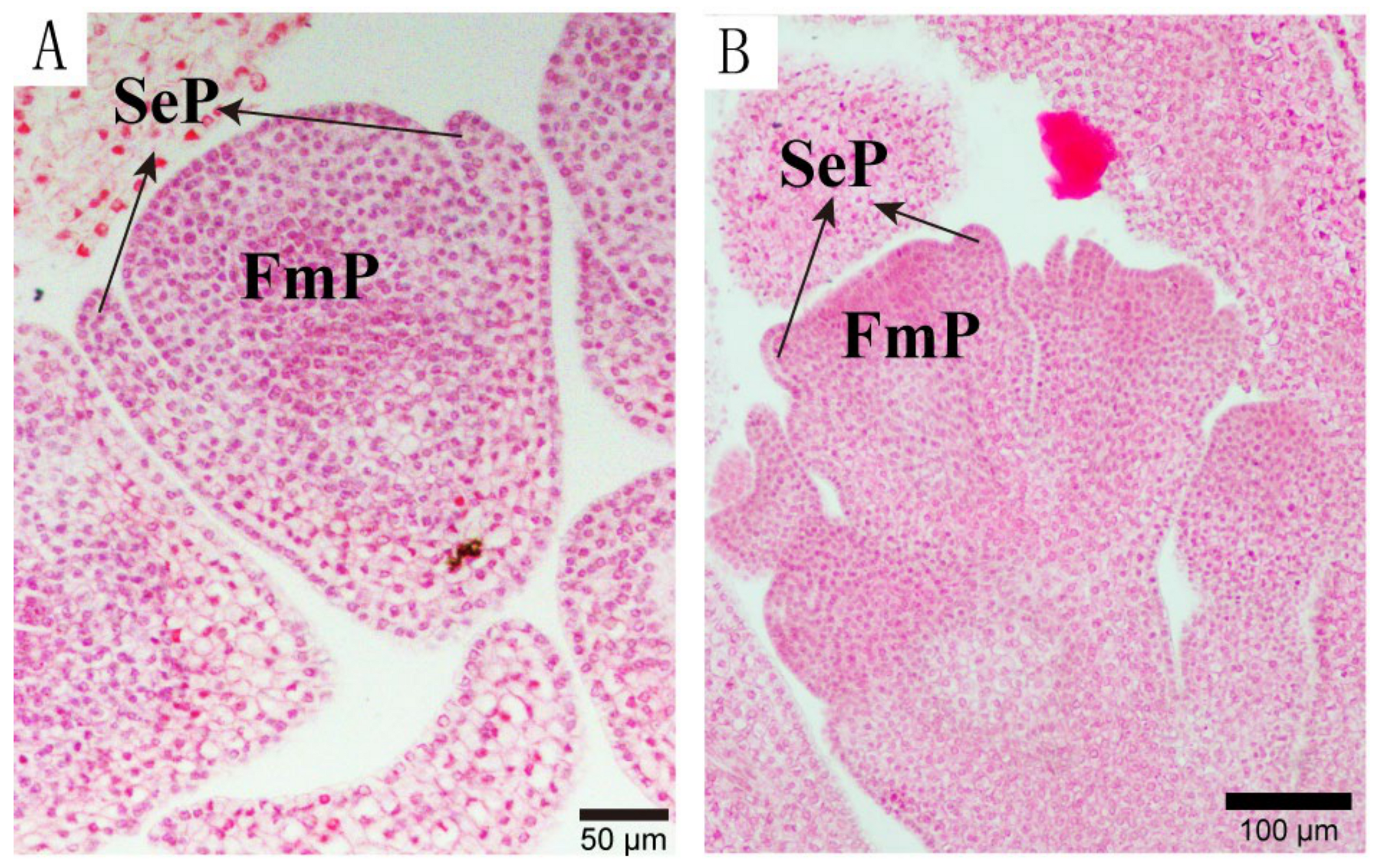
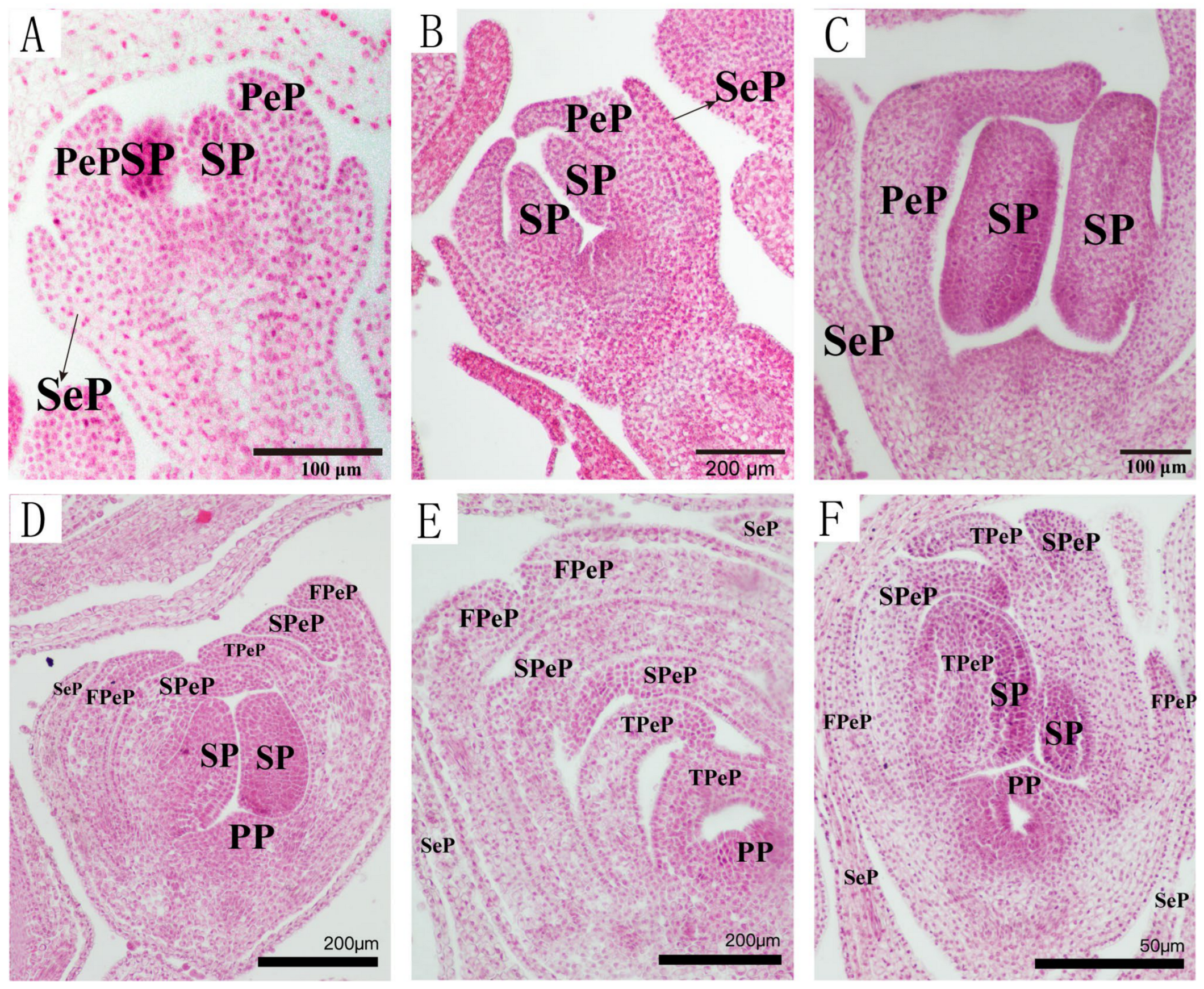
3.1.3. Anatomical Characteristics of Single and Double Flower Buds at Different Differentiation Stages
- (i)
- Bract Primordium Differentiation Stage:
- (ii)
- Inflorescence and Floret Primordium Differentiation Stage:
- (iii)
- Sepal Primordium Differentiation Stage:
- (iv)
- Petal Primordium Differentiation Stage:
- (v)
- Stamen Primordium Differentiation Stage:
- (vi)
- Carpel Primordium Differentiation Stage:
3.2. Transcriptomic and Metabolomic Differences in the Differentiation Processes of Single and Double Flower Buds
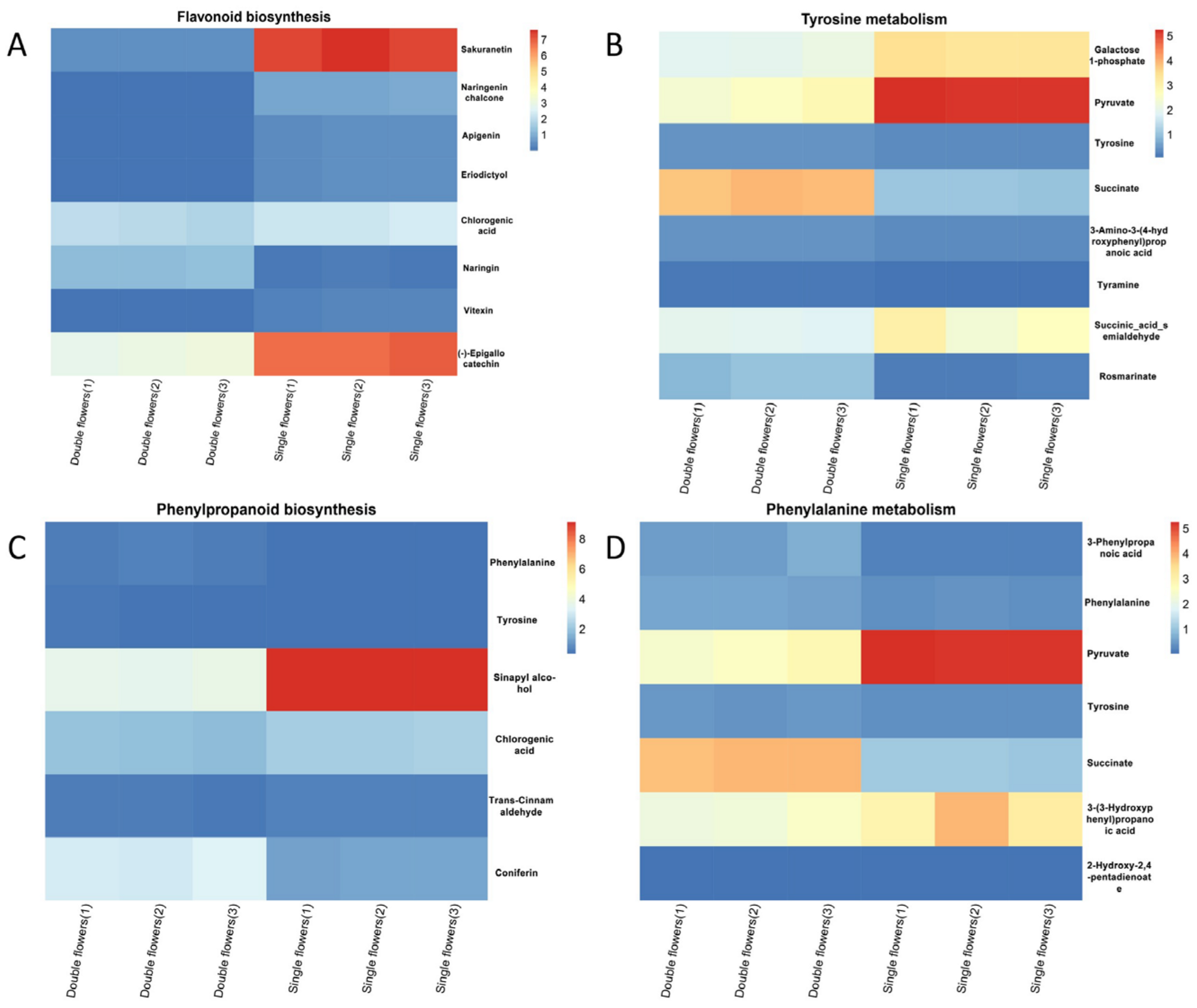
3.3. Expression Characteristics of Genes and Metabolites Related to Flower Color Formation
3.4. Association Between Carbohydrate Metabolites and Petal Structure Formation During Floral Bud Differentiation in S. vulgaris
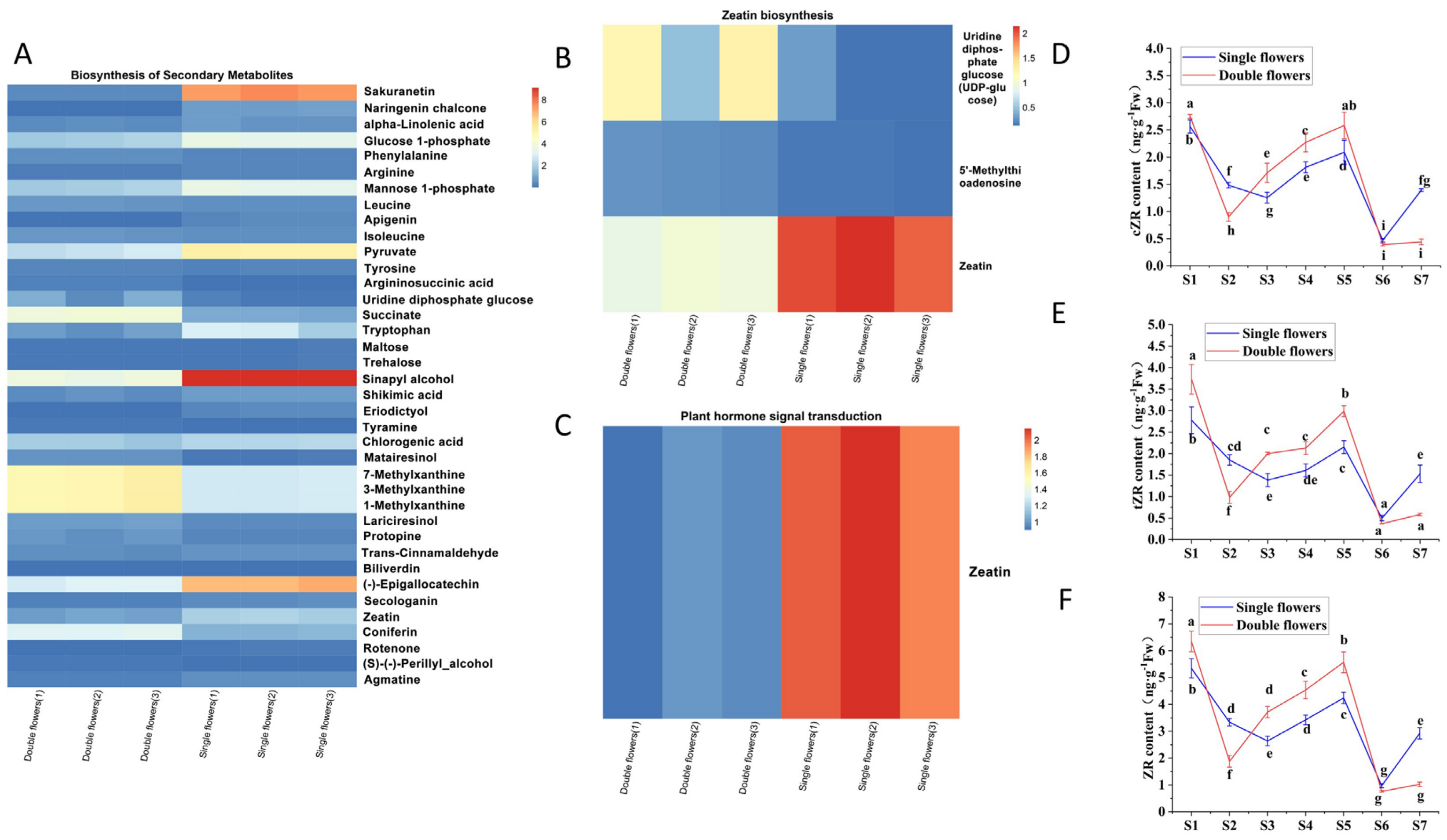
3.5. Expression Characteristics of Genes and Metabolites Associated with Petal Layer Formation
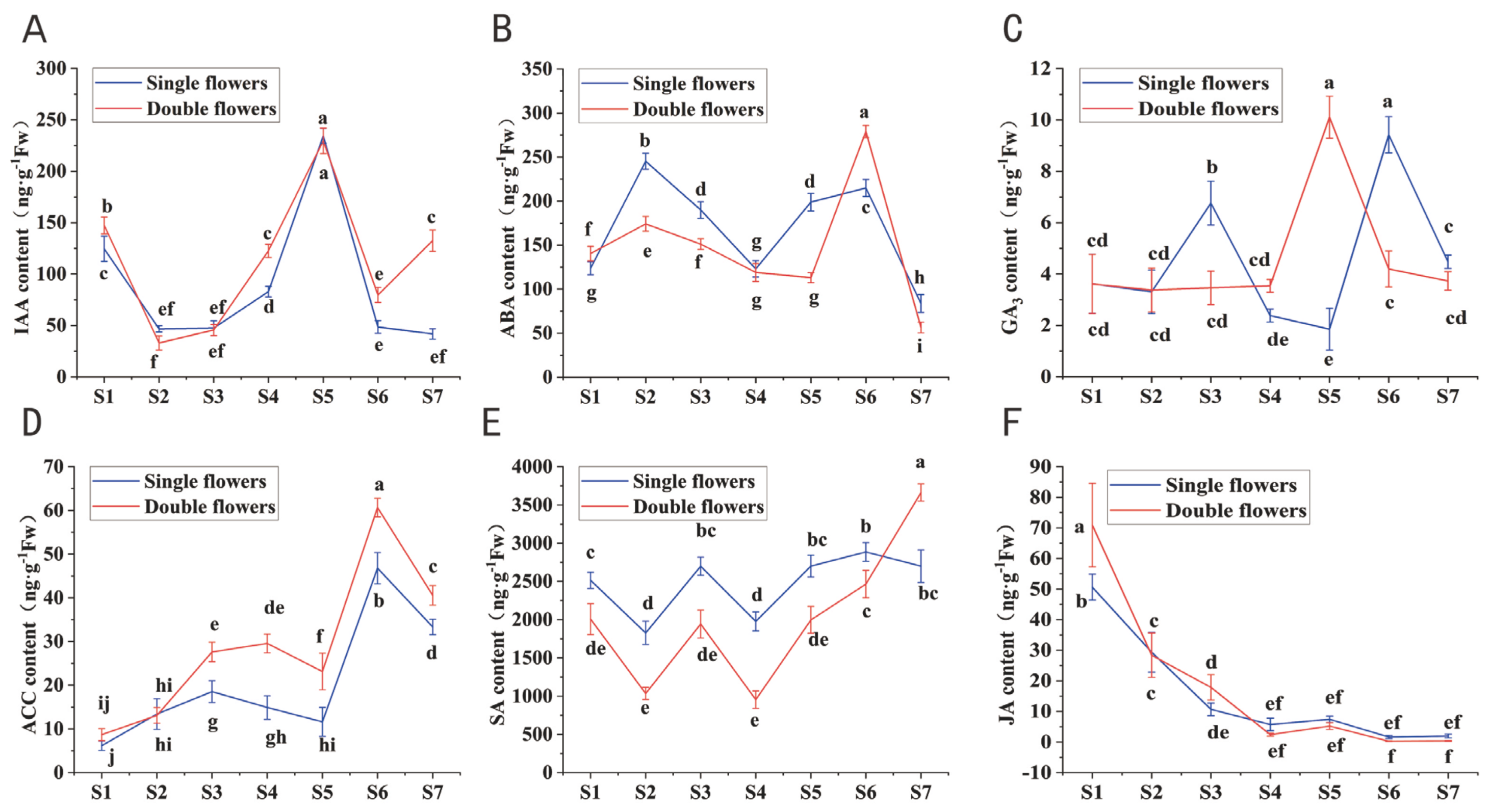
3.6. Differences in Endogenous Hormone Levels Between Single and Double Flowers of S. vulgaris During Floral Bud Differentiation
- (i)
- ABA/GA3 Ratio:
- (ii)
- IAA/GA3 Ratio:
- (iii)
- ABA/IAA Ratio:
- (iv)
- ZR/GA3 Ratio:
- (v)
- IAA/ZR Ratio:
- (vi)
- SA/ACC Ratio:
3.7. Relationship Between the ABC Transporter System and Antioxidant Enzyme Activity During Floral Bud Differentiation in Single and Double Flowers of S. vulgaris
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Appendix AMS2 Name | KEGG COMPOUND ID | Class | Subclass |
|---|---|---|---|
| Sakuranetin | C09833 | Flavonoids | Flavanones |
| 3-Phenylpropanoic acid | C05629 | ||
| Naringenin chalcone | C06561 | Flavonoids | Chalcones |
| 4-Hydroxyphenethylalcohol | C06044 | Phenylethanoids | Phenylethanoids |
| Esculin | C09264 | Coumarins | Simple coumarins |
| gamma-Linolenic acid | C06426 | Fatty acids and conjugates | Unsaturated fatty acids |
| alpha-Linolenic acid | C06427 | Fatty acids and conjugates | Unsaturated fatty acids |
| Malic acid | C00149 | Fatty acids and conjugates | Dicarboxylic acids |
| Glucose 1-phosphate | C00103 | Saccharides | Monosaccharides |
| Guanosine | C00387 | Nucleosides | Purine nucleos(t)ides |
| Phenylalanine | C00079 | Small peptides | Amino acids |
| Galactose 1-phosphate | C00446 | Saccharides | Monosaccharides |
| Dodecanedioic acid | C02678 | Fatty acids and conjugates | Dicarboxylic acids |
| 6-Hydroxyluteolin 7-glucoside | C17763 | Flavonoids | Flavones |
| Arginine | C00062 | Small peptides | Amino acids |
| LPC(18:2/0:0) | C04100 | ||
| Norleucine | C01933 | Fatty acids and conjugates | Amino fatty acids |
| 9-Oxo-10(E),12(E)-octadecadienoic acid | C14766 | Fatty acids and conjugates | Unsaturated fatty acids |
| Mannose 1-phosphate | C00636 | Saccharides | Monosaccharides |
| Leucine | C00123 | Small peptides | Amino acids |
| Apigenin | C01477 | Flavonoids | Flavones |
| 5,7-Dihydroxychromone | C09001 | Chromanes | Chromones |
| Methylmalonic acid | C02170 | Fatty acids and conjugates | Dicarboxylic acids |
| Uracil | C00106 | Pseudoalkaloids | Purine alkaloids |
| Isoleucine | C00407 | Small peptides | Amino acids |
| 2′-Deoxyguanosine 5′-monophosphate (dGMP) | C00362 | Nucleosides | Purine nucleos(t)ides |
| Nicotinamide | C00153 | Nicotinic acid alkaloids | Pyridine alkaloids |
| Pyruvate | C00022 | Fatty acids and conjugates | Oxo fatty acids |
| Tyrosine | C00082 | Small peptides | Amino acids |
| Argininosuccinic acid | C03406 | Small peptides | Amino acids |
| Uridine diphosphate glucose (UDP-glucose) | C00029 | Amino sugars and aminoglycosides | Amino sugars |
| Succinate | C00042 | Fatty acids and conjugates | Dicarboxylic acids |
| UDP-galactose | C00052 | Amino sugars and aminoglycosides | Amino sugars |
| Euscaphic acid | C17890 | Triterpenoids | Ursane and taraxastane triterpenoids |
| Lumichrome | C01727 | Pseudoalkaloids | Pteridine alkaloids |
| Tryptophan | C00078 | Small peptides | Amino acids |
| Maltose | C00208 | Saccharides | Disaccharides |
| Melibiose | C05402 | Saccharides | Disaccharides |
| Trehalose | C01083 | Saccharides | Disaccharides |
| Sinapyl alcohol | C02325 | Phenylpropanoids (C6–C3) | Cinnamic acids and derivatives |
| Shikimic acid | C00493 | Phenolic acids (C6–C1) | Shikimic acids and derivatives |
| Homoarginine | C01924 | Small peptides | Amino acids |
| Eriodictyol | C05631 | Flavonoids | Flavanones |
| Arabinono-1,4-lactone | C01114 | Saccharides | Monosaccharides |
| Daltogen | C06771 | ||
| 3-Amino-3-(4-hydroxyphenyl)propanoic acid | C04368 | Small peptides | Amino acids |
| Isomaltose | C00252 | Saccharides | Disaccharides |
| Tyramine | C00483 | Tyrosine alkaloids | Phenylethylamines |
| Glycerophosphocholine | C00670 | ||
| Cellobiose | C00185 | Saccharides | Disaccharides |
| 2-Hydroxystearic acid | C03045 | Fatty acids and conjugates | Hydroxy fatty acids |
| Galactaric acid | C00879 | Fatty acids and conjugates | Hydroxy fatty acids |
| Chlorogenic acid | C00852 | Phenylpropanoids | Cinnamic acids and derivatives |
| Matairesinol | C10682 | Lignans | Dibenzylbutyrolactone lignans |
| 7-Methylxanthine | C16353 | Pseudoalkaloids | Purine alkaloids |
| Emodin | C10343 | Polycyclic aromatic polyketides | Anthraquinones and anthrones |
| Methylimidazoleacetic acid | C05828 | Histidine alkaloids | Imidazole alkaloids |
| Palmitic acid | C00249 | Fatty acids and conjugates | Branched fatty acids |
| 3-(3-Hydroxyphenyl)propanoic acid | C11457 | ||
| 3-(2-Hydroxyphenyl)propanoic acid | C01198 | ||
| Dimethyl phthalate | C11233 | Phenolic acids | Simple phenolic acids |
| Naringin | C09789 | Flavonoids | Flavanones |
| Adenosine | C00212 | Nucleosides | Purine nucleos(t)ides |
| Vanillic acid | C06672 | Phenolic acids | Cinnamic acids and derivatives |
| Glutamine | C00064 | Small peptides | Amino acids |
| 5′-Methylthioadenosine | C00170 | Nucleosides | Purine nucleos(t)ides |
| 3-Methylxanthine | C16357 | Pseudoalkaloids | Purine alkaloids |
| 1-Methylxanthine | C16358 | Pseudoalkaloids | Purine alkaloids |
| Pioglitazone | C07675 | ||
| Lariciresinol | C10646 | Lignans | Furanoid lignans |
| Pravastatin | C01844 | Fatty acyls | Fatty alcohols |
| Psoralidin | C10523 | Isoflavonoids | Coumestan |
| Piceatannol | C05901 | Stilbenoids | Monomeric stilbenes |
| Luteolin 7-glucoside | C03951 | Flavonoids | Flavones |
| Vitexin | C01460 | Flavonoids | Flavones |
| Succinic_acid_semialdehyde | C00232 | Fatty acyls | Fatty aldehydes |
| 3-O-Feruloylquinic acid | C02572 | Phenylpropanoids | Cinnamic acids and derivatives |
| 1,3-Dicaffeoylquinic acid | C10445 | Phenylpropanoids | Cinnamic acids and derivatives |
| Salicin | C01451 | Phenolic acids | Simple phenolic acids |
| Cuminaldehyde | C06577 | Monoterpenoids | Menthane monoterpenoids |
| Acetophenone | C07113 | ||
| Syringic acid | C10833 | Phenolic acids | Simple phenolic acids |
| 3,4-Dihydroxybenzaldehyde | C16700 | Phenolic acids | Shikimic acids and derivatives |
| Hyperoside | C10073 | Flavonoids | Flavonols |
| Quercitrin | C01750 | Flavonoids | Flavonols |
| Protopine | C05189 | Tyrosine alkaloids | Isoquinoline alkaloids |
| trans-Cinnamaldehyde | C00903 | Phenylpropanoids | Cinnamic acids and derivatives |
| Biliverdin | C00500 | ||
| Nandrolone | C07254 | Steroids | Estrane steroids |
| (−)-Epigallocatechin | C12136 | Flavonoids | Flavan-3-ols |
| 3-Hydroxy-4-methoxycinnamic acid | C10470 | Phenylpropanoids | Cinnamic acids and derivatives |
| Glabridin | C10421 | Isoflavonoids | Pterocarpan |
| Secologanin | C01852 | Monoterpenoids | Secoiridoid monoterpenoids |
| Pitavastatin | C13334 | ||
| Etoposide | C01576 | Lignans | Arylnaphthalene and aryltetralin lignans |
| Zeatin | C00371 | Pseudoalkaloids | Purine alkaloids |
| Nizatidine | C07270 | ||
| Morin | C10105 | Flavonoids | Flavonols |
| coniferin | C00761 | Phenylpropanoids | Cinnamic acids and derivatives |
| Neolinustatin | C08336 | ||
| Arctiin | C16915 | Lignans | Dibenzylbutyrolactone lignans |
| Rhein | C10401 | Polycyclic aromatic polyketides | Anthraquinones and anthrones |
| Rotenone | C07593 | Isoflavonoids | Rotenoids |
| Procyanidin B2 | C17639 | Flavonoids | Proanthocyanins |
| (S)-(−)-Perillyl_alcohol | C02452 | Monoterpenoids | Menthane monoterpenoids |
| Rosmarinate | C01850 | Phenylpropanoids | Cinnamic acids and derivatives |
| Agmatine | C00179 | Ornithine alkaloids | Polyamines |
| Stearoyl-CoA | C00412 | Fatty esters | Fatty acyl CoAs |
| Oleoyl-CoA | C00510 | Fatty esters | Fatty acyl CoAs |
| Nopaline | C01682 | Small peptides | Amino acids |
| 2-Hydroxy-2,4-pentadienoate | C00596 | Fatty acids and conjugates | Unsaturated fatty acids |
| Guanine | C00242 | Pseudoalkaloids | Pteridine alkaloids |
| 2-Furoate | C01546 |
References
- Eason, J.R.; Webster, D. Development and senescence of Sandersonia aurantiaca (Hook.) flowers. Sci. Hortic. 1995, 63, 113–121. [Google Scholar] [CrossRef]
- Önder, S.; Tonguç, M.; Önder, D.; Erbaş, S.; Mutlucan, M. Flower color and carbohydrate metabolism changes during the floral development of Rosa damascena. S. Afr. J. Bot. 2023, 156, 234–243. [Google Scholar] [CrossRef]
- Wang, H.; Lu, Y.; Zhang, T.; Liu, Z.; Cao, L.; Chang, Q.; Liu, Y.; Lu, X.; Yu, S.; Li, H.; et al. The double flower variant of yellowhorn is due to a LINE1 transposon-mediated insertion. Plant Physiol. 2023, 191, 1122–1137. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, X.; Lin, S.; Yang, S.; Yan, X.; Bendahmane, M.; Bao, M.; Fu, X. Mapping a double flower phenotype-associated gene DcAP2L in Dianthus chinensis. J. Exp. Bot. 2020, 71, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Jiang, W.; Lin, Z.; Lin, Q.; Ye, Q.; Wang, W.; Xie, Q.; He, X.; Luo, C.; Chen, Q. SMRT and Illumina RNA-Seq identifies potential candidate genes related to the double flower phenotype and unveils SsAP2 as a key regulator of the double-flower trait in Sagittaria sagittifolia. Int. J. Mol. Sci. 2022, 23, 2240. [Google Scholar] [CrossRef]
- Zhu, H.; Shi, Y.; Zhang, J.; Bao, M.; Zhang, J. Candidate genes screening based on phenotypic observation and transcriptome analysis for double flower of Prunus mume. BMC Plant Biol. 2022, 22, 499. [Google Scholar] [CrossRef] [PubMed]
- Dadpour, M.R.; Naghiloo, S.; Peighambardoust, S.H.; Panahirad, S.; Aliakbari, M.; Movafeghi, A. Comparison of floral ontogeny in wild-type and double-flowered phenotypes of Syringa vulgaris L. (Oleaceae). Sci. Hortic. 2011, 127, 535–541. [Google Scholar] [CrossRef]
- Wang, Q.; Dan, N.; Zhang, X.; Lin, S.; Bao, M.; Fu, X. Identification, Characterization and Functional Analysis of C-Class Genes Associated with Double Flower Trait in Carnation (Dianthus caryphyllus L.). Plants 2020, 9, 87. [Google Scholar] [CrossRef]
- Xia, Y.; Shi, M.; Chen, W.; Hu, R.; Jing, D.; Wu, D.; Wang, S.; Li, Q.; Deng, H.; Guo, Q.; et al. Expression Pattern and Functional Characterization of PISTILLATA Ortholog Associated with the Formation of Petaloid Sepals in Double-Flower Eriobotrya japonica (Rosaceae). Front. Plant Sci. 2019, 10, 1685. [Google Scholar] [CrossRef]
- Joan Reynolds, J.T. Double flowers a scientific study; Pembridge Press: London, UK, 1983; Volume 59. [Google Scholar]
- Meyerowitz, E.M.; Smyth, D.R.; Bowman, J.L. Abnormal flowers and pattern formation in floral development. Development 1989, 106, 209–217. [Google Scholar] [CrossRef]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef] [PubMed]
- Thei, G.; Saedler, H. Plant biology: Floral quartets. Nature 2001, 409, 469–472. [Google Scholar]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.I. Effects of temperature and nutrition on growth and flower-bud initiation in apricots. N. Z. J. Agric. Res. 1970, 13, 726–734. [Google Scholar] [CrossRef][Green Version]
- Sanyal, D.; Bangerth, F. Stress induced ethylene evolution and its possible relationship to auxin-transport, cytokinin levels, and flower bud induction in shoots of apple seedlings and bearing apple trees. Plant Growth Regul. 1998, 24, 127–134. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2016, 67, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Meijón, M.; Cañal, M.J.; Fernández, H.; Rodríguez, A.; Fernández, B.; Rodríguez, R.; Feito, I. Hormonal profile in vegetative and floral buds of azalea: Levels of polyamines, gibberellins, and cytokinins. J. Plant Growth Regul. 2011, 30, 74–82. [Google Scholar] [CrossRef]
- Wan, C.; Mi, L.; Chen, B.; Li, J.; Huo, H.; Xu, J.; Chen, X. Effects of nitrogen during nursery stage on flower bud differentiation and early harvest after transplanting in strawberry. Braz. J. Bot. 2018, 41, 1–10. [Google Scholar] [CrossRef]
- Garrod, J.F.; Harris, G.P. Studies on the glasshouse carnation: Effects of temperature and growth substances on petal number. Ann. Bot. 1974, 38, 1025–1031. [Google Scholar] [CrossRef]
- Guo, X.; Yu, C.; Luo, L.; Wan, H.; Zhen, N.; Xu, T.; Tan, J.; Pan, H.; Zhang, Q. Transcriptome of the floral transition in Rosa chinensis ‘Old Blush’. BMC Genom. 2017, 18, 199. [Google Scholar] [CrossRef]
- Alabadí, D.; Blázquez, M.A.; Carbonell, J.; Ferrandiz, C.; Perez-Amador, M.A. Instructive roles for hormones in plant development. Int. J. Dev. Biol. 2009, 53, 1597–1608. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Liu, Y.; Shi, B.; Yuan, H. Exogenous IAA improves the seedling growth of Syringa villosa via regulating the endogenous hormones and enhancing the photosynthesis. Sci. Hortic. 2023, 308, 111585. [Google Scholar] [CrossRef]
- Singh, V.K.; Garg, R.; Jain, M. A global view of transcriptome dynamics during flower development in chickpea by deep sequencing. Plant Biotechnol. J. 2013, 11, 691–701. [Google Scholar] [CrossRef]
- He, C.; Liu, X.; da Silva, J.A.T.; Liu, N.; Zhang, M.; Duan, J. Transcriptome sequencing and metabolite profiling analyses provide comprehensive insight into molecular mechanisms of flower development in Dendrobium officinale (Orchidaceae). Plant Mol. Biol. 2020, 104, 529–548. [Google Scholar] [CrossRef]
- Pei, L.; Gao, Y.; Feng, L.; Zhang, Z.; Liu, N.; Yang, B.; Zhao, N. Phenolic acids and flavonoids play important roles in flower bud differentiation in Mikania micrantha: Transcriptomics and metabolomics. Int. J. Mol. Sci. 2023, 24, 16550. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zou, Q.; Guo, Q.; Yang, F.; Wu, L.; Zhang, W. Widely targeted metabolomics analysis reveals the effect of flooding stress on the synthesis of flavonoids in Chrysanthemum morifolium. Molecules 2019, 24, 3695. [Google Scholar] [CrossRef] [PubMed]
- Schmitzer, V.; Osterc, G.; Veberic, R.; Stampar, F. Correlation between chromaticity values and major anthocyanins in seven Acer palmatum Thunb. cultivars. Sci. Hortic. 2009, 119, 442–446. [Google Scholar] [CrossRef]
- Schmitzer, V.; Veberic, R.; Osterc, G.; Stampar, F. Changes in the phenolic concentration during flower development of rose ‘KORcrisett’. J. Am. Soc. Hortic. Sci. 2009, 134, 491–496. [Google Scholar] [CrossRef]
- Lewandowska, N.; Klimowicz, A. Antioxidant properties of selected parts of Syringa vulgaris L. Pomeranian J. Life Sci. 2022, 68, 64–74. [Google Scholar]
- Woźniak, M.; Michalak, B.; Wyszomierska, J.; Dudek, M.K.; Kiss, A.K. Effects of Phytochemically Characterized Extracts From Syringa vulgaris and Isolated Secoiridoids on Mediators of Inflammation in a Human Neutrophil Model. Front Pharmacol. 2018, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Jędrzejuk, A.; Szlachetka, W.I. Effect of forcing term on morphology of flower buds and quality of panicles in lilac (Syringa vulgaris L.) ‘Mme Florent Stepman’. Ann. Wars. Agric. Univ. Hortic. Landsc. Archit. 2003, 24, 41–49. [Google Scholar]
- Jedrzejuk, A.; Szlachetka, W. Development of flower organs in common lilac (Syringa vulgaris L.) Cv. Mme florent stepman. Acta Biol. Cracoviensia Ser. Bot. 2005, 42, 41–52. [Google Scholar]
- Jêdrzejuk, A. Ultrastructure of Pollen Grains from Forced and Unforced Shrubs of Common Lilac. J. Plant Growth Regul. 2005, 24, 83–92. [Google Scholar] [CrossRef]
- Jêdrzejuk, A.; Łukaszewska, A.J. High temperatures applied at fall forcing disturb ovule development in Syringa vulgaris L. “Mme Florent Stepman”. Acta Physiol. Plant. 2008, 30, 673–678. [Google Scholar] [CrossRef]
- Hanganu, D.; Niculae, M.; Ielciu, I.; Olah, N.K.; Munteanu, M.; Burtescu, R.; Ștefan, R.; Olar, L.; Pall, E.; Andrei, S.; et al. Chemical Profile, Cytotoxic Activity and Oxidative Stress Reduction of Different Syringa vulgaris L. Extracts. Mol. 2021, 26, 3104. [Google Scholar] [CrossRef] [PubMed]
- Gąsecka, M.; Krzymińska-Bródka, A.; Magdziak, Z.; Czuchaj, P.; Bykowska, J. Phenolic Compounds and Organic Acid Composition of Syringa vulgaris L. Flowers and Infusions. Molecules 2023, 28, 5159. [Google Scholar] [CrossRef]
- Varga, E.; Barabás, C.; Tóth, A.; Boldizsár, I.; Noszál, B.; Tóth, G. Phenolic composition, antioxidant and antinociceptive activities of Syringa vulgaris L. bark and leaf extracts. Nat. Prod. Res. 2019, 33, 1664–1669. [Google Scholar] [CrossRef]
- Tóth, G.; Barabás, C.; Tóth, A.; Kéry, Á.; Béni, S.; Boldizsár, I.; Varga, E.; Noszál, B. Characterization of antioxidant phenolics in Syringa vulgaris L. flowers and fruits by HPLC-DAD-ESI-MS. Biomed. Chromatogr. BMC 2016, 30, 923–932. [Google Scholar] [CrossRef]
- Shang, L.; Ma, D.; Hong, S.; Zhao, Y.; Zhang, G.; Ma, Q.; Gu, C. Observation on Flower Bud Differentiation of Crape Myrtle in Red Soil Environment. Phyton-Int. J. Exp. Bot. 2022, 91, 2607–2617. [Google Scholar] [CrossRef]
- Cardiff, R.D.; Miller, C.H.; Munn, R.J. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harbor Protocols 2014, 2014, pdb-prot073411. [Google Scholar] [CrossRef] [PubMed]
- Benito, P.; Celdrán, M.; Bellón, J.; Arbona, V.; González-Guzmán, M.; Porcel, R.; Mulet, J.M. The combination of a microbial and a non-microbial biostimulant increases yield in lettuce (Lactuca sativa) under salt stress conditions by up-regulating cytokinin biosynthesis. J. Integr. Plant Biology. 2022, 66, 2140–2157. [Google Scholar] [CrossRef]
- Yao, Y.; Nan, L.; Wang, K.; Xia, J.; Ma, B.; Cheng, J. Integrative leaf anatomy structure, physiology, and metabolome analyses revealed the response to drought stress in sainfoin at the seedling stage. Phytochem. Analysis. 2024, 35, 1174–1185. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Li, J.; Zhang, L.; Chen, K.; Shao, J.; Zheng, B.; Zhu, L. Polyethyleneimine-coated MXene quantum dots improve cotton tolerance to Verticillium dahliae by maintaining ROS homeostasis. Nat. Commun. 2023, 14, 7392. [Google Scholar] [CrossRef]
- Zhou, G.; Yin, H.; Chen, F.; Wang, Y.; Gao, Q.; Yang, F.; Wan, Y. The genome of Areca catechu provides insights into sex determination of monoecious plants. New Phytol. 2022, 236, 2327–2343. [Google Scholar] [CrossRef] [PubMed]
- Owen, S.J.; Abrams, S.R. Measurement of plant hormones by liquid chromatography-mass spectrometry. In Plant Hormones. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2009; pp. 39–51. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Regev, A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Cánovas, R.; Moffat, A.; Turpin, A. CSAM: Compressed SAM format. Bioinformatics 2016, 32, 3709–3716. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; Brand, T.v.D. Create Elegant Data Visualisations Using the Grammar of Graphics. R Package Version 3.3.2. Available online: https://cran.r-project.org/web/packages/ggplot2 (accessed on 21 February 2025).
- Li, X.; Cai, K.; Fan, Z.; Wang, J.; Wang, L.; Wang, Q.; Wang, L.; Pei, X.; Zhao, X. Dissection of Transcriptome and Metabolome Insights into the Isoquinoline Alkaloid Biosynthesis during stem development in Phellodendron amurense (Rupr.). Plant Sci. 2022, 325, 111461. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, Y.; Han, R.; Liu, L.; Pei, X.; Zhao, X. Widely targeted metabolomic profiling combined with transcriptome analysis provides new insights into lipid biosynthesis in seed kernels of Pinus koraiensis. Int. J. Mol. Sci. 2023, 24, 12887. [Google Scholar] [CrossRef] [PubMed]
- Xing, B.; Wan, S.; Su, L.; Riaz, M.W.; Li, L.; Ju, Y.; Shao, Q. Two polyamines-responsive WRKY transcription factors from Anoectochilus roxburghii play opposite functions on flower development. Plant Sci. 2023, 327, 111566. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Y.; Zhang, Y.; Liu, G.; Yu, S.; Zheng, Z. The overall regulatory network and contributions of ABC(D)E model genes in yellowhorn flower development. BMC Plant Biol. 2024, 24, 1081. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, L.; Yin, Q.; Liu, X.; Masabni, J.; Xiong, H.; Zou, F. Transcription Factor and Zeatin Co-Regulate Mixed Catkin Differentiation of Chinese Chestnut (Castanea mollissima). Forests 2023, 14, 2057. [Google Scholar] [CrossRef]
- Sun, T.P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arab. Book 2008, 6, e0103. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Yang, H.; Yang, J.; Wang, Y.; Ye, T.; Xiang, L.; Chan, Z.; Wang, Y. Tulip transcription factor TgWRKY75 activates salicylic acid and abscisic acid biosynthesis to synergistically promote petal senescence. J. Exp. Bot. 2024, 75, 2435–2450. [Google Scholar] [CrossRef]
- Gong, F.; Jing, W.; Jin, W.; Liu, H.; Zhang, Y.; Wang, R.; Wei, Y.; Tang, K.; Jiang, Y.; Gao, J.; et al. RhMYC2 controls petal size through synergistic regulation of jasmonic acid and cytokinin signaling in rose. Plant J. Cell Mol. Biol. 2024, 120, 459–472. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szécsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C.; Bendahmane, M. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Brioudes, F.; Joly, C.; Szécsi, J.; Varaud, E.; Leroux, J.; Bellvert, F.; Bertrand, C.; Bendahmane, M. Jasmonate controls late development stages of petal growth in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2009, 60, 1070–1080. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Benítez, M.; Corvera-Poiré, A.; Cador, Á.C.; de Folter, S.; de Buen, A.G.; Sánchez-Corrales, Y.E. Flower development. Arab. Book/Am. Soc. Plant Biol. 2010, 8, e0127. [Google Scholar] [CrossRef]
- Krizek, B.A.; Anderson, J.T. Control of flower size. J. Exp. Bot. 2013, 64, 1427–1437. [Google Scholar] [CrossRef]
- Cai, K.; Zhu, S.; Jiang, Z.; Xu, K.; Sun, X.; Li, X. Biological macromolecules mediated by environmental signals affect flowering regulation in plants: A comprehensive review. Plant Physiol. Biochem. PPB 2024, 214, 108931. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, T.; Saito, S.; Yamanashi, T.; Kato, M.; Takebayashi, K.; Hamamoto, S.; Tsujii, M.; Takagi, T.; Nagata, N.; Ikeda, H.; et al. The HKT1 Na + transporter protects plant fertility by decreasing Na+ content in stamen filaments. Sci. Adv. 2023, 9, 5495. [Google Scholar] [CrossRef]
- Han, M.; Zhao, Y.; Meng, J.; Yin, J.; Li, H. Analysis of physicochemical and antioxidant properties of Malus spp. petals reveals factors involved in flower color change and market value. Sci. Hortic. 2023, 310, 111688. [Google Scholar] [CrossRef]
- Ding, B. The roles of R2R3-MYBs in regulating complex pigmentation patterns in flowers. Hortic. Plant Journal. 2023, 9, 1067–1078. [Google Scholar] [CrossRef]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lv, S.; Zhao, L.; Gao, T.; Yu, C.; Hu, J.; Ma, F. Advances in the study of the function and mechanism of the action of flavonoids in plants under environmental stresses. Planta 2023, 257, 108. [Google Scholar] [CrossRef]
- Li, X.; Zheng, M.; Gan, Q.; Long, J.; Fan, H.; Wang, X.; Guan, Z. The formation and evolution of flower coloration in Brassica crops. Front. Genet. 2024, 15, 1396875. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, Y.; Zhang, M.; Wang, Y.; Deng, X.; Sun, H.; Yang, D.; Xu, L.; Song, H.; Yang, M. Color fading in lotus (Nelumbo nucifera) petals is manipulated both by anthocyanin biosynthesis reduction and active degradation. Plant Physiol. Biochem. PPB 2022, 179, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The defective in anther dehiscence1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Huang, X.; Liu, L.; Qiang, X.; Meng, Y.; Li, Z.; Huang, F. Integrative Metabolomic and Transcriptomic Analysis Elucidates That the Mechanism of Phytohormones Regulates Floral Bud Development in Alfalfa. Plants 2024, 13, 1078. [Google Scholar] [CrossRef]







| Gene ID | Forward Primer (5′ to 3′) | Reverse Primer (3′ to 5′) |
|---|---|---|
| TRINITY_DN50647_c0_g1 | AGCCTTCTGGTGAGCATGGA | TGAAGCTGTCGACAGCAGTA |
| TRINITY_DN36080_c0_g1 | CAGGCTACCGTCTTGTTGGA | GTGCTAGCAGTCCTTGGTCT |
| TRINITY_DN34830_c0_g1 | TGCTGACGGTGACTGATGTT | AGCTTGTGACCGTTGTAGCG |
| TRINITY_DN14054_c0_g1 | ATGCTTGGAGAGCTGTGTGC | CGATGAGTCCTGAGGCTTGT |
| TRINITY_DN32532_c0_g1 | GGTGATCGTAGTGTGAGCCA | CTTGGCATGTTGAGGCTGAT |
| TRINITY_DN62324_c0_g1 | TGGAGCTTCTGAGGTGGTTA | CGTACTGCTGATCAGGATGC |
| TRINITY_DN42129_c0_g1 | CGTGAAGTGTGACTGGCTGA | GCTTGAAGCTGAGCCTGATA |
| TRINITY_DN36412_c0_g2 | TGAAGCTGTCAGGACGTGTT | CTTGAGTGCAGTGTCTGAGC |
| TRINITY_DN34320_c0_g2 | AGTGGTCGATCAGTGGCTGT | TGACTGACTGAGGCTGTGAG |
| Stage of Flower Bud Differentiation | Single Flowers | Double Flowers | |||||
|---|---|---|---|---|---|---|---|
| Date and Duration | Longitudinal Diameter (mm) | Transverse Diameter (mm) | Two-Layer Corolla | Three-Layer Corolla | Longitudinal Diameter (mm) | Transverse Diameter (mm) | |
| Date and Duration | |||||||
| Spathe primordium differentiation stage | 21 May– 11 June | 1.6403 ± 0.3231 mn | 1.7596 ± 0.3478 mn | 21 May– 11 June | 21 May– 11 June | 1.8328 ± 0.4437 m | 1.5135 ± 0.2912 n |
| 21 | 21 | 21 | |||||
| Inflorescence and small floral primordium stage | 11 June– 9 July | 2.6529 ± 0.7669 l | 2.7831 ± 0.7523 kl | 11 June– 16 July | 11 June– 16 July | 3.329 ± 0.6747 j | 2.893 ± 0.9193 k |
| 28 | 35 | 35 | |||||
| Sepal primordium differentiation stage | 9 July– 23 July | 4.0315 ± 0.4481 h | 4.0001 ± 0.3342 h | 16 July– 23 July | 16 July– 23 July | 4.2289 ± 0.3736 h | 3.75 ± 0.1971 i |
| 14 | 7 | 7 | |||||
| Petal primordium differentiation stage | 23 July– 14 August | 4.4865 ± 0.4554 g | 4.2144 ± 0.3222 h | 23 July– 4 September | 23 July– 11 September | 6.949 ± 0.6065 c | 4.2258 ± 0.3898 h |
| 22 | 43 | 50 | |||||
| Stamen primordium differentiation stage | 14 August– 11 September | 5.6236 ± 0.4786 e | 4.5532 ± 0.4696 g | 4 September– 11 September | 8.0392 ± 0.3034 b | 4.6314 ± 0.2645 g | |
| 28 | 7 | ||||||
| Pistil primordium differentiation stage | 11 September –25 September | 6.3373 ± 0.413 d | 5.188 ± 0.3846 f | 11 September– 16 October | 11 September– 16 October | 8.4374 ± 0.4859 a | 5.0727 ± 0.4019 f |
| 14 | 35 | 35 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Li, J.; Qi, Y.; Ma, Y.; Han, W.; Tian, L.; Sun, P.; Siqin, T.; Li, C.; Bai, H. Physiological and Molecular Insights into the Development of Single and Double Flowers in Syringa vulgaris L. Forests 2025, 16, 409. https://doi.org/10.3390/f16030409
Wang Z, Li J, Qi Y, Ma Y, Han W, Tian L, Sun P, Siqin T, Li C, Bai H. Physiological and Molecular Insights into the Development of Single and Double Flowers in Syringa vulgaris L. Forests. 2025; 16(3):409. https://doi.org/10.3390/f16030409
Chicago/Turabian StyleWang, Zhaoning, Jing Li, Yu Qi, Yuandong Ma, Wenzhe Han, Linping Tian, Peilin Sun, Tuya Siqin, Chunming Li, and Hui Bai. 2025. "Physiological and Molecular Insights into the Development of Single and Double Flowers in Syringa vulgaris L." Forests 16, no. 3: 409. https://doi.org/10.3390/f16030409
APA StyleWang, Z., Li, J., Qi, Y., Ma, Y., Han, W., Tian, L., Sun, P., Siqin, T., Li, C., & Bai, H. (2025). Physiological and Molecular Insights into the Development of Single and Double Flowers in Syringa vulgaris L. Forests, 16(3), 409. https://doi.org/10.3390/f16030409





