3.3. Working Example
In order to demonstrate the practicality and accuracy of the forestPSD package, this study provides a working example that illustrates the entire process of dynamic analysis of forest population structure and quantity, which can be completed in eight steps, as detailed below:
Step 1: Install and load the package.
# Install.packages(“forestPSD”)
library(forestPSD)
Step 2: Organize the data into a data format suitable for population structure analysis.
number=c(8283,5238,1921,1425,926,659,479,228,57,24,10)
Npop =Ntable(ax=number)
Taking the age class data in
Table 1 as an example, we utilize the Ntable() function to organize the data into a data format (Npop,
Table 4) that is suitable for population structure analysis. Here, a
x represents the number of tree individuals corresponding to each age class as the age class progresses.
Step 3: Plot for population structure analysis.
# Install.packages(“ggplot2”)
library(ggplot2)
fpop.p<-ggplot()+geom_bar(aes(x=rank,y=ax,group=ageclass),data=Npop,stat = “identity”)+
scale_x_continuous(breaks=1:11)+
scale_x_discrete(limits=Npop$ageclass)+
xlab(“Age class”)+ylab(“Number of individuals”)
fpop.p
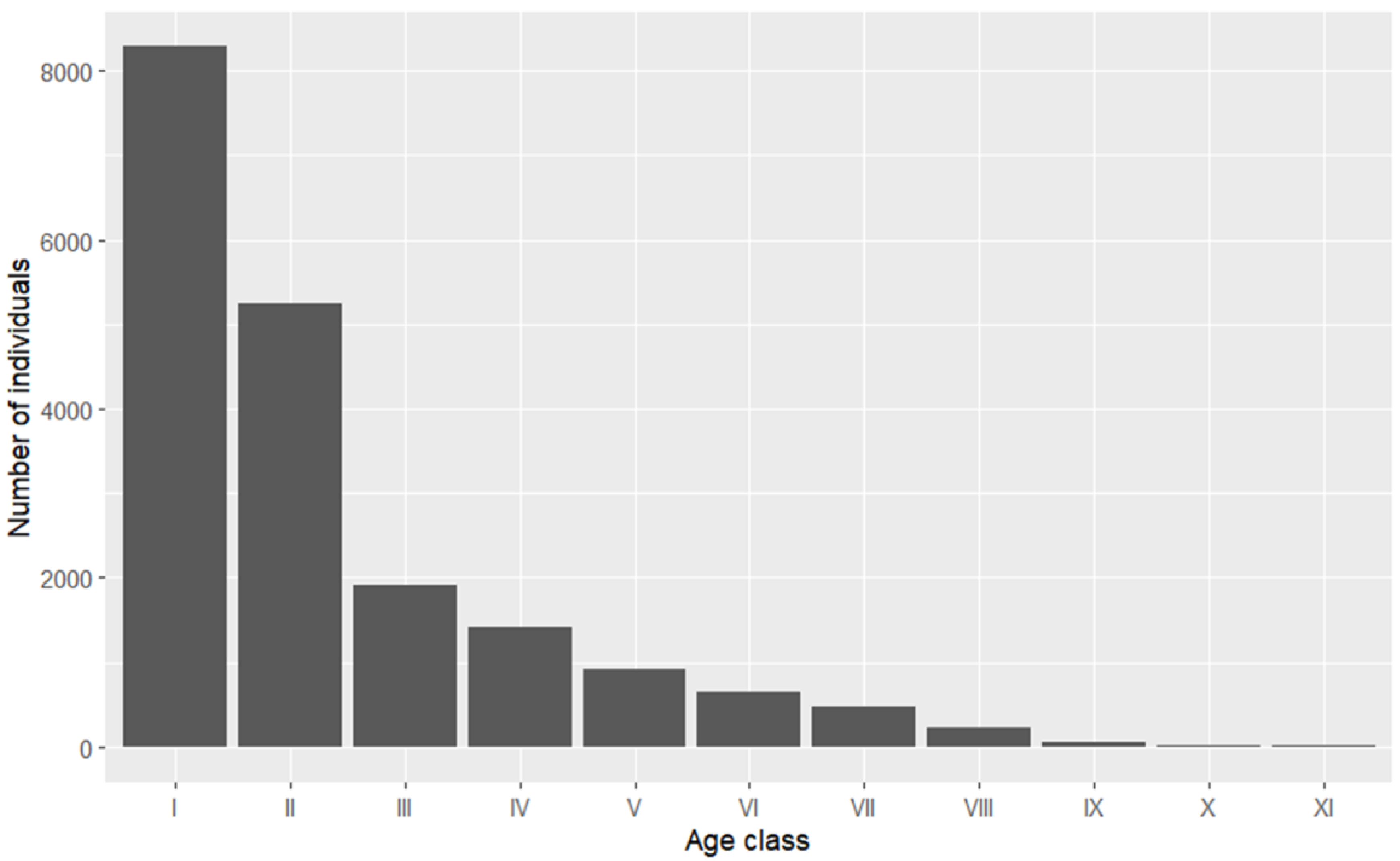
We employed the ggplot2 package to create the plot for the forest population structure (
Figure 2). As the age advances, the number of individuals in this forest population gradually decreases. The age structure of the population presents an inverted “J”-shaped distribution, which clearly demonstrates that the population is a progressive one.
We calculated the population dynamic index using the dyn() function, and the key results are presented in
Table 5. The results demonstrate that the dynamic indices for each age class are all greater than 0, which indicates that the population can adapt effectively to the local environment and maintain relative stability over a certain period. When external disturbances are ignored, the dynamic index for the entire population
, suggesting that the population is a growing population. When the influence of external disturbances is considered,
, which is close to 0, indicating that the population shows a tendency of transition from a growing population to a stable one.
We generated the life table by using the lifetable() function, and the key results are presented in
Table 6.
Step 6: Regression analysis for survival curves.
psd_D1<-psdfun(ax=Npop$ax,index=“Deevey1”)
psd_D1
psd_D2<-psdfun(ax=Npop$ax,index=“Deevey2”)
psd_D2
psd_D3<-psdfun(ax=Npop$ax,index=“Deevey3”)
psd_D3
The results of fitting the survival curve using the Deevey formulas were obtained by using the psdfun() function. The key results are presented in
Table 7. These results suggest that among the three Deevey formulas, Deevey II yields the best fitting effect, with an R
2 value reaching 0.987.
Step 7: Plot for regression analysis of survival curves.
# Install.packages(“ggplot2”)
library(ggplot2)
psdnls.p<-ggplot()+geom_bar(aes(x=age,y=ax,group=ageclass),data=psd_D2$Data,stat = “identity”)+
geom_line(aes(x=age,y=predict),color=“blue”,linewidth=1,data=psd_D2$Data)+
geom_text(aes(x=9,y=7700),label=expression(paste(italic(y),”=15040exp(-0.5828”,italic(x),”)”)))+
geom_text(aes(x=10,y=7300),label=expression(paste(R^2,”=0.987”)))+
scale_x_continuous(breaks=1:11)+
scale_x_discrete(limits=psd_D2$Data$ageclass)+
xlab(“Age class”)+ylab(“Number of individuals”)
psdnls.p
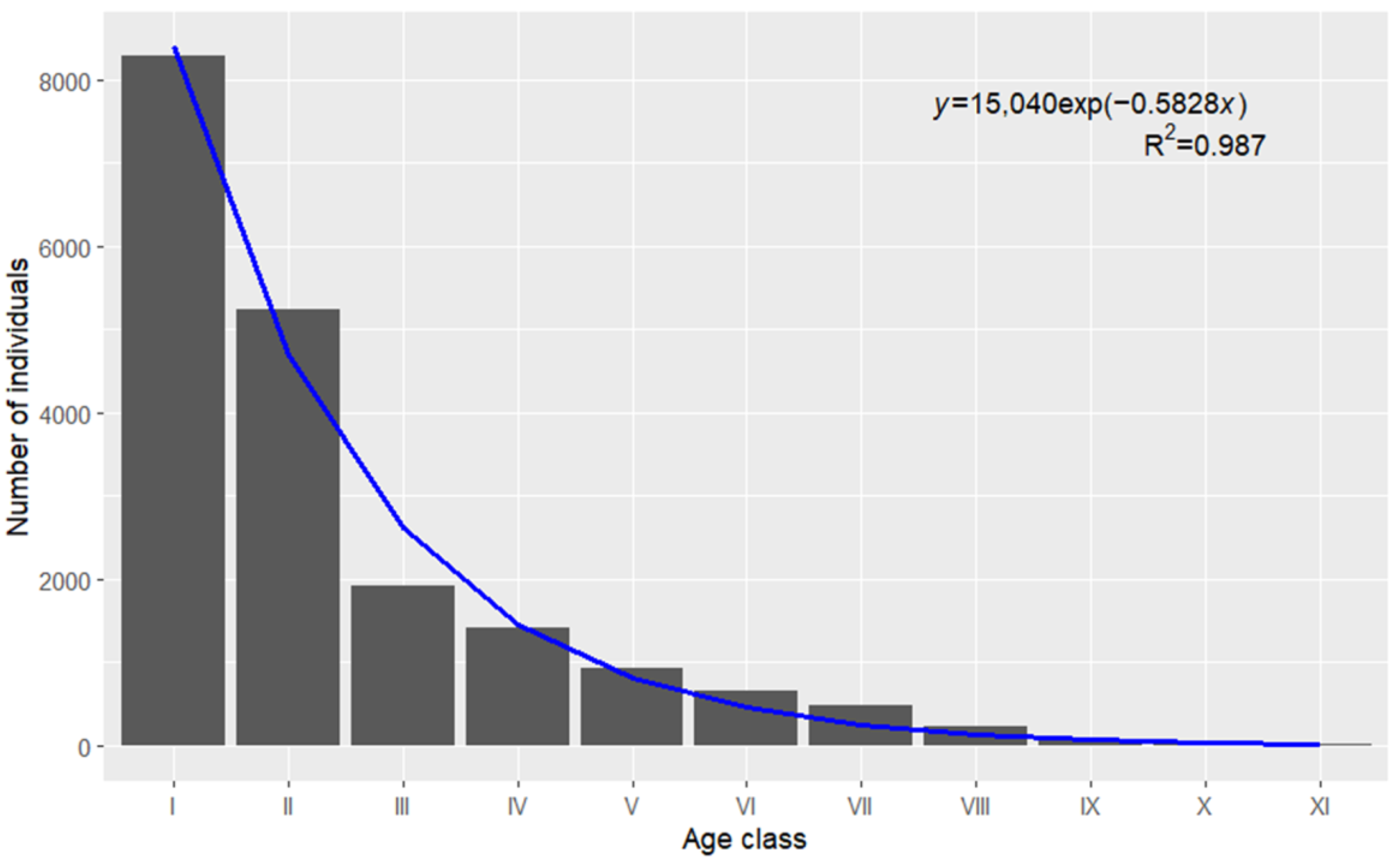
We utilized the ggplot2 package to generate a plot for the regression analysis of survival curves (
Figure 3). Meanwhile, the reshape2 package was employed to process the data. The results show that the Deevey II formula exhibits a satisfactory fitting effect.
Step 8: Time series prediction for the population dynamic changes.
Mdata<-Mpre(ax=Npop$ax,n=c(2,3,5,6,8,10))
# Install.packages(“reshape2”)
library(reshape2)
Mdata.melt<-reshape2::melt(Mdata,id=c(“rank”,”ageclass”))
Mdata.melt$ageclass<-factor(Mdata.melt$ageclass,levels=unique(Mdata.melt$ageclass))
# Install.packages(“ggplot2”)
library(ggplot2)Mpre.p<-ggplot()+geom_line(aes(x=ageclass,y=value,color=variable,group=variable),
linewidth=0.5,data=Mdata.melt)+
xlab(“Age class”)+ylab(“Number of individuals”)+labs(color=“ “)
Mpre.p
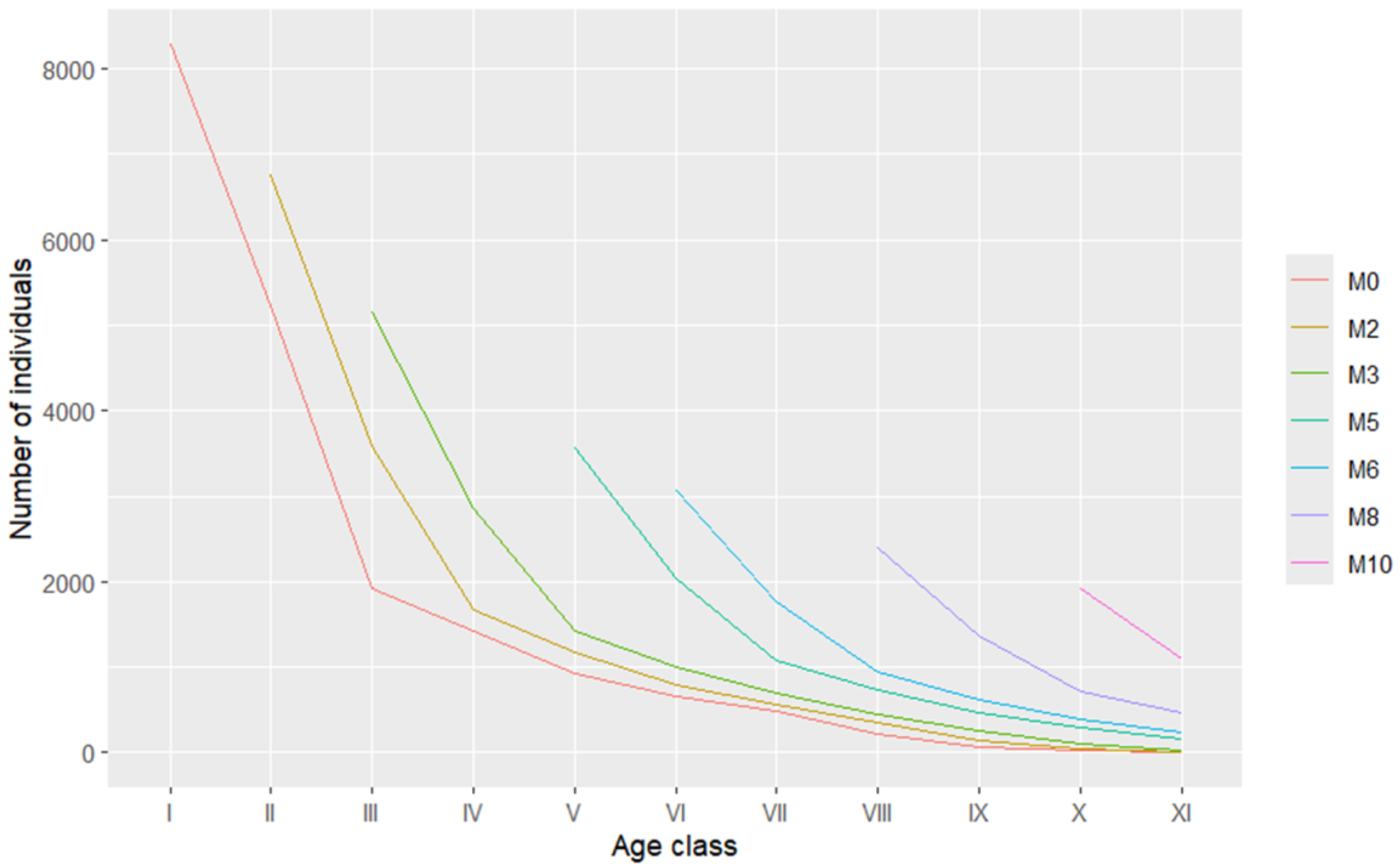
We predicted the time series by using the Mpre() function and generated a table and a figure for the time series prediction of the population dynamic changes (
Table 8 and
Figure 4). The results indicate that, in the prediction series, after time intervals corresponding to 2, 3, 5, 6, 8, and 10 age classes in the future, the peaks of the individual numbers in each age class of the population shift backward successively.
3.4. Presentation of a Comprehensive Case
In this section, leveraging the data from our prior publication [
13], we offer a comprehensive and in-depth demonstration of the research process related to the structure and numerical dynamics of forest populations. The study area was a secondary forest situated in a typical karst region of Guizhou Province, China. This forest has undergone more than 30 years of natural restoration after being abandoned. Through the establishment of representative sample plots and systematic vegetation surveys, we conducted a comparative analysis of the structural and numerical dynamic characteristics of five dominant populations. Our aim was to establish scientific theoretical underpinnings that can serve as reliable references for the protection and restoration of karst secondary forests. In this study, a total of 41 species and 3251 individuals of woody plants were surveyed. Among them, there were 1038 individuals of Betula luminifera (BL, accounting for 31.9%), 793 of Platycarya strobilacea (PS, 24.4%), 344 of Pinus massoniana (PM, 10.6%), 198 of Liquidambar formosana (LF, 6.1%), and 183 of Populus davidiana (PD, 5.6%). These five populations, with a combined total of 2556 individuals, constituted 78.6% of the surveyed plants and were identified as the dominant populations within the community. The age class data of these five dominant populations are presented in
Table 9. The detailed criteria for age class division and relevant basic information are comprehensively elaborated in our published article [
13]. Moreover, the R codes utilized in this section are available in the
Supplementary File.
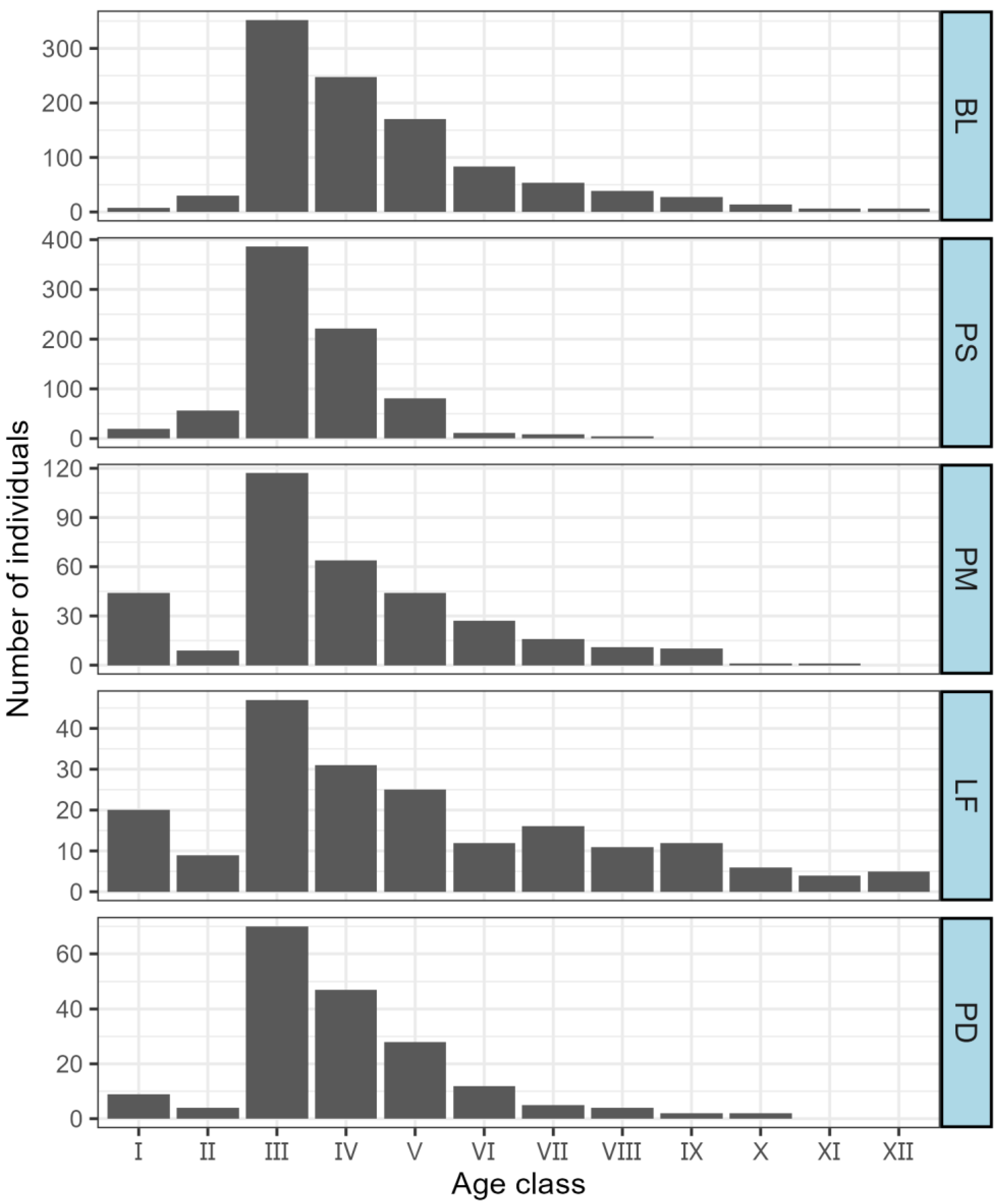
As depicted in
Figure 5, the age class structures of the five dominant populations are relatively comprehensive. In general, for each population, the number of individuals exhibits a unimodal pattern, initially rising and subsequently declining as the age class advances. Nevertheless, owing to the relatively scarce number of individuals in age classes I—II of each dominant population and the substantial fluctuations observed in these early age classes, there is an overall deficiency in population renewal. This ultimately leads to suboptimal population stability.
The dynamic indices show (
Table 10) that the numeric dynamics of the five dominant populations all exhibit a trend of first decline and then growth. The overall dynamic indices V
pi and V′
pi of the five dominant populations are both greater than zero, indicating that the five dominant populations are generally in a growth trend. The growth potential is ranked as PM > PS > PD > BL > LF. When affected by the external environment, the Pinus massoniana population has the highest probability of being disturbed, while the Betula luminifera population has the lowest. In addition, the insufficient number of individuals in age classes I, II may hinder the stable and sustainable development of the population.
Through the compilation of static life tables for the five dominant populations (
Table 11), it becomes feasible to ascertain the survival status of different populations across various growth stages. This approach allows for a comprehensive understanding of the competitive relationships among populations and a clear elucidation of the disparities in aspects such as resource acquisition and space occupation. Consequently, it offers vital information for the study of niche differentiation within the community and the dynamic development of populations.
The results of fitting the survival curves of the five dominant populations (
Table 12) indicate that, among the three Deevey formulas, the Deevey II formula yields the best fitting effect, with a larger R
2 value. Evidently, the survival curves of the five dominant populations are all closer to the Deevey II type.
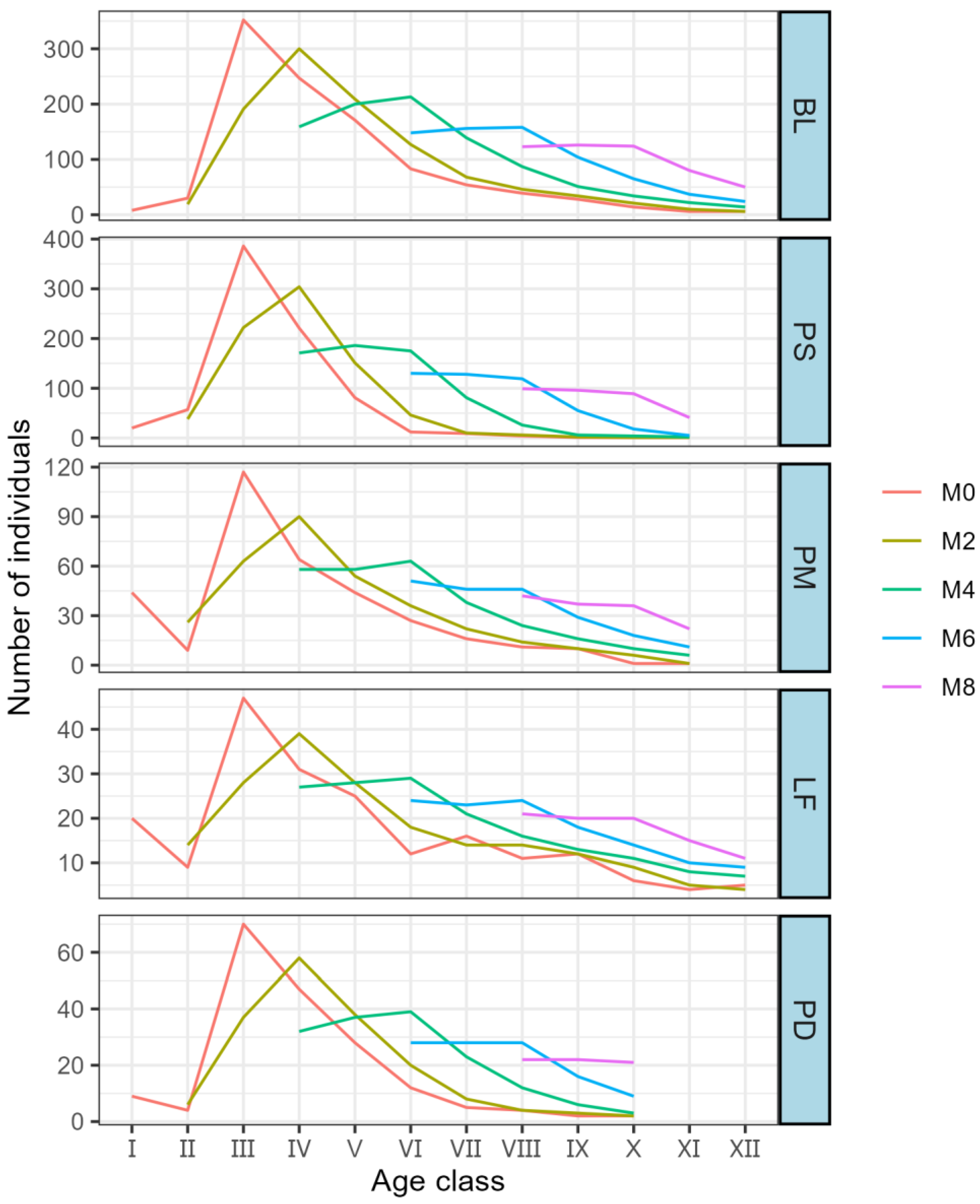
The time series prediction results indicate (
Figure 6) that the five dominant populations exhibit roughly the same trend. Specifically, the population size experiences a sharp decline in the early stage and then a stable increase in the later stage. In the future age classes, the advantage in the number of individuals in each dominant population weakens. Subsequently, as the age class increases, the decline in the number of individuals gradually disappears and then shows an increasing trend. The most significant increase in the number of individuals occurs in the VI age class. After six age classes, the structures of various populations gradually become more reasonable, presenting a stable growth trend.
In summary, notable disparities exist in the structure and numerical dynamics of the five dominant populations. Nevertheless, overall, they display a stable growth tendency. The limited number of individuals in the I–II age classes stands out as the primary factor impeding the stable development of the populations. It is advisable to improve the stand quality and the quantity of regeneration through a combination of enclosure protection and artificial tending measures. This integrated approach is expected to foster the stable and sustainable development of the secondary forest community.