Assessment of Genetic Variation in Natural Populations of Hippophae rhamnoides L. from Kazakhstan Using Retrotransposon-Based Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. DNA Extraction and iPBS Genotyping
2.3. Genetic Diversity Assessment
2.4. PCR Reaction and Data Analysis
2.5. Data Analysis
3. Results
3.1. Analysis of iPBS Loci Variability in H. rhamnoides
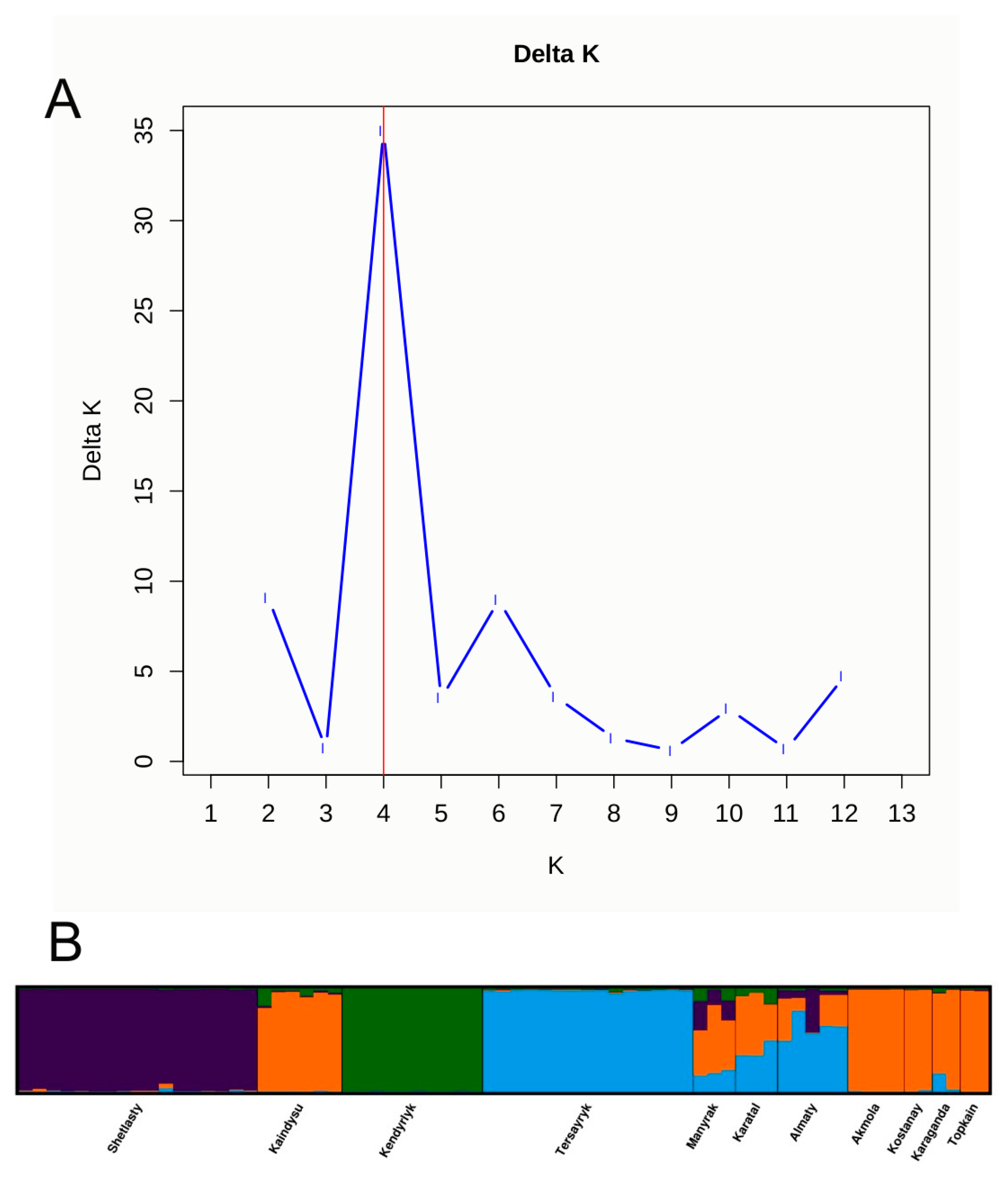
3.2. Genetic Differentiation of H. rhamnoides Populations Revealed by iPBS Markers
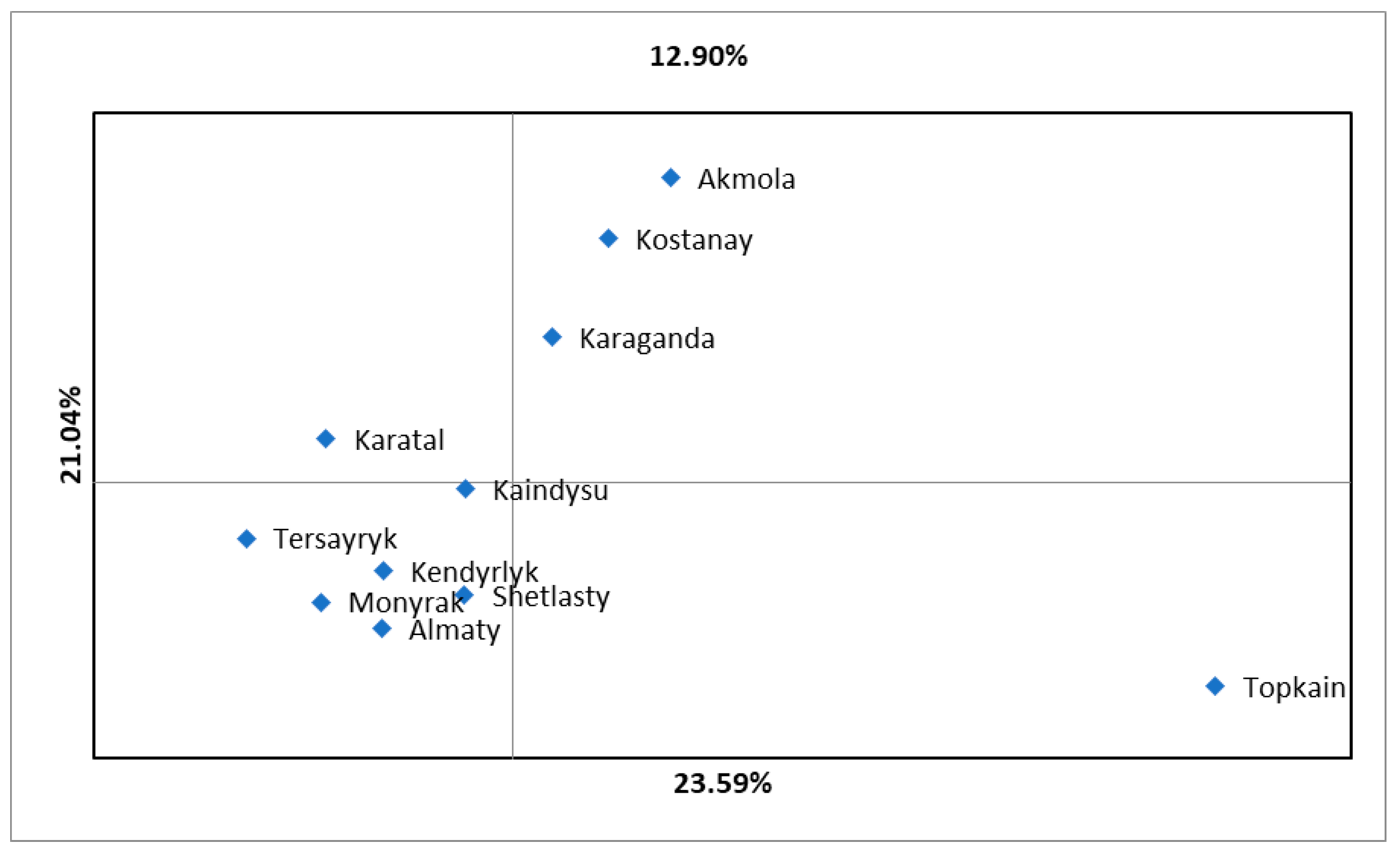
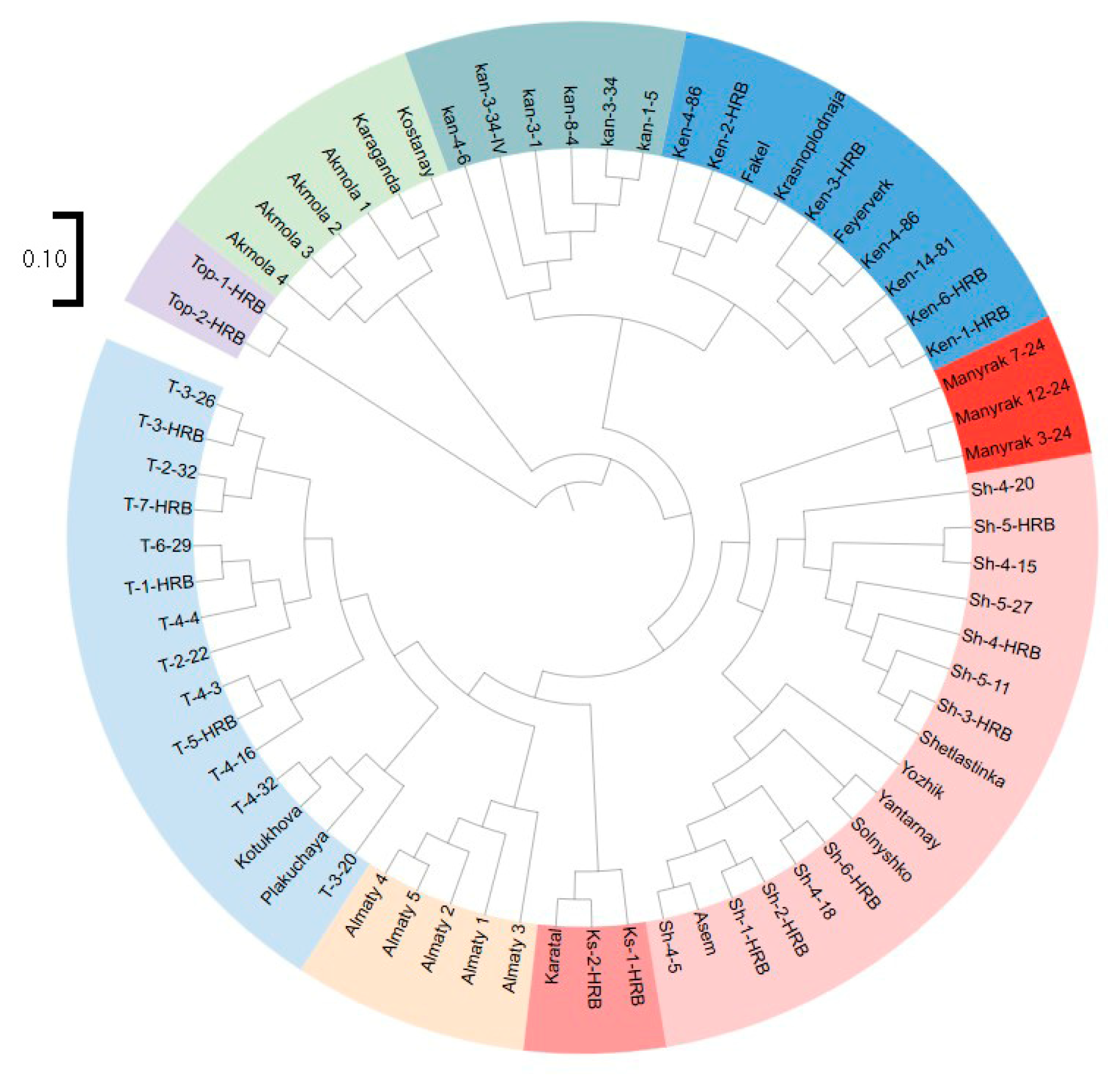
3.3. PCoA Analysis and Clustering for the Assessment of the Genetic Structure of H. rhamnoides Germplasm Based on iPBS Profiling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFLP | Amplified fragment length polymorphism |
| LTR | Long terminal repeat |
| PBS | Primer-binding site |
| PCoA | Principal coordinate analysis |
| SAMPL | Selective amplification of microsatellite polymorphic loci |
| SCAR | Sequence-characterised amplified region |
| SE | Standard error |
| SNP | Single-nucleotide polymorphisms |
| SSR | Simple sequence repeats |
References
- Royal Botanic Gardens, Kew. Plants of the World Online: Hippophae L. 2025. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:14480-1 (accessed on 17 June 2024).
- Gu, W.; Zhang, T.; Liu, S.Y.; Tian, Q.; Yang, C.X.; Lu, Q.; Fu, X.G.; Kates, H.R.; Stull, G.W.; Soltis, P.S.; et al. Phylogenomics, reticulation, and biogeographical history of Elaeagnaceae. Plant Divers. 2024, 46, 683–697. [Google Scholar] [CrossRef]
- Mihal, M.; Roychoudhury, S.; Sirotkin, A.V.; Kolesarova, A. Sea buckthorn, its bioactive constituents, and mechanism of action: Potential application in female reproduction. Front. Endocrinol. 2023, 14, 1244300. [Google Scholar] [CrossRef]
- Panteleeva, E.I. Sea Buckthorn (Hippophae rhamnoides L.); Siberian Branch of the Russian Academy of Sciences: Barnaul, Russia, 2006; 249p. [Google Scholar]
- Yue, X.F.; Shang, X.; Zhang, Z.J.; Zhang, Y.N. Phytochemical composition and antibacterial activity of the essential oils from different parts of sea buckthorn (Hippophae rhamnoides L.). J. Food Drug Anal. 2017, 25, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.F.; Chen, H.Y.; Pan, Y.; Feng, H.; Fang, D.M.; Yang, J.; Wang, Y.Y.; Sahu, S.K.; Liu, J.L.; Xing, Y.E.; et al. Genome of provides insights into a conserved molecular mechanism in actinorhizal and rhizobial symbioses. New Phytol. 2022, 235, 276–291. [Google Scholar] [CrossRef] [PubMed]
- Bartish, I.V. An ancient medicinal plant at the crossroads of modern horticulture and genetics: Genetic resources and biotechnology of sea buckthorn (Hippophae L., Elaeagnaceae). In Gene Pool Diversity and Crop Improvement; Rajpal, V.R., Rao, S.R., Raina, S.N., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 1, pp. 415–446. [Google Scholar] [CrossRef]
- Vdovina, T. Intraspecific Diversity of Wild Sea Buckthorn (Hippophae rhamnoides L.) in the East Kazakhstan Region and Prospects for Its Introduction. Ph.D. Thesis, Al-Farabi Kazakh National University, Almaty, Kazakhstan, 2010; p. 207. [Google Scholar]
- Kubentayev, S.; Kotukhov, Y.A.; Danilova, A.; Suleimenov, A.; Sumbembayev, A.A.; Nanoscience, T. Phytocoenotic structure and stocks of main medical plants in southern part of Altai Mountain System (East Kazakhstan). J. Comput. Theor. Nanosci. 2019, 16, 2822–2834. [Google Scholar] [CrossRef]
- Akiyanova, F.; Atalikhova, A.; Jussupova, Z.; Simbatova, A.; Nazhbiev, A. Current State of Ecosystems and Their Recreational Use of the Burabai National Park (Northern Kazakhstan). Eurasian J. Biosci. 2019, 13, 1231–1243. [Google Scholar]
- Beveridge, T.; Li, T.S.; Oomah, B.D.; Smith, A. Sea buckthorn products: Manufacture and composition. J. Agric. Food Chem. 1999, 47, 3480–3488. [Google Scholar] [CrossRef]
- Ma, Q.G.; He, N.X.; Huang, H.L.; Fu, X.M.; Zhang, Z.L.; Shu, J.C.; Wang, Q.Y.; Chen, J.; Wu, G.; Zhu, M.N.; et al. Hippophae rhamnoides L.: A comprehensive review on the botany, traditional uses, phytonutrients, health benefits, quality markers, and applications. J. Agric. Food Chem. 2023, 71, 4769–4788. [Google Scholar] [CrossRef]
- Wang, K.; Xu, Z.; Liao, X. Bioactive compounds, health benefits and functional food products of sea buckthorn: A review. Crit. Rev. Food Sci. Nutr. 2022, 62, 6761–6782. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Yang, W.; Kallio, H.; Yang, B. Health promoting properties and sensory characteristics of phytochemicals in berries and leaves of sea buckthorn (Hippophae rhamnoides). Crit. Rev. Food Sci. Nutr. 2022, 62, 3798–3816. [Google Scholar] [CrossRef]
- Singh, B. Indian sea buckthorn. In New Age Herbals; Singh, B., Peter, K.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 29–54. [Google Scholar] [CrossRef]
- Chen, A.; Feng, X.W.; Dorjsuren, B.; Chimedtseren, C.; Damda, T.A.; Zhang, C.H. Traditional food, modern food and nutritional value of Sea buckthorn (Hippophae rhamnoides L.): A review. J. Futur. Foods 2023, 3, 191–205. [Google Scholar] [CrossRef]
- Liu, S.; Xiao, P.; Kuang, Y.; Hao, J.; Huang, T.; Liu, E. Flavonoids from sea buckthorn: A review on phytochemistry, pharmacokinetics and role in metabolic diseases. J. Food Biochem. 2021, 45, e13724. [Google Scholar] [CrossRef]
- Dhyani, D.; Maikhuri, R.K.; Misra, S.; Rao, K.S. Endorsing the declining indigenous ethnobotanical knowledge system of seabuckthorn in Central Himalaya, India. J. Ethnopharmacol. 2010, 127, 329–334. [Google Scholar] [CrossRef]
- Singh, I.P.; Ahmad, F.; Gore, D.D.; Tikoo, K.; Bansal, A.; Jachak, S.M.; Jena, G. Therapeutic potential of seabuckthorn: A patent review (2000–2018). Expert Opin. Ther. Pat. 2019, 29, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.X.; Gao, A.X.; Dong, T.T.X.; Tsim, K.W.K. Flavonoids from Seabuckthorn (Hippophae rhamnoides L.) mimic neurotrophic functions in inducing neurite outgrowth in cultured neurons: Signaling via PI3K/Akt and ERK pathways. Phytomedicine 2023, 115, 154832. [Google Scholar] [CrossRef] [PubMed]
- Pop, R.M.; Socaciu, C.; Pintea, A.; Buzoianu, A.D.; Sanders, M.G.; Gruppen, H.; Vincken, J.P. UHPLC/PDA–ESI/MS analysis of the main berry and leaf flavonol glycosides from different Carpathian Hippophae rhamnoides L. varieties. Phytochem. Anal. 2013, 24, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Gong, X.; Li, X.; Wang, C.; Li, M.J.M. Advanced research on the antioxidant activity and mechanism of polyphenols from Hippophae species-a review. Molecules 2020, 25, 917. [Google Scholar] [CrossRef]
- Kim, J.S.; Kwon, Y.S.; Sa, Y.J.; Kim, M.J. Isolation and identification of sea buckthorn (Hippophae rhamnoides) phenolics with antioxidant activity and alpha-glucosidase inhibitory effect. J. Agric. Food Chem. 2011, 59, 138–144. [Google Scholar] [CrossRef]
- Michel, T.; Destandau, E.; Le Floch, G.; Lucchesi, M.E.; Elfakir, C. Antimicrobial, antioxidant and phytochemical investigations of sea buckthorn (Hippophae rhamnoides L.) leaf, stem, root and seed. Food Chem. 2012, 131, 754–760. [Google Scholar] [CrossRef]
- Zielińska, A.; Nowak, I. Abundance of active ingredients in sea-buckthorn oil. Lipids Health Dis. 2017, 16, 95. [Google Scholar] [CrossRef]
- Gęgotek, A.; Jastrząb, A.; Jarocka-Karpowicz, I.; Muszyńska, M.; Skrzydlewska, E.J.A. The effect of sea buckthorn (Hippophae rhamnoides L.) seed oil on UV-induced changes in lipid metabolism of human skin cells. Antioxidants 2018, 7, 110. [Google Scholar] [CrossRef]
- Zhou, W.; Ouyang, J.; Hu, N.; Li, G.; Wang, H. Protective effect of two alkaloids from Hippophae rhamnoides Linn. against doxorubicin-induced toxicity in H9c2 cardiomyoblasts. Molecules 2021, 26, 1946. [Google Scholar] [CrossRef]
- Suryakumar, G.; Gupta, A. Medicinal and therapeutic potential of Sea buckthorn (Hippophae rhamnoides L.). J. Ethnopharmacol. 2011, 138, 268–278. [Google Scholar] [CrossRef]
- Fatima, T.; Kesari, V.; Watt, I.; Wishart, D.; Todd, J.F.; Schroeder, W.R.; Paliyath, G.; Krishna, P. Metabolite profiling and expression analysis of flavonoid, vitamin C and tocopherol biosynthesis genes in the antioxidant-rich sea buckthorn (Hippophae rhamnoides L.). Phytochemistry 2015, 118, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Ciesarová, Z.; Murkovic, M.; Cejpek, K.; Kreps, F.; Tobolková, B.; Koplík, R.; Belajová, E.; Kukurová, K.; Daško, Ľ.; Panovská, Z.; et al. Why is sea buckthorn (Hippophae rhamnoides L.) so exceptional? A review. Food Res. Int. 2020, 133, 109170. [Google Scholar] [CrossRef]
- Cho, H.; Cho, E.; Jung, H.; Yi, H.C.; Lee, B.; Hwang, K.T. Antioxidant activities of sea buckthorn leaf tea extracts compared with green tea extracts. Food Sci. Biotechnol. 2014, 23, 1295–1303. [Google Scholar] [CrossRef]
- Tkacz, K.; Wojdylo, A.; Turkiewicz, I.P.; Ferreres, F.; Moreno, D.A.; Nowicka, P. UPLC-PDA-Q/TOF-MS profiling of phenolic and carotenoid compounds and their influence on anticholinergic potential for AChE and BuChE inhibition and on-line antioxidant activity of selected Hippophae rhamnoides L. cultivars. Food Chem. 2020, 309, 125766. [Google Scholar] [CrossRef]
- Nybom, H.; Ruan, C.; Rumpunen, K. The systematics, reproductive biology, biochemistry, and breeding of sea buckthorn—A review. Genes 2023, 14, 2120. [Google Scholar] [CrossRef]
- Vdovina, T.A.; Isakova, E.A.; Lagus, O.A.; Sumbembayev, A.A. Selection assessment of promising forms of natural Hippophae rhamnoides (Elaeagnaceae) populations and their offspring in the Kazakhstan Altai Mountains. Biodiversitas J. Biol. Divers. 2024, 25, 1809–1822. [Google Scholar] [CrossRef]
- Besschetnov, V.P.; Kentbaev, E.Z. Experience of green cuttings of sea buckthorn in the conditions of south-east Kazakhstan. News of higher educational institutions. For. Mag. 2018, 56–62. [Google Scholar] [CrossRef]
- Kentbaev, E.J.; Kentbaeva, B.A. Features of sea buckthorn varieties and forms by seed size. Fruit Grow. Seed Prod. Introd. Woody Plants 2019, 22, 75–78. [Google Scholar]
- Shadmanova, L.S.; Mukan, G.; Akhatov, K.Z.; Yeszhanova, A.; Kanapin, C.B.; Sitpaeva, G.T. Current state and ecological features of Hippophae rhamnoides L. cenopopulations in Northern Kazakhstan. Fundam. Exp. Biol. 2024, 11629, 101–107. [Google Scholar]
- Li, H.; Ruan, C.; Ding, J.; Li, J.; Wang, L.; Tian, X. Diversity in sea buckthorn (Hippophae rhamnoides L.) accessions with different origins based on morphological characteristics, oil traits, and microsatellite markers. PLoS ONE 2020, 15, e0230356. [Google Scholar] [CrossRef]
- Yao, Y.; Tigerstedt, P.M.J.G.R.; Evolution, C. Isozyme studies of genetic diversity and evolution in Hippophae. Genet. Resour. Crop Evol. 1993, 40, 153–164. [Google Scholar] [CrossRef]
- Bartish, I.V.; Jeppsson, N.; Nybom, H. Population genetic structure in the dioecious pioneer plant species Hippophae rhamnoides investigated by random amplified polymorphic DNA (RAPD) markers. Mol. Ecol. 1999, 8, 791–802. [Google Scholar] [CrossRef]
- Ruan, C.J.; Rumpunen, K.; Nybom, H. Advances in improvement of quality and resistance in a multipurpose crop: Sea buckthorn. Crit. Rev. Biotechnol. 2013, 33, 126–144. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.M.; An, L.Z.; Chen, T.; Xu, S.J.; Liu, G.X.; Zheng, X.L.; Pu, L.L.; Liu, Y.J.; Lian, Y.S. Analysis of the genetic diversity and relationships among and within species of Hippophae (Elaeagnaceae) based on RAPD markers’. Plant Syst. Evol. 2006, 260, 25–37. [Google Scholar] [CrossRef]
- Nybom, H.; Bartish, I.V. Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect. Plant Ecol. Evol. Syst. 2000, 3, 93–114. [Google Scholar] [CrossRef]
- Raina, S.N.; Jain, S.; Sehgal, D.; Kumar, A.; Dar, T.H.; Bhat, V.; Pandey, V.; Vaishnavi, S.; Bhargav, A.; Singh, V.; et al. Diversity and relationships of multipurpose seabuckthorn (Hippophae L.) germplasm from the Indian Himalayas as assessed by AFLP and SAMPL markers. Genet. Resour. Crop. Evol. 2012, 59, 1033–1053. [Google Scholar] [CrossRef]
- Muhammad, A.N.; Krutovsky, K.V.; Markus, M.; Oliver, G.; Asif Ali, K.; Andreas, B.; Martin, W. Morphological and Genetic Diversity of Sea Buckthorn (Hippophae rhamnoides L.) in the Karakoram Mountains of Northern Pakistan. Diversity 2018, 10, 76. [Google Scholar] [CrossRef]
- Li, H.; Ruan, C.J.; Wang, L.; Ding, J.A.; Tian, X.J. Development of RNA-Seq SSR markers and application to genetic relationship analysis among sea buckthorn germplasm. J. Am. Soc. Hortic. Sci. 2017, 142, 200–208. [Google Scholar] [CrossRef]
- Liu, Q.; Ye, G.; Ma, Y. Development and application of microsatellite markers in Hippophae rhamnoides subsp. sinensis Rousi (Hippophae rhamnoides L.) based on transcriptome sequencing. Front. Genet. 2024, 15, 1373028. [Google Scholar] [CrossRef]
- Wang, Y.H.; Jiang, H.; Peng, S.M.; Korpelainen, H. Genetic structure in fragmented populations of Hippophae rhamnoides ssp. sinensis in China investigated by ISSR and cpSSR markers. Plant Syst. Evol. 2011, 295, 97–107. [Google Scholar] [CrossRef]
- Das, K.; Ganie, S.H.; Mangla, Y.; Dar, T.U.H.; Chaudhary, M.; Thakur, R.K.; Tandon, R.; Raina, S.N.; Goel, S. ISSR markers for gender identification and genetic diagnosis of Hippophae rhamnoides ssp. turkestanica growing at high altitudes in Ladakh region (Jammu and Kashmir). Protoplasma 2017, 254, 1063–1077. [Google Scholar] [CrossRef]
- Bone, K.; Bocharkina, I.; Karlov, G.; Razumova, O.V. The use of ISSR markers for the analysis of polymorphism in sea buckthorn (Hippophae rhamnoides L.) of different origins. Mosc. Univ. Biol. Sci. Bull. 2020, 75, 36–40. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Z.; Wu, C.; Ma, Z.; Ding, G.; Cao, G.; Ruan, S.; Lin, S. Genetic diversity and variation of Chinese fir from Fujian Province and Taiwan, China, based on ISSR markers. PLoS ONE 2017, 12, e0175571. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R. Comprehensive web-based platform for advanced pcr design, genotyping, synthetic biology, molecular diag-nostics and sequence analysis. Mol. Ther. Nucleic Acids 2025, 36, 102716. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, J.; Tian, Z.; Norbu, N.; Chen, Y.; Chen, J.; Zhang, W.; Qiong, L. Development of sex-specific molecular markers for early sex identification in Hippophae gyantsensis based on whole-genome resequencing. BMC Plant Biol. 2024, 24, 1187. [Google Scholar] [CrossRef]
- Zeng, Z.; Wang, R.; Wang, J.; Chen, Y.; Wang, Y.; Song, Z.; Zhang, W.; Qiong, L. Development and validation of sex-linked molecular markers for rapid and accurate identification of male and female Hippophae tibetana plants. Sci. Rep. 2024, 14, 19243. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.; Pereira, G.; Garrido, I.; Tavares-de-Sousa, M.M.; Espinosa, F. Comparison of RAPD, ISSR, and AFLP molecular markers to reveal and classify orchardgrass (Dactylis glomerata L.) germplasm variations. PLoS ONE 2016, 11, e0152972. [Google Scholar] [CrossRef] [PubMed]
- Arvas, Y.E.; Kocaçalışkan, İ.; Ordu, E.; Erişen, S. Comparative retrotransposon analysis of mutant and non-mutant rice varieties grown at different salt concentrations. Biotechnol. Biotechnol. Equip. 2022, 36, 25–34. [Google Scholar] [CrossRef]
- Arvas, Y.E.; Marakli, S.; Kaya, Y.; Kalendar, R. The power of retrotransposons in high-throughput genotyping and sequencing. Front. Plant Sci. 2023, 14, 1174339. [Google Scholar] [CrossRef]
- Belyayev, A.; Josefiová, J.; Jandová, M.; Kalendar, R.; Krak, K.; Mandák, B. Natural history of a satellite DNA family: From the ancestral genome component to species-specific sequences, concerted and non-concerted evolution. Int. J. Mol. Sci. 2019, 20, 1201. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Kalendar, R.; Kairov, U. Genome-wide tool for sensitive de novo identification and visualisation of interspersed and tan-dem repeats. Bioinform. Biol. Insights 2024, 18, 11779322241306391. [Google Scholar] [CrossRef]
- Khapilina, O.; Raiser, O.; Danilova, A.; Shevtsov, V.; Turzhanova, A.; Kalendar, R.J.P. DNA profiling and assessment of genetic diversity of relict species Allium altaicum Pall. on the territory of Altai. PeerJ 2021, 9, e10674. [Google Scholar] [CrossRef]
- Turzhanova, A.; Kubentaev, S.; Magzumova, S.; Sarkytbayeva, A.; Khapilina, O. The impact of local environmental differences on the phenotypic plasticity and genetic variation of Kazakhstani populations of Paeonia anomala. Front. Ecol. Evol. 2025, 13, 1608776. [Google Scholar] [CrossRef]
- Abdollahi Mandoulakani, B.; Yaniv, E.; Kalendar, R.; Raats, D.; Bariana, H.S.; Bihamta, M.R.; Schulman, A.H. Development of IRAP- and REMAP-derived SCAR markers for marker-assisted selection of the stripe rust resistance gene yr15 derived from wild emmer wheat. Theor. Appl. Genet. 2015, 128, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Benoit, M.; Catoni, M.; Drost, H.G.; Brestovitsky, A.; Oosterbeek, M.; Paszkowski, J. Sensitive detection of pre-integration intermediates of long terminal repeat retrotransposons in crop plants. Nat. Plants 2019, 5, 26–33. [Google Scholar] [CrossRef]
- Papolu, P.K.; Ramakrishnan, M.; Mullasseri, S.; Kalendar, R.; Wei, Q.; Zou, L.H.; Ahmad, Z.; Vinod, K.K.; Yang, P.; Zhou, M.B. Retrotransposons: How the continuous evolutionary front shapes plant genomes for response to heat stress. Front. Plant Sci. 2022, 13, 1064847. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Zhang, Z.; Mullasseri, S.; Kalendar, R.; Ahmad, Z.; Sharma, A.; Liu, G.; Zhou, M.; Wei, Q. Epigenetic stress memory: A new approach to study cold and heat stress responses in plants. Front. Plant Sci. 2022, 13, 1075279. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Khapilina, O.N.; Turzhanova, A.S.; Erbay, M.; Magzumova, S.; Mamirova, A. Genetic polymorphism in the Amaranthaceae species in the context of stress tolerance. Plants 2023, 12, 3470. [Google Scholar] [CrossRef]
- Terletskaya, N.V.; Turzhanova, A.S.; Khapilina, O.N.; Zhumagul, M.Z.; Meduntseva, N.D.; Kudrina, N.O.; Korbozova, N.K.; Kubentayev, S.A.; Kalendar, R. Genetic diversity in natural populations of Rhodiola species of different adaptation strategies. Genes 2023, 14, 794. [Google Scholar] [CrossRef] [PubMed]
- Hosid, E.; Brodsky, L.; Kalendar, R.; Raskina, O.; Belyayev, A. Diversity of Long Terminal Repeat Retrotransposon Genome Distribution in Natural Populations of the Wild Diploid Wheat Aegilops speltoides. Genetics 2012, 190, 263–274. [Google Scholar] [CrossRef]
- Villano, C.; Carputo, D.; Frusciante, L.; Santoro, X.; Aversano, R. Use of SSR and retrotransposon-based markers to interpret the population structure of native grapevines from southern Italy. Mol. Biotechnol. 2014, 56, 1011–1020. [Google Scholar] [CrossRef]
- Kubis, S.E.; Castilho, A.M.M.F.; Vershinin, A.V.; Heslop-Harrison, J.S.P. Retroelements, transposons and methylation status in the genome of oil palm (Elaeis guineensis) and the relationship to somaclonal variation. Plant Mol. Biol. 2003, 52, 69–79. [Google Scholar] [CrossRef]
- Melnikova, N.V.; Arkhipov, A.A.; Zubarev, Y.A.; Novakovskiy, R.O.; Turba, A.A.; Pushkova, E.N.; Zhernova, D.A.; Mazina, A.S.; Dvorianinova, E.M.; Sigova, E.A.; et al. Genetic diversity of Hippophae rhamnoides varieties with different fruit characteristics based on whole-genome sequencing. Front. Plant Sci. 2025, 16, 1542552. [Google Scholar] [CrossRef]
- Vdovina, T.; Lagus, O.; Vinokurov, A.; Aimenova, Z.; Sumbembayev, A. Assessment of Biochemical Composition of Fruits of Hippophae rhamnoides (Elaeagnaceae juss.), Viburnum opulus (Viburnaceae raf.) and Lonicera caerulea subsp. altaica (Caprifoliaceae juss.). Metabolites 2025, 15, 256. [Google Scholar] [CrossRef]
- Givnish, T.J.; Evans, T.M.; Zjhra, M.L.; Patterson, T.B.; Berry, P.E.; Sytsma, K.J. Molecular evolution, adaptive radiation, and geographic diversification in the amphiatlantic family Rapateaceae: Evidence from ndhF sequences and morphology. Evolution 2000, 54, 1915–1937. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Ivanov, K.I.; Samuilova, O.; Kairov, U.; Zamyatnin, A.A., Jr. Isolation of high-molecular-weight DNA for long-read sequencing using a high-salt gel electroelution trap. Anal. Chem. 2023, 95, 17818–17825. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Ivanov, K.I.; Akhmetollayev, I.; Kairov, U.; Samuilova, O.; Burster, T.; Zamyatnin, A.A., Jr. An improved method and device for nucleic acid isolation using a high-salt gel electroelution trap. Anal. Chem. 2024, 96, 15526–15530. [Google Scholar] [CrossRef] [PubMed]
- Kalendar, R.; Boronnikova, S.; Seppänen, M. Isolation and purification of DNA from complicated biological samples. Methods Mol. Biol. 2021, 2222, 57–67. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Chen, S.; Dick, J.; Owen, A.B. Consistency of Markov chain quasi-Monte Carlo on continuous state spaces. Ann. Stat. 2011, 39, 673–701. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Vdovina, T.; Lagus, O.; Aimenova, J. Biochemical evaluation of promising seedlings and forms of sea buckthorn in the introduction population (Altai Botanical Garden). Eurasian J. Appl. Biotechnol. 2024, 3S, 14. [Google Scholar] [CrossRef]
- Sun, K.; Chen, W.; Ma, R.; Chen, X.; Li, A.; Ge, S. Genetic variation in Hippophae rhamnoides ssp. sinensis (Elaeagnaceae) revealed by RAPD markers. Biochem. Genet. 2006, 44, 186–197. [Google Scholar] [CrossRef]
- Belyayev, A.; Kalendar, R.; Josefiová, J.; Paštová, L.; Habibi, F.; Mahelka, V.; Mandák, B.; Krak, K. Telomere sequence variability in genotypes from natural plant populations: Unusual block-organized double-monomer terminal telomeric arrays. BMC Genom. 2023, 24, 572. [Google Scholar] [CrossRef]
- Tian, C.; Nan, P.; Shi, S.; Chen, J.; Zhong, Y. Molecular genetic variation in Chinese populations of three subspecies of Hippophae rhamnoides. Biochem. Genet. 2004, 42, 259–267. [Google Scholar] [CrossRef] [PubMed]

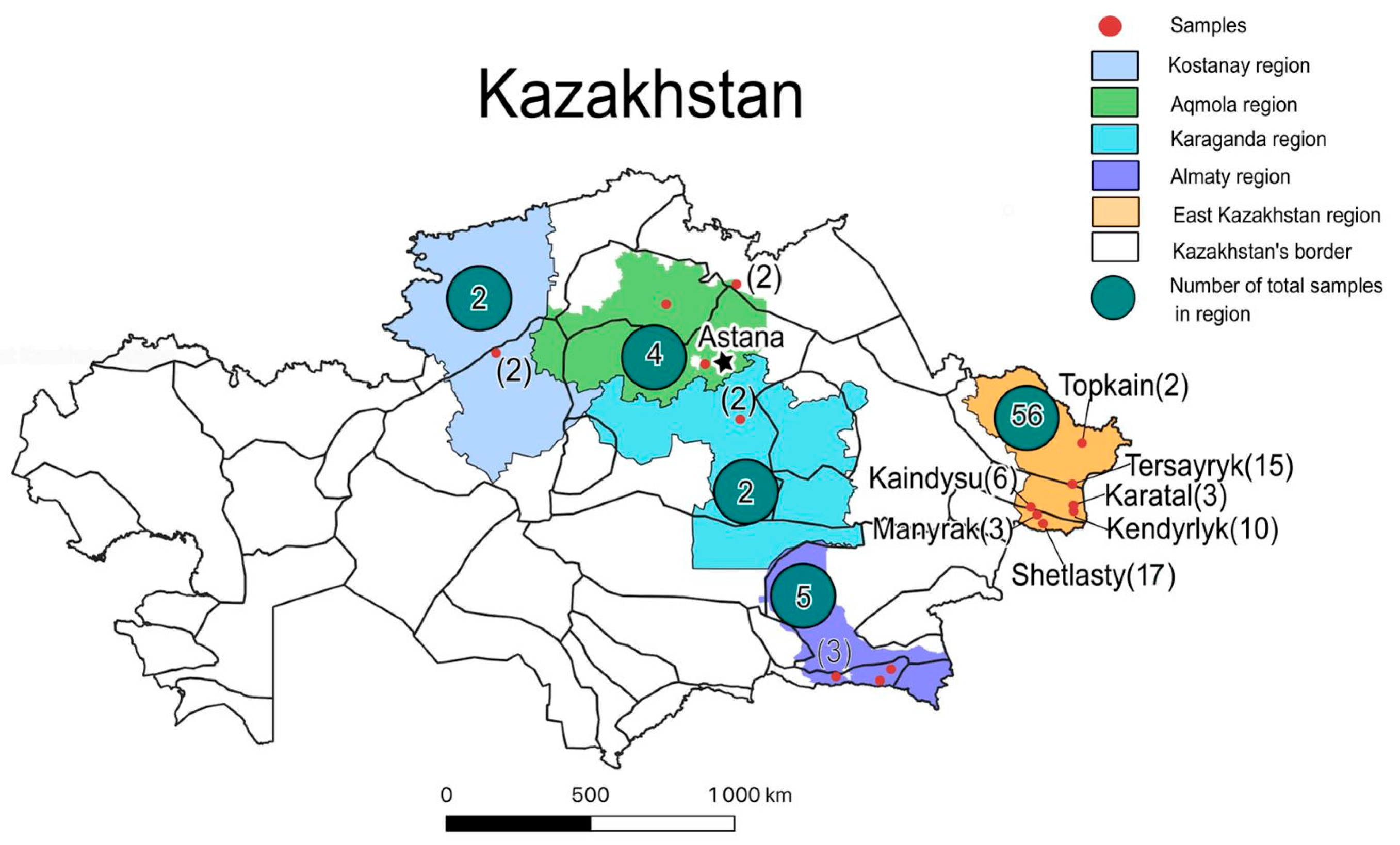
| Populations | Location | Sample Type | Sample Accession | Original Number |
|---|---|---|---|---|
| Kendyrlyk | 47.47830 85.21473 | Varieties | Feyerverk | K-14-82 (3−24) III |
| Fakel | K-14-81(3−25) III | |||
| Krasnoplodnaja | K-14-81(4−27) | |||
| Wild population | Ken-14-86 | K-14-86(4−18) | ||
| Ken-4-86 | K-4-86(4−28) | |||
| Ken-14-81 | K-14-81(3−38) | |||
| Herbarium | Ken-1-HRB | Kendyrlyk-84−1 | ||
| Ken-6-HRB | Kendyrlyk-84−6 | |||
| Ken-2-HRB | Kendyrlyk-84−2 | |||
| Ken-3-HRB | Kendyrlyk-8−-3 | |||
| Shetlasty | 47.16724 84.09574 | Varieties | Asem | Sh-9-81 (3−27) |
| Shetlastinka | Sh-7 (2−24) III | |||
| Jantarnaja | Sh-12 05 (2−1) III | |||
| Solnyshko | Sh-12-81 (1−18) | |||
| Yozhik | Sh-3−84 | |||
| Wild population | Sh-4-18 | Sh-9-81(4−18) IV | ||
| Sh-4-20 | Sh-9-81(4−20) IV | |||
| Sh-4-15 | Sh-9-81(4−15) | |||
| Sh-5-27 | Sh-20-84(5−27) | |||
| Sh-5-11 | Sh-16-86(5−11) | |||
| Sh-4-5 | Sh-9-81(4−5) | |||
| Herbarium | Sh-1-HRB | Shetlasty-4−84 | ||
| Sh-2-HRB | Shetlasty-3−84 | |||
| Sh-3-HRB | Shetlasty-7−84 | |||
| Sh-4-HRB | Shetlasty-13−84 | |||
| Sh-5-HRB | Shetlasty-9−84 | |||
| Sh-6-HRB | Shetlasty-2 −84 | |||
| Kaindysu | 47.585513 83.64861 | Wild population | Kan-3-34 IV | Kan-2-84 (3−34) IV |
| Kan-4-6 | Kan-9-8 (4−6) IV | |||
| Kan-1-5 | Kan-3-82 (1−5) | |||
| Kan-3-1 | Kan-3-82 (3−1) | |||
| Kan-8-4 | Kan-8-81 (8−4) | |||
| Kan-3-34 | Kan-2-84 (3−34) | |||
| Tersayryk | 48.16005 85.16727 | Varieties | Plakuchaja | T-14-82 (2−32) |
| Jubilejnaja Kotuhova | T-2-82 (2−22) III | |||
| Wild population | T-3-26 | T-14-86 (3−26) | ||
| T-2-32 | T-2-82 (2−32) | |||
| T-6-29 | T-14-82 (6−29) | |||
| T-4-3 | T-3-83 (4−3) | |||
| T-4-4 | T-3-83 (4−4) | |||
| T-3-20 | T-14-86 (3−20) | |||
| T-4-16 | T-17-82 (4−16) | |||
| T-2-32 | T-14-82 (2−32) | |||
| T-4-32 | T-7-86 (4−32) | |||
| Herbarium | T-3-HRB | Tersayryk 3-86 | ||
| T-7-HRB | Tersayryk 7-86 | |||
| T-1-HRB | Tersayryk 1-86 | |||
| T-5-HRB | Tersayryk 5-86 | |||
| Karatal | 47.62766 85.20537 | Wild population | Ks-1-24 | Karatal KP 1-24 |
| Herbarium | Ks-1-HRB | Karatal K-3 | ||
| Ks-2-HRB | Karatal KP | |||
| Topkain | 49.17389 85.51889 | Herbarium | Top-1-HRB | Topkain 3-84 |
| Top-2-HRB | Topkain 5-84 | |||
| Manyrak | 47.39027 83.87944 | Wild population | Man-3-24 | Manyrak 3-24 |
| Man-12-24 | Manyrak 12-24 | |||
| Man-7-24 | Manyrak 7-24 | |||
| Almaty | 43.32902 78.54828 | Wild population | ALM-1 | Almaty-24-1 |
| 43.132895 76.545421 | ALM-2 | Almaty-24-2 | ||
| 43.064783 76.544676 | ALM-3 | Almaty-24-3 | ||
| 43.043301 76.542341 | ALM-4 | Almaty-24-4 | ||
| 43.02056 78.14644 | ALM-5 | Almaty-24-5 | ||
| Akmola | 52.912467 72.912467 | Wild population | AKML-1 | Akmola-24-1 |
| 52.912467 72.922467 | AKML-2 | Akmola-24-2 | ||
| 51.074516 71.765893 | Wild population | AST-1 | Astana-24-11 | |
| 52.46106 70.34146 | Herbarium | AKML-1-HRB | AKML-17-1 | |
| Kostanay | 51.33319 64.13590 | Herbarium | Kos-1-HRB | Kostanay-23-1 |
| Herbarium | Kos-2-HRB | Kostanay-23-2 | ||
| Karaganda | 49.753249 73.043323 | Herbarium | Kar-1-HRB | Karaganda-23-1 |
| Herbarium | Kar-2-HRB | Karaganda-23-2 |
| ID | Sequence | Tm | GC (%) | LC (%) |
|---|---|---|---|---|
| 2225 | AGCATAGCTTTGATACCA | 54.1 | 38.9 | 80 |
| 2232 | AGAGAGGCTCGGATACCA | 60.9 | 55.6 | 83 |
| 2237 | CCCCTACCTGGCGTGCCA | 69.6 | 72.2 | 77 |
| 2239 | ACCTAGGCTCGGATGCCA | 65.0 | 61.1 | 91 |
| 2240 | AACCTGGCTCAGATGCCA | 63.2 | 55.6 | 88 |
| 2247 | AACCTGGCTCTGATACCA | 55.9 | 50 | 83 |
| 2252 | TCATGGCTCATGATACCA | 57.1 | 44.4 | 80 |
| 2300 | CACCGGGCTCTGATACCA | 64.0 | 61.1 | 90 |
| 2374 | CCCAGCAAACCA | 48.2 | 58.1 | 79 |
| Population | Na | Ne | I | He | uHe | PB, % | Number of Private Bands |
|---|---|---|---|---|---|---|---|
| Shetlasty | 1.322 | 1.320 | 0.296 | 0.194 | 0.20 | 60.0 | 1 |
| Kaindysu | 0.965 | 1.234 | 0.202 | 0.135 | 0.148 | 37.4 | 0 |
| Kendyrlyk | 1.017 | 1.199 | 0.189 | 0.122 | 0.129 | 41.7 | 0 |
| Tersayryk | 1.304 | 1.258 | 0.258 | 0.164 | 0.169 | 59.1 | 5 |
| Manyrak | 0.722 | 1.208 | 0.162 | 0.113 | 0.135 | 26.1 | 0 |
| Karatal | 0.617 | 1.116 | 0.099 | 0.067 | 0.08 | 17.4 | 0 |
| Almaty | 0.783 | 1.189 | 0.158 | 0.108 | 0.12 | 26.9 | 0 |
| Akmola | 0.704 | 1.131 | 0.109 | 0.075 | 0.085 | 18.3 | 0 |
| Kostanay | 0.774 | 1.172 | 0.147 | 0.101 | 0.134 | 24.4 | 0 |
| Karaganda | 0.843 | 1.221 | 0.189 | 0.13 | 0.173 | 31.3 | 0 |
| Topkain | 0.548 | 1.061 | 0.053 | 0.036 | 0.048 | 8.7 | 2 |
| Mean | 0.873 | 1.192 | 0.169 | 0.113 | 0.129 | 31.9 | |
| SE | 0.024 | 0.009 | 0.007 | 0.005 | 0.006 | 4.9 |
| Variability | Df | SS | MS | Est. Var. | % | PhiPT | p Value |
|---|---|---|---|---|---|---|---|
| Among populations | 10 | 544.953 | 54.495 | 7.417 | 40% | 0.401 | 0.001 |
| Within populations | 58 | 642.09 | 11.071 | 11.071 | 60% | ||
| General | 68 | 1187.043 | 18.487 | 100% |
| Shetlasty | Kaindysu | Kendyrlyk | Tersayryk | Monyrak | Karakatal | Almaty | Akmola | Kostanay | Karaganda | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0.145 | 0 | Kaindysu | ||||||||
| 0.174 | 0.130 | 0 | Kendyrlyk | |||||||
| 0.103 | 0.171 | 0.137 | 0 | Tersayryk | ||||||
| 0.148 | 0.213 | 0.215 | 0.143 | 0 | Monyrak | |||||
| 0.238 | 0.179 | 0.182 | 0.140 | 0.228 | 0 | Karakatal | ||||
| 0.135 | 0.223 | 0.248 | 0.147 | 0.259 | 0.252 | 0 | Almaty | |||
| 0.261 | 0.242 | 0.31 | 0.268 | 0.327 | 0.248 | 0.342 | 0 | Akmola | ||
| 0.193 | 0.186 | 0.256 | 0.211 | 0.252 | 0.234 | 0.288 | 0.126 | 0 | Kostanay | |
| 0.202 | 0.202 | 0.221 | 0.174 | 0.304 | 0.241 | 0.237 | 0.231 | 0.131 | 0 | Karaganda |
| 0.314 | 0.354 | 0.375 | 0.428 | 0.42 | 0.436 | 0.401 | 0.412 | 0.384 | 0.384 | Topkain |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tumenbayeva, A.; Turzhanova, A.; Magzumova, S.; Vdovina, T.; Sumbembayev, A.; Satekov, Y.; Shevtsov, V.; Raiser, O.; Tagimanova, D.; Khapilina, O. Assessment of Genetic Variation in Natural Populations of Hippophae rhamnoides L. from Kazakhstan Using Retrotransposon-Based Markers. Forests 2025, 16, 1593. https://doi.org/10.3390/f16101593
Tumenbayeva A, Turzhanova A, Magzumova S, Vdovina T, Sumbembayev A, Satekov Y, Shevtsov V, Raiser O, Tagimanova D, Khapilina O. Assessment of Genetic Variation in Natural Populations of Hippophae rhamnoides L. from Kazakhstan Using Retrotransposon-Based Markers. Forests. 2025; 16(10):1593. https://doi.org/10.3390/f16101593
Chicago/Turabian StyleTumenbayeva, Asem, Ainur Turzhanova, Saule Magzumova, Tatiana Vdovina, Aidar Sumbembayev, Yeskendir Satekov, Vladislav Shevtsov, Olesya Raiser, Damelya Tagimanova, and Oxana Khapilina. 2025. "Assessment of Genetic Variation in Natural Populations of Hippophae rhamnoides L. from Kazakhstan Using Retrotransposon-Based Markers" Forests 16, no. 10: 1593. https://doi.org/10.3390/f16101593
APA StyleTumenbayeva, A., Turzhanova, A., Magzumova, S., Vdovina, T., Sumbembayev, A., Satekov, Y., Shevtsov, V., Raiser, O., Tagimanova, D., & Khapilina, O. (2025). Assessment of Genetic Variation in Natural Populations of Hippophae rhamnoides L. from Kazakhstan Using Retrotransposon-Based Markers. Forests, 16(10), 1593. https://doi.org/10.3390/f16101593






