Genome-Wide Identification and Expression Profiling of MYC Transcription Factor Family in Toona sinensis Under Abiotic and Hormonal Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Identification of the TsMYC Family Members
2.3. Multiple Sequence Alignment and Phylogenetic Analysis
2.4. Chromosomal Localization and Synteny Analysis
2.5. Gene Structure and Conserved Motif Analysis
2.6. Cis-Regulatory Elements Analysis of Promoters Regions of TsMYC Genes
2.7. Prediction of Protein–Protein Interaction Networks
2.8. RNA Extraction and qRT-PCR Analysis
2.9. The Subcellular Localization Analysis of TsMYC17
2.10. Statistical Analysis
3. Results
3.1. Identification and Physicochemical Property Analysis of TsMYC Family Members
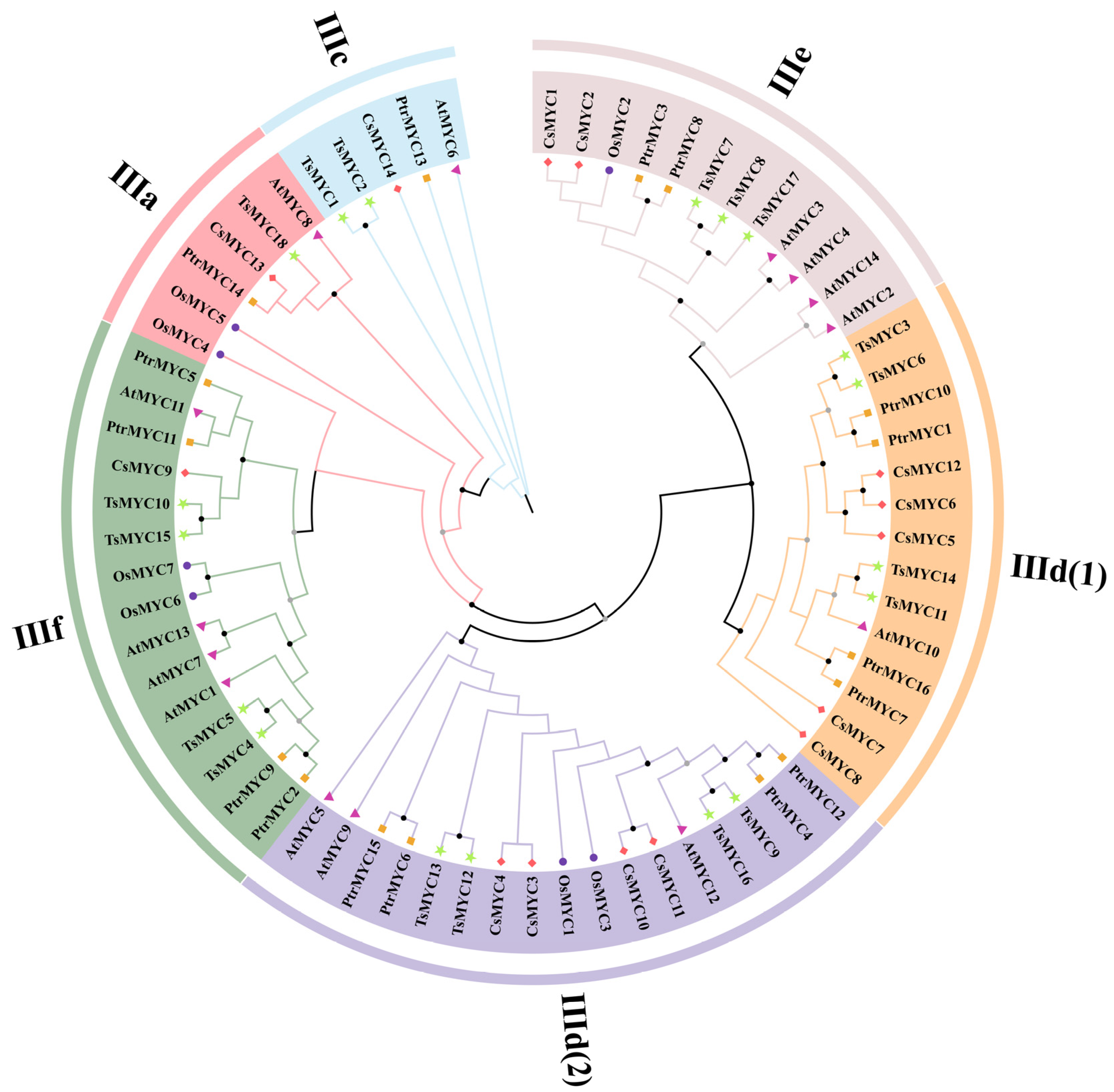
3.2. Phylogenetic and Classification Analysis of the TsMYC Gene Family
3.3. Chromosomal Localization and Synteny Analysis of TsMYC Genes
3.4. Gene Structure and Conserved Motifs Analysis of TsMYC Genes
3.5. Cis-Acting Elements Analysis of TsMYC Promoters
3.6. Interaction Network Prediction of the TsMYCs
3.7. Expression Profiling of TsMYC Genes in Different Tissue
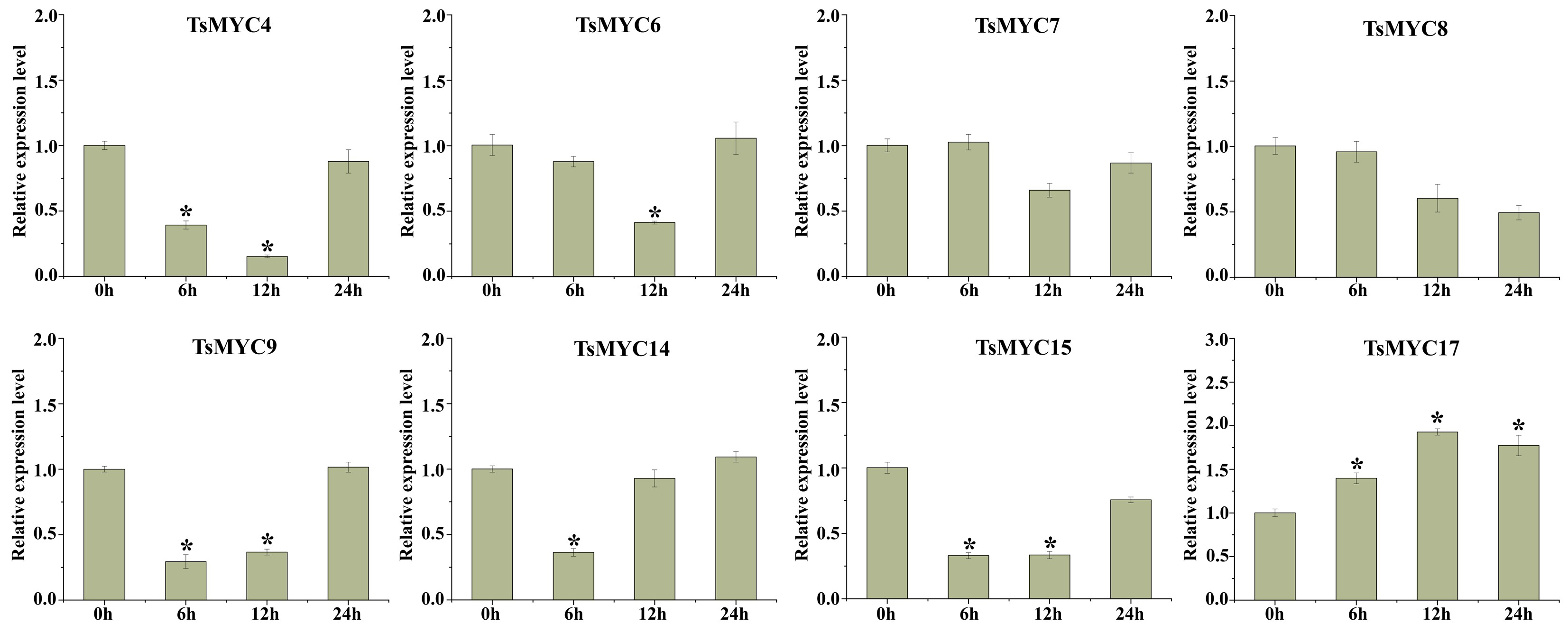
3.8. Expression Profiles of TsMYC Genes Under MeJA Treatment
3.9. The Phytohormone Response Pattern of TsMYCs
3.10. Expression Profiles of TsMYC Genes Under Salt Stress
3.11. Subcellular Localization of TsMYC17
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, W.; Liu, Y.; Hu, M.; Zhang, M.; Yang, J.; Liang, F.; Wu, C. Toona. sinensis: A comprehensive review on its traditional usages, phytochemistry, pharmacology, and toxicology. Rev. Bras. Farmacogn. 2019, 29, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, I.; Banks, J.C. Variation in phenology, growth, and wood anatomy of Toona sinensis and Toona ciliata in relation to different environmental conditions. Int. J. Plant Sci. 2006, 167, 831–841. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Cao, Y.; Gu, Q.; Yang, H.; Tam, J.P. Quantitative analysis and comparison of four major flavonol glycosides in the leaves of Toona sinensis (A. Juss.) Roemer (Chinese Toon) from various origins by high-performance liquid chromatography-diode array detector and hierarchical clustering analysis. Pharmacogn. Mag. 2016, 12, S270–S276. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, H.; Wang, R.; Lan, S.; Wang, Y.; Zhang, Y.; Li, W. Traditional Uses, Chemical Constituents and Pharmacological Activities of the Toona sinensis. Plant Mol. 2024, 29, 718. [Google Scholar] [CrossRef]
- Xiang, L.; Jian, D.; Zhang, F.; Yang, C.; Bai, G.; Lan, X.; Liao, Z. The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J. Exp. Bot. 2019, 70, 4835–4848. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Chai, X.; Lv, J.; Hu, L.; Wang, J.; Liu, Z. Genome-wide identification and expression analysis of the MYC transcription factor family and its response to sulfur stress in cabbage (Brassica oleracea L.). Gene 2022, 814, 146116. [Google Scholar] [CrossRef]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Wang, J.; Liu, B.; Qi, T.; Xie, D. MYC5 is involved in jasmonate-regulated plant growth, leaf senescence, and defense responses. Plant Cell Physiol. 2017, 58, 1752–1763. [Google Scholar] [CrossRef]
- Wang, Q.; Li, B.; Qiu, Z.; Lu, Z.; Hang, Z.; Wu, F.; Zhu, X. Genome-wide identification of MYC transcription factors and their potential functions in the growth and development regulation of tree peony (Paeonia suffruticosa). Plants 2024, 13, 437. [Google Scholar] [CrossRef]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signaling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Pérez, A.C.; Goossens, A. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Sohn, S.I.; Pandian, S.; Rakkammal, K.; Largia, M.J.V.; Thamilarasan, S.K.; Balaji, S.; Ramesh, M. Jasmonates in plant growth and development and elicitation of secondary metabolites: An updated overview. Front. Plant Sci. 2022, 13, 942789. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, H.; Luo, T.; Liu, Y.; Nie, X.; Li, H. Characteristics and expression pattern of MYC genes in Triticum aestivum, Oryza. sativa, and Brachypodium distachyon. Plants 2019, 8, 274. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Calvo, P.; Chini, A.; Fernández-Barbero, G.; Chico, J.M.; Gimenez-Ibanez, S.; Geerinck, J.; Solano, R. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 2011, 23, 701–715. [Google Scholar] [CrossRef]
- Zhang, M.; Jin, X.; Chen, Y.; Wei, M.; Liao, W.; Zhao, S.; Yu, L. TcMYC2a, a basic helix-loop-helix transcription factor, transduces JA-signals and regulates taxol biosynthesis in Taxus chinensis. Front. Plant Sci. 2018, 9, 863. [Google Scholar] [CrossRef]
- Xu, Y.H.; Liao, Y.C.; Lv, F.F.; Zhang, Z.; Sun, P.W.; Gao, Z.H.; Wei, J.H. Transcription factor AsMYC2 controls the jasmonate-responsive expression of ASS1 regulating sesquiterpene biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017, 58, 1924–1933. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB Homologs in Drought and Abscisic Acid-Regulated Gene Expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar] [CrossRef]
- Valenzuela, C.E.; Acevedo-Acevedo, O.; Miranda, G.S.; Vergara-Barros, P.; Holuigue, L.; Figueroa, C.R.; Figueroa, P.M. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4209–4220. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef]
- Yang, Z.; Li, Y.; Gao, F.; Jin, W.; Li, S.; Kimani, S.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Lv, B.; Shao, S.; Zhao, Y.; Yang, M.; Zuo, A.; Ma, P. The SmMYC2-SmMYB36 complex is involved in methyl jasmonate-mediated tanshinones biosynthesis in Salvia miltiorrhiza. Plant J. 2024, 119, 746–761. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, D.D.; Fang, X.; Chen, X.Y.; Mao, Y.B. Plant specialized metabolism regulated by jasmonate signaling. Plant Cell Physiol. 2019, 60, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.Y.; Vorst, O.; Fiers, M.W.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Bateman, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002, 1, 2–3. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying, and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Paterson, A.H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and collinearity in plant genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; von Mering, C. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [CrossRef]
- Ren, L.; Wan, W.; Yin, D.; Deng, X.; Ma, Z.; Gao, T.; Cao, X. Genome-wide analysis of WRKY transcription factor genes in Toona sinensis: An insight into evolutionary characteristics and terpene synthesis. Front. Plant Sci. 2022, 13, 1063850. [Google Scholar] [CrossRef]
- Yan, P.; Zeng, Y.; Shen, W.; Tuo, D.; Li, X.; Zhou, P. Nimble cloning: A simple, versatile, and efficient system for standardized molecular cloning. Front. Bioeng. Biotechnol. 2020, 7, 460. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, Z.; Wang, C.; Zhou, X.; Tang, N.; Zhang, W.; Ye, J. Genome-wide identification of HD-ZIP gene family and screening of genes related to prickle development in Zanthoxylum armatum. Plant Genome 2023, 16, e20295. [Google Scholar] [CrossRef]
- Fay, J.C.; Wu, C.I. Sequence divergence, functional constraint, and selection in protein evolution. Annu. Rev. Genom. Hum. Genet. 2003, 4, 213–235. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Muiño, J.M.; de Bruijn, S.; Pajoro, A.; Geuten, K.; Vingron, M.; Angenent, G.C.; Kaufmann, K. Evolution of DNA-binding sites of a floral master regulatory transcription factor. Mol. Biol. Evol. 2016, 33, 185–200. [Google Scholar] [CrossRef]
- Chini, A.; Fonseca, S.G.D.C.; Fernandez, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; Solano, R. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef]
- Niu, Y.; Figueroa, P.; Browse, J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 2011, 62, 2143–2154. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Kong, Y.; Zhang, X.; Liao, Z.; He, Y.; Li, L.; Hong, G. Structural and functional organization of the MYC transcriptional factors in Camellia sinensis. Planta 2021, 253, 93. [Google Scholar] [CrossRef] [PubMed]
- Song, R.F.; Li, T.T.; Liu, W.C. Jasmonic acid impairs Arabidopsis seedling salt stress tolerance through MYC2-mediated repression of CAT2 expression. Front. Plant Sci. 2021, 12, 730228. [Google Scholar] [CrossRef]
- Chung, B.Y.; Simons, C.; Firth, A.E.; Brown, C.M.; Hellens, R.P. Effect of 5’UTR introns on gene expression in Arabidopsis thaliana. BMC Genom. 2006, 7, 120. [Google Scholar] [CrossRef]
- Yang, J.; Gao, M.; Huang, L.; Wang, Y.; van Nocker, S.; Wan, R.; Gao, H. Identification and expression analysis of the apple (Malus × domestica) basic helix-loop-helix transcription factor family. Sci. Rep. 2017, 7, 28. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, Z.; Lei, T.; Zhang, Y.; Xiao, Q.; Yu, Z.; Wang, J. A likely autotetraploidization event shaped the Chinese mahogany (Toona. sinensis) genome. Hortic. Plant J. 2023, 9, 306–320. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-regulatory elements: Molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. The interplay between light and jasmonate signaling during defence and development. J. Exp. Bot. 2011, 62, 4087–4100. [Google Scholar] [CrossRef]
- Fukazawa, J.; Mori, K.; Ando, H.; Mori, R.; Kanno, Y.; Seo, M.; Takahashi, Y. Jasmonate inhibits plant growth and reduces gibberellin levels via microRNA5998 and transcription factor MYC2. Plant Physiol. 2023, 193, 2197–2214. [Google Scholar] [CrossRef]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Garrido-Bigotes, A.; Figueroa, N.E.; Figueroa, P.M.; Figueroa, C.R. Jasmonate signalling pathway in strawberry: Genome-wide identification, molecular characterization and expression of JAZs and MYCs during fruit development and ripening. PLoS ONE 2018, 13, e0197118. [Google Scholar] [CrossRef]
- Clarke, S.M.; Cristescu, S.M.; Miersch, O.; Harren, F.J.; Wasternack, C.; Mur, L.A. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2009, 182, 175–187. [Google Scholar] [CrossRef]
- Yue, H.; Wang, M.; Liu, S.; Du, X.; Song, W.; Nie, X. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genom. 2016, 17, 343. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, G.; Zeng, S.; Cui, X.; Liao, Y.; Zhang, W.; Xu, F.; Jiang, D. Genome-Wide Identification and Expression Profiling of MYC Transcription Factor Family in Toona sinensis Under Abiotic and Hormonal Stresses. Forests 2025, 16, 1756. https://doi.org/10.3390/f16121756
Zhou G, Zeng S, Cui X, Liao Y, Zhang W, Xu F, Jiang D. Genome-Wide Identification and Expression Profiling of MYC Transcription Factor Family in Toona sinensis Under Abiotic and Hormonal Stresses. Forests. 2025; 16(12):1756. https://doi.org/10.3390/f16121756
Chicago/Turabian StyleZhou, Guoquan, Sirui Zeng, Xinru Cui, Yongling Liao, Weiwei Zhang, Feng Xu, and Daoju Jiang. 2025. "Genome-Wide Identification and Expression Profiling of MYC Transcription Factor Family in Toona sinensis Under Abiotic and Hormonal Stresses" Forests 16, no. 12: 1756. https://doi.org/10.3390/f16121756
APA StyleZhou, G., Zeng, S., Cui, X., Liao, Y., Zhang, W., Xu, F., & Jiang, D. (2025). Genome-Wide Identification and Expression Profiling of MYC Transcription Factor Family in Toona sinensis Under Abiotic and Hormonal Stresses. Forests, 16(12), 1756. https://doi.org/10.3390/f16121756





