Specific Function and Assembly of Crucial Microbes for Dendroctonus armandi Tsai et Li
Abstract
1. Introduction
2. Materials and Methods
2.1. Pine Logs and Bark Beetle Collection
2.2. Metagenomic Data Acquiring and Uploading
2.3. Gut and Cuticular Microbial Composition Analyses
2.4. Gut and Cuticular Microbial Functional Analyses
2.5. Gut and Cuticular Microbial Community Assembly Analyses
3. Results
3.1. Metagenome Sequencing of Microbes
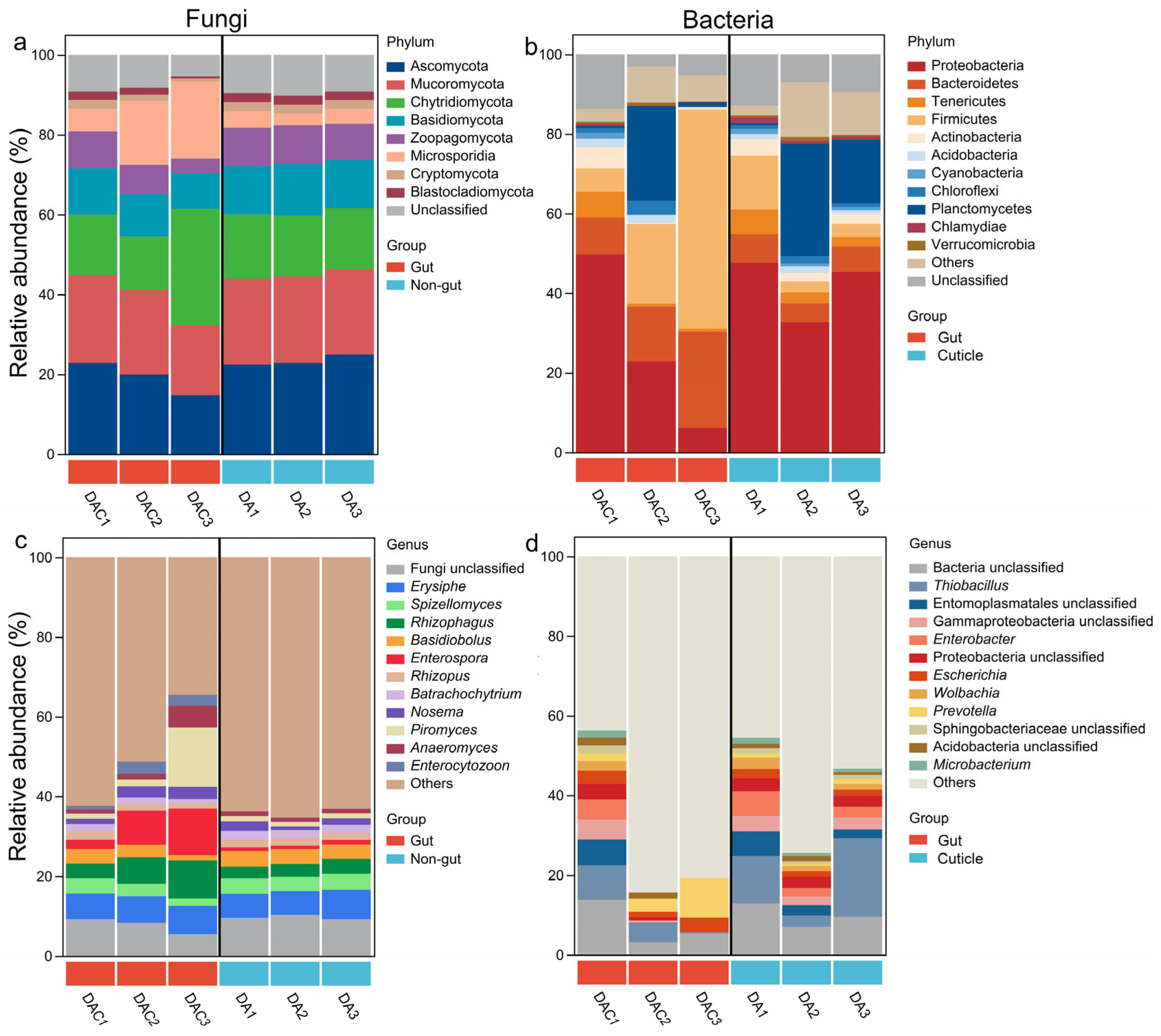
3.2. Gut and Cuticle-Symbionts Community Structures
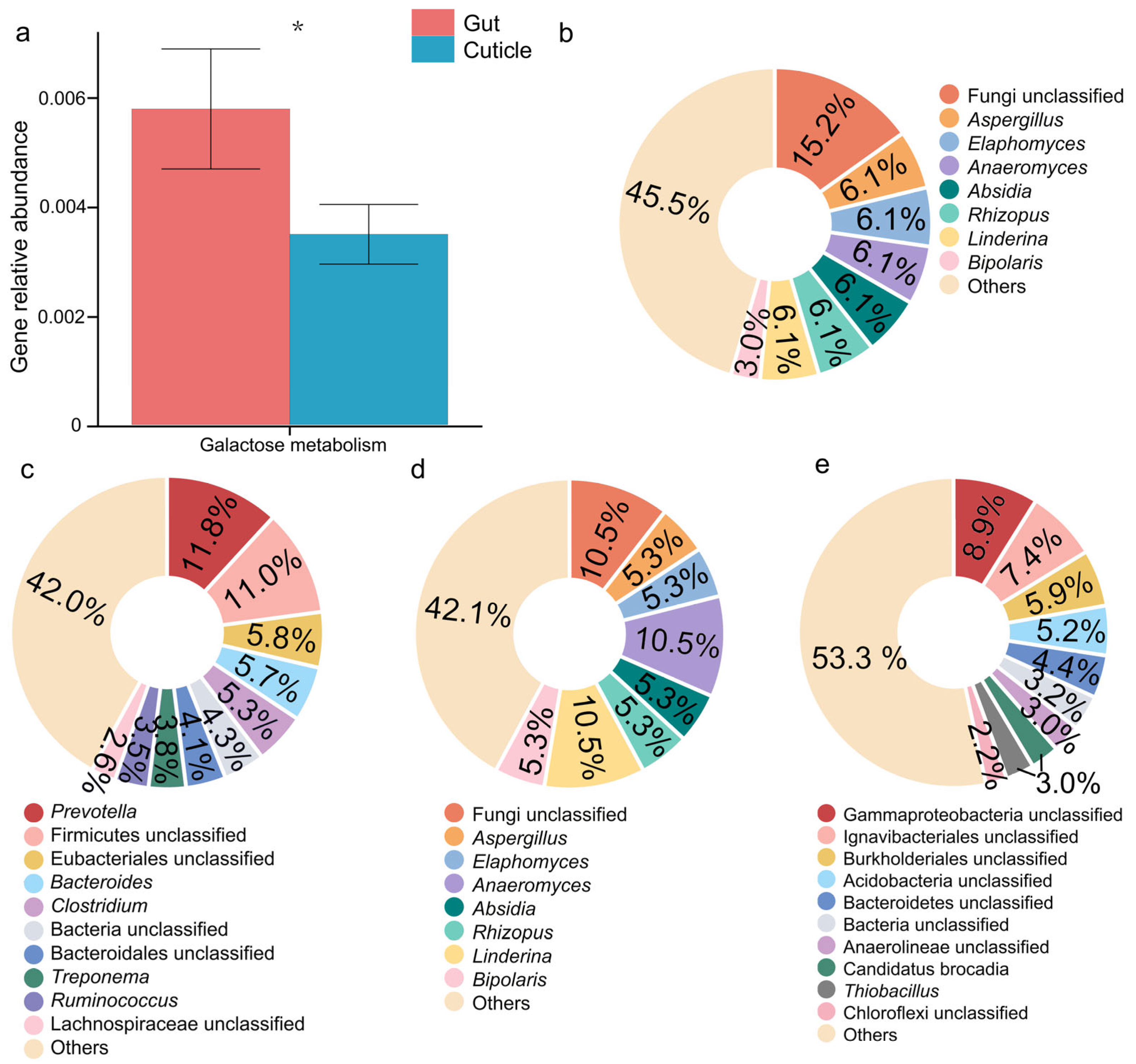
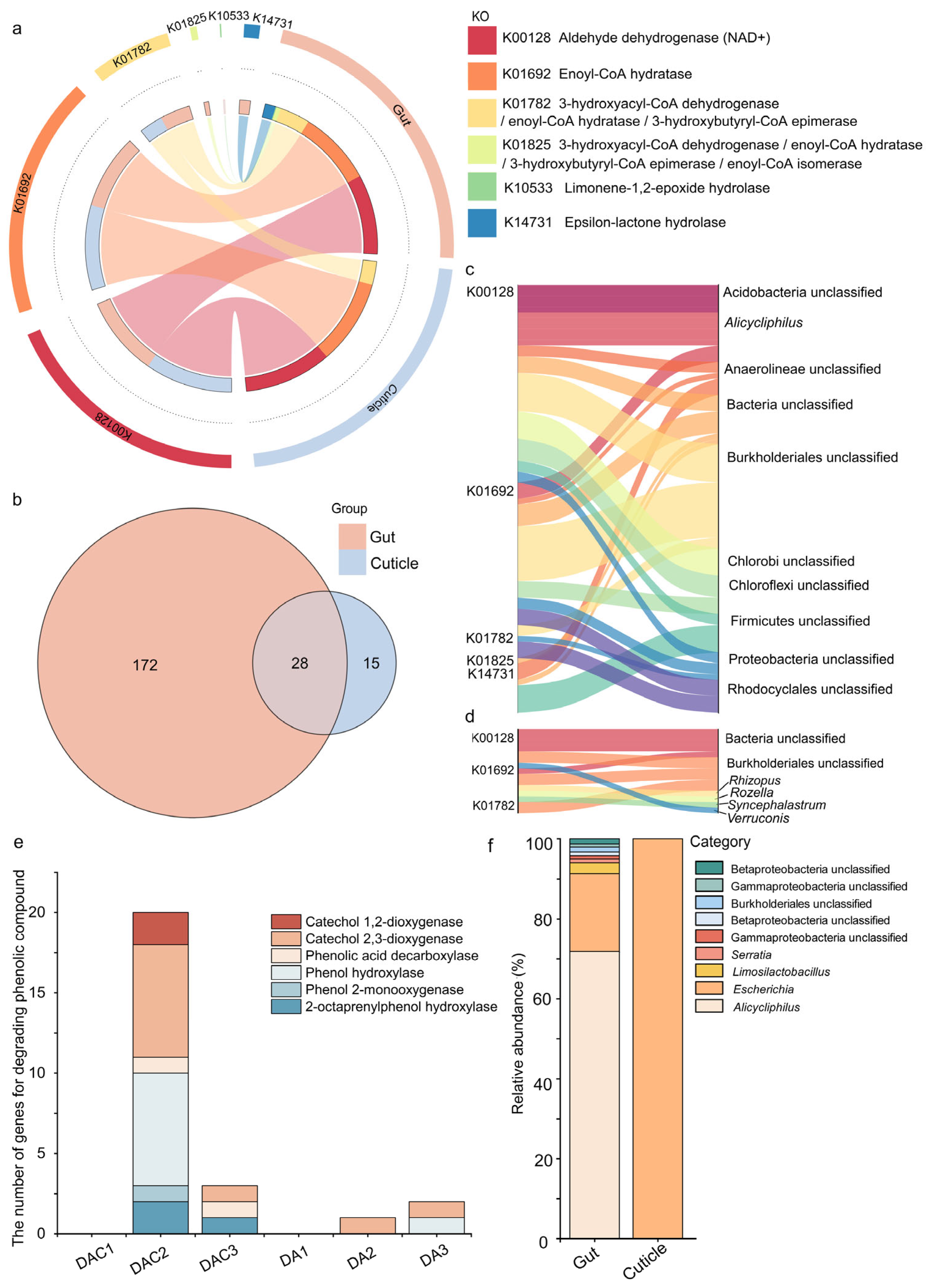
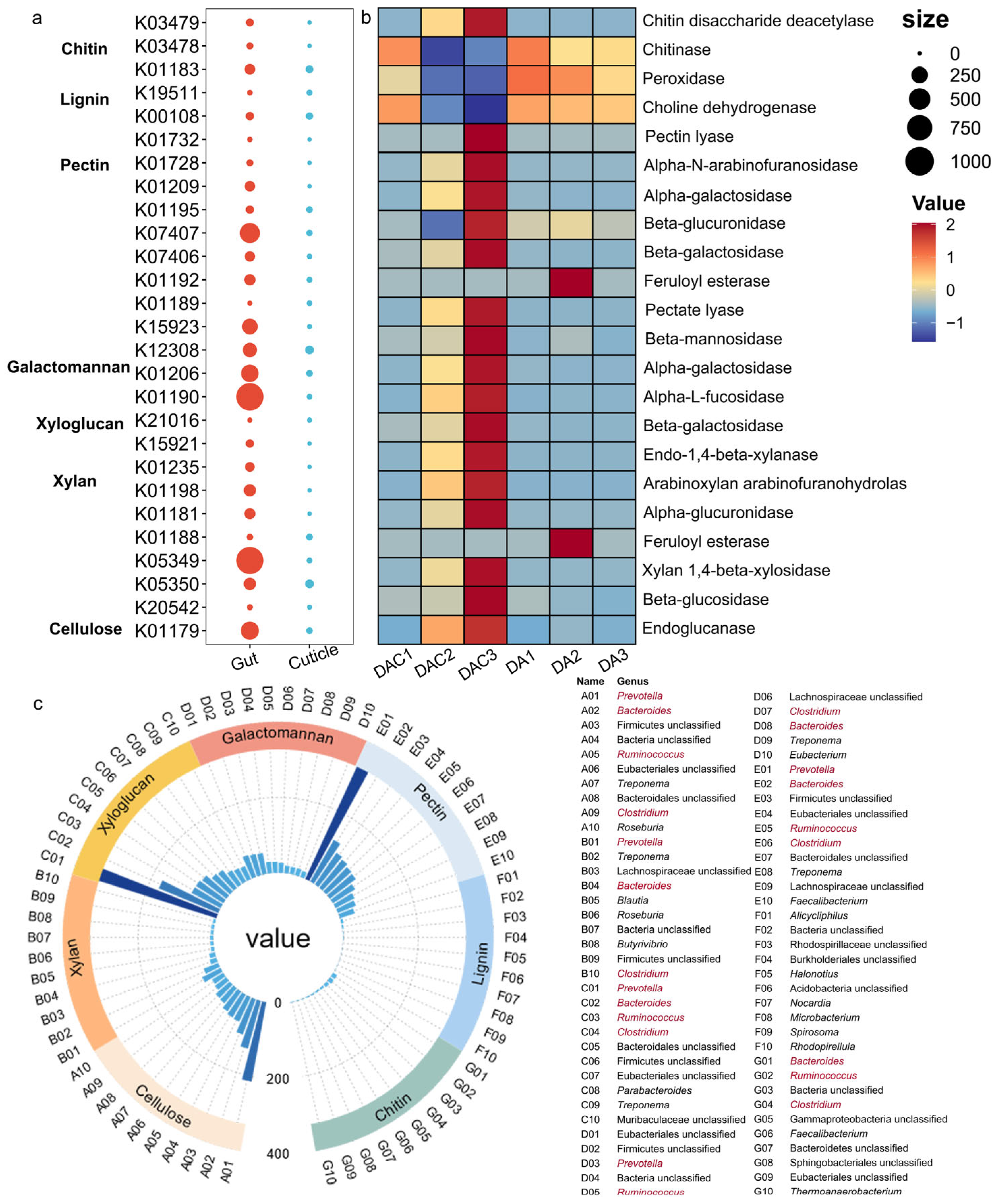
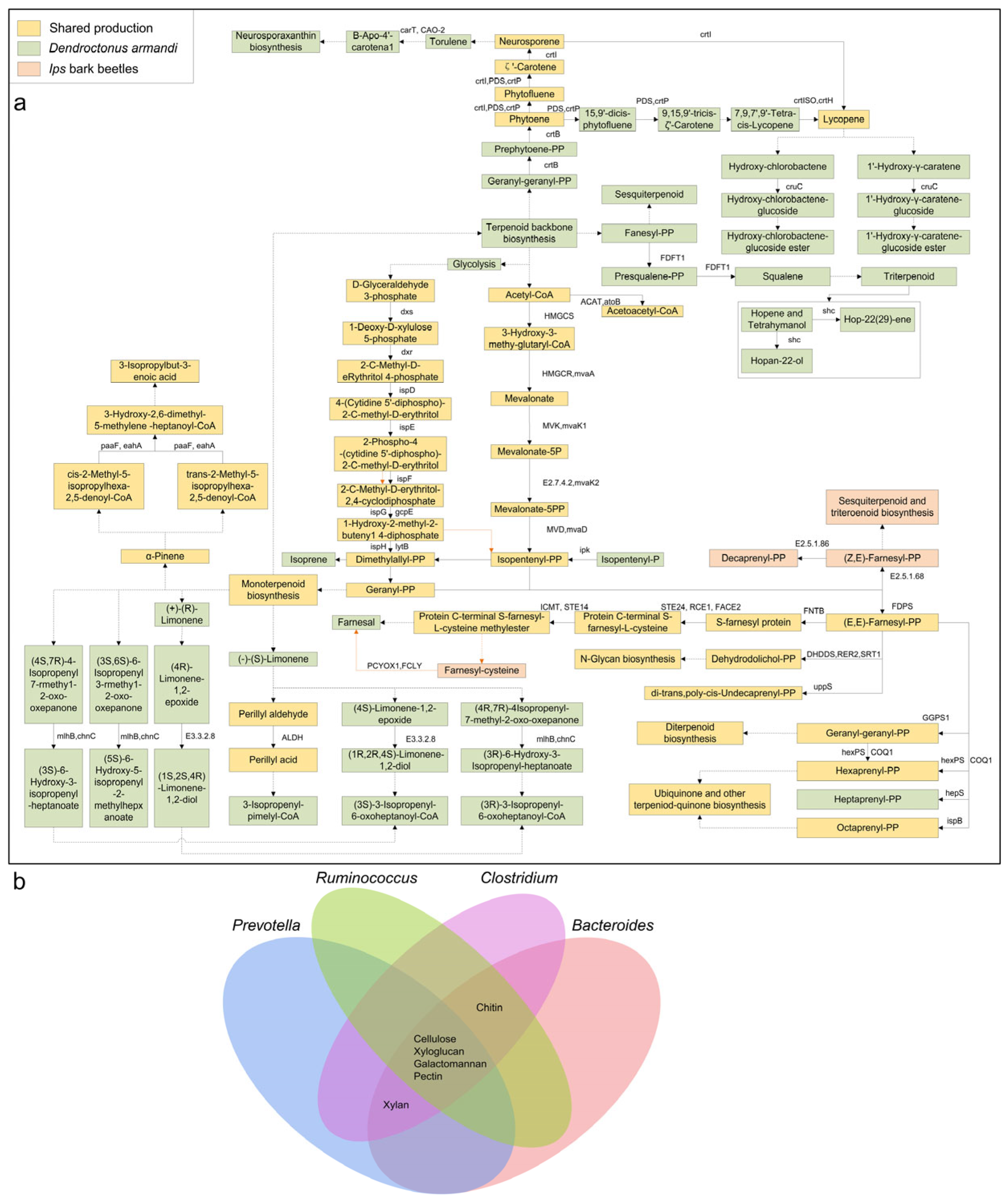
3.3. Gut and Cuticular Microbial Functional Characteristics
3.4. Crucial Drivers of Gut and Cuticular Microbial Assemblies
4. Discussion
4.1. The Higher Gut Bacterial Community Diversity
4.2. Crucial Gut Bacteria in Degrading Complex Compounds
4.3. Stochasticity for Driving Core Bacterial Community Assemblies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LEfSe | Linear discriminant analysis Effect Size |
| NR | Non-redundant |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KOs | KEGG Orthologies |
| CAZy | Carbohydrate-Active enZYmes |
| HS | heterogeneous selection |
| HOS | homogeneous selection |
| βNTI | beta-nearest taxon index |
| ML | maximum likelihood |
References
- Farrell, B.D.; Sequeira, A.S.; O’Meara, B.C.; Normark, B.B.; Chung, J.H.; Jordal, B.H. The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae). Evolution 2001, 55, 2011–2027. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Kandasamy, D.; Krokene, P.; Chen, J.; Gershenzon, J.; Hammerbacher, A. Fungal associates of the tree-killing bark beetle, Ips typographus, vary in virulence, ability to degrade conifer phenolics and influence bark beetle tunneling behavior. Fungal Ecol. 2019, 38, 71–79. [Google Scholar] [CrossRef]
- DiGuistini, S.; Wang, Y.; Liao, N.Y.; Taylor, G.; Tanguay, P.; Feau, N.; Henrissat, B.; Chan, S.K.; Hesse-Orce, U.; Alamouti, S.M.; et al. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc. Nat. Acad. Sci. USA 2011, 108, 2504–2509. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kautz, M.; Trowbridge, A.M.; Hammerbacher, A.; Raffa, K.F.; Adams, H.D.; Goodsman, D.W.; Xu, C.; Meddens, A.J.H.; Kandasamy, D.; et al. Tree defence and bark beetles in a drying world: Carbon partitioning, functioning and modelling. New Phytol. 2020, 225, 26–36. [Google Scholar] [CrossRef]
- Hammerbacher, A.; Paetz, C.; Wright, L.P.; Fischer, T.C.; Bohlmann, J.; Davis, A.J.; Fenning, T.M.; Gershenzon, J.; Schmidt, A. Flavan-3-ols in Norway spruce: Biosynthesis, accumulation, and function in response to attack by the bark beetle-associated fungus Ceratocystis polonica. Plant Physiol. 2014, 164, 2107–2122. [Google Scholar] [CrossRef]
- Kaltenpoth, M. Actinobacteria as mutualists: General healthcare for insects. Trends Microbiol. 2009, 17, 529–535. [Google Scholar] [CrossRef]
- Glass, N.L.; Schmoll, M.; Cate, J.H.D.; Coradetti, S. Plant cell wall deconstruction by Ascomycete fungi. Annu. Rev. Microbiol. 2013, 67, 477–498. [Google Scholar] [CrossRef]
- Morales-Jiménez, J.; Zúñiga, G.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb. Ecol. 2009, 58, 879–891. [Google Scholar] [CrossRef]
- Morales-Jiménez, J.; Zúñiga, G.; Ramírez-Saad, H.C.; Hernández-Rodríguez, C. Gut associated bacteria throughout the life cycle of the bark beetle Dendroctonus rhizophagus Thomas and Bright (Curculionidae: Scolytinae) and their cellulolytic activities. Microb. Ecol. 2012, 64, 268–278. [Google Scholar] [CrossRef]
- Six, D.L. The bark beetle holobiont: Why microbes matter. J. Chem. Ecol. 2013, 39, 989–1002. [Google Scholar] [CrossRef]
- García-Fraile, P. Roles of bacteria in the bark beetle holobiont—How do they shape this forest pest. Ann. Appl. Biol. 2018, 172, 111–125. [Google Scholar] [CrossRef]
- Douglas, A.E. Multiorganismal insects: Diversity and function of resident Microorganisms. Annu. Rev. Entomol. 2015, 60, 17–34. [Google Scholar] [CrossRef]
- Cao, Q.; Zhao, Y.; Koski, T.M.; Li, H.; Sun, J. Effects of simulated gut pH environment on bacterial composition and pheromone production of Dendroctonus valens. Insect Sci. 2024, 31, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microb. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, W.S.; Ziganshina, E.E.; Shagimardanova, E.I.; Gogoleva, N.E.; Ziganshin, A.M. Comparison of intestinal bacterial and fungal communities across various xylophagous beetle larvae (Coleoptera: Cerambycidae). Sci. Rep. 2018, 8, 10073. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Deng, J.; Zhou, F.; Cheng, C.; Zhang, L.; Zhang, J.; Lu, M. Gut microbiota in an invasive bark beetle infected by a pathogenic fungus accelerates beetle mortality. J. Pest Sci. 2019, 92, 343–351. [Google Scholar] [CrossRef]
- Torres-Banda, V.; Obregón-Molina, G.; Soto-Robles, L.V.; Albores-Medina, A.; López, M.F.; Zúñiga, G. Gut transcriptome of two bark beetle species stimulated with the same kairomones reveals molecular differences in detoxification pathways. Comput Struct Biotec. 2022, 20, 3080–3095. [Google Scholar] [CrossRef]
- Gupta, S.; Chakraborty, A.; Roy, A. Prospects for deploying microbes against tree-killing beetles (Coleoptera) in Anthropocene. Front. For. Glob. Change 2023, 6, 1182834. [Google Scholar] [CrossRef]
- Kostovcik, M.; Bateman, C.C.; Kolarik, M.; Stelinski, L.L.; Jordal, B.H.; Hulcr, J. The ambrosia symbiosis is specific in some species and promiscuous in others: Evidence from community pyrosequencing. ISME J. 2015, 9, 126–138. [Google Scholar] [CrossRef]
- Hulcr, J.; Rountree, N.R.; Diamond, S.E.; Stelinski, L.L.; Fierer, N.; Dunn, R.R. Mycangia of ambrosia beetles host communities of bacteria. Microb. Ecol. 2012, 64, 784–793. [Google Scholar] [CrossRef]
- Cardoza, Y.J.; Klepzig, K.D.; Raffa, K.F. Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol. Entomol. 2006, 31, 636–645. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Zhang, S.; Deng, J.; Lu, M.; Zhang, L.; Zhang, J. Comparative analysis of the immune system of an invasive bark beetle, Dendroctonus valens, infected by an entomopathogenic fungus. Dev. Comp. Immunol. 2018, 88, 65–69. [Google Scholar] [CrossRef]
- Wang, T.T. Study on Diversity and Pathogenicity of Ophiostomatoid Fungi Associated with the Dendroctonus armandi in Pinus armandii; Chinese Academy of Forestry: Beijing, China, 2020. [Google Scholar]
- Wang, Z.; Zhou, Q.; Zheng, G.; Fang, J.; Han, F.; Zhang, X.; Lu, Q. Abundance and diversity of Ophiostomatoid fungi associated with the Great Spruce Bark Beetle (Dendroctonus micans) in the Northeastern Qinghai-Tibet Plateau. Front. Microbiol. 2021, 12, 721395. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, L.; Wang, H.; Decock, C.; Lu, Q. Ophiostomatoid fungi associated with Ips bark beetles in China. Fungal Divers. 2024, 129, 283–364. [Google Scholar] [CrossRef]
- Hu, X.; Li, M.; Chen, H. Community structure of gut fungi during different developmental stages of the Chinese white pine beetle (Dendroctonus armandi). Sci. Rep. 2015, 5, 8411. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Li, M.; Zhang, F.; Chen, H. Influence of starvation on the structure of gut-associated bacterial communities in the Chinese white pine beetle (Dendroctonus armandi). Forests 2016, 7, 126. [Google Scholar] [CrossRef]
- Wang, J.; Chen, H.; Tang, M. Community structure of gut bacteria of Dendroctonus armandi (Coleoptera: Curculionidae: Scolytinae) larvae during overwintering stage. Sci. Rep. 2017, 7, 14242. [Google Scholar] [CrossRef]
- Cheng, T.; Veselská, T.; Křížková, B.; Švec, K.; Havlíček, V.; Stadler, M.; Kolařík, M. Insight into the genomes of dominant yeast symbionts of European spruce bark beetle, Ips typographus. Front. Microbiol. 2023, 14, 1108975. [Google Scholar] [CrossRef]
- Hu, X.; Yu, J.M.; Wang, C.Y.; Chen, H. Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae). Forests 2014, 5, 455–465. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Y.; Wang, T.; Liu, D.; Lu, Q. Pathophysiology and transcriptomic responses of Pinus armandii defenses to ophiostomatoid fungi. Tree Physiol. 2024, 44, tpae056. [Google Scholar] [CrossRef]
- Liu, F.; Ye, F.; Cheng, C.; Kang, Z.; Kou, H.; Sun, J. Symbiotic microbes aid host adaptation by metabolizing a deterrent host pine carbohydrate d-pinitol in a beetle-fungus invasive complex. Sci. Adv. 2022, 8, eadd5051. [Google Scholar] [CrossRef] [PubMed]
- Khara, A.; Chakraborty, A.; Modlinger, R.; Synek, J.; Roy, A. Comparative metagenomic study unveils new insights on bacterial communities in two pine-feeding Ips beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 2024, 15, 1400894. [Google Scholar] [CrossRef] [PubMed]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Chen, R.; Stegen, J.C.; Guo, Z.; Zhang, J.; Li, Z.; Lin, X. Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Mol. Ecol. 2018, 27, 5238–5251. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Yang, R.; Wang, X.Y.; Wen, T.; Gong, M.H.; Shen, Y.; Xu, J.Y.; Zhao, D.S.; Du, Y.Z. Gut microbiota composition in the sympatric and diet-sharing Drosophila simulans and Dicranocephalus wallichii bowringi shaped largely by community assembly processes rather than regional species pool. iMeta 2022, 1, e57. [Google Scholar] [CrossRef]
- Zhu, Y.X.; Yang, T.Y.; Deng, J.H.; Yin, Y.; Song, Z.R.; Du, Y.Z. Stochastic processes drive divergence of bacterial and fungal communities in sympatric wild insect species despite sharing a common diet. mSphere 2024, 9, e00386-24. [Google Scholar] [CrossRef]
- Liu, C.; Wang, H.; Wang, Z.; Liang, L.; Li, Y.; Liu, D.; Lu, Q. Distinct assembly processes of intestinal and non-intestinal microbes of bark beetles from clues of metagenomic insights. Sci. Rep. 2025, 15, 7910. [Google Scholar] [CrossRef]
- Niederdorfer, R.; Fragner, L.; Yuan, L.; Hausherr, D.; Wei, J.; Magyar, P.; Joss, A.; Lehmann, M.F.; Ju, F.; Bürgmann, H. Distinct growth stages controlled by the interplay of deterministic and stochastic processes in functional anammox biofilms. Water Res. 2021, 200, 117225. [Google Scholar] [CrossRef]
- Six, D.L.; Bracewell, R. Dendroctonus. Chapter 8. In Bark Beetles; Academic Press: Cambridge, MA, USA, 2015; pp. 305–350. [Google Scholar]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. biotechn. 2017, 35, 1026–1028. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Res. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, X.B.; Shi, L.Y.; Zhu, W.Y.; Ma, L.; Wang, J.J.; Xu, S.; Yang, R.; Zhang, X.X.; Han, Y.F.; et al. Calculation method for stochastic and deterministic assembly processes of prokaryotic communities. Bioprotocol 2021, 101, e2003400. (In Chinese) [Google Scholar]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef]
- Zhou, F.; Lou, Q.; Wang, B.; Xu, L.; Cheng, C.; Lu, M.; Sun, J. Altered carbohydrates allocation by associated bacteria-fungi interactions in a bark beetle-microbe symbiosis. Sci. Rep. 2016, 6, 20135. [Google Scholar]
- Chakraborty, A.; Ashraf, M.Z.; Modlinger, R.; Synek, J.; Schlyter, F.; Roy, A. Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): Identifying core bacterial assemblage and their ecological relevance. Sci. Rep. 2020, 10, 18572. [Google Scholar] [CrossRef]
- Chakraborty, A.; Modlinger, R.; Ashraf, M.Z.; Synek, J.; Schlyter, F.; Roy, A. Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 2020, 11, 568853. [Google Scholar] [CrossRef]
- Vega, F.E.; Biedermann, P.H.W. On interactions, associations, mycetangia, mutualists and symbiotes in insect-fungus symbioses. Fungal Ecol. 2020, 44, 100909. [Google Scholar] [CrossRef]
- Baños-Quintana, A.P.; Gershenzon, J.; Kaltenpoth, M. The Eurasian spruce bark beetle Ips typographus shapes the microbial communities of its offspring and the gallery environment. Front. Microbiol. 2024, 15, 1367127. [Google Scholar] [CrossRef]
- Paine, T.D.; Raffa, K.F.; Harrington, T.C. Interactions among scolytid bark beetles, their associated fungi, and live host conifers. Annu. Rev. Entomol. 1997, 42, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Fossdal, C.G.; Nagy, N.E.; Johnsen, O.; Dalen, L.S. Local and systemic stress responses in Norway spruce: Similarities in gene expression between a compatible pathogen interaction and drought stress. Physiol. Mol. Plant P. 2007, 70, 161–173. [Google Scholar] [CrossRef]
- Vazquez-Ortiz, K.; Pineda-Mendoza, R.M.; González-Escobedo, R.; Davis, T.S.; Salazar, K.F.; Rivera-Orduña, F.N.; Zúñiga, G. Metabarcoding of mycetangia from the Dendroctonus frontalis species complex (Curculionidae: Scolytinae) reveals diverse and functionally redundant fungal assemblages. Front. Microbiol. 2022, 13, 969230. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Kim, J.J.; Lu, M.; Breuil, C. Determining fungal diversity on Dendroctonus ponderosae and Ips pini affecting lodgepole pine using cultural and molecular methods. Fungal Divers. 2006, 19, 79–94. [Google Scholar]
- Hernández-García, J.A.; Gonzalez-Escobedo, R.; Briones-Roblero, C.I.; Cano-Ramírez, C.; Rivera-Orduña, F.N.; Zúñiga, G. Gut bacterial communities of Dendroctonus valens and D. mexicanus (Curculionidae: Scolytinae): A metagenomic analysis across different geographical locations in Mexico. Int. J. Mol. Sci. 2018, 19, 2578. [Google Scholar] [CrossRef]
- Xu, L.; Shi, Z.; Wang, B.; Lu, M.; Sun, J. Pine defensive monoterpene α-Pinene influences the feeding behavior of Dendroctonus valens and its gut bacterial community structure. Int. J. Mol. Sci. 2016, 17, 1734. [Google Scholar] [CrossRef]
- Li, S.H.; Nagy, N.E.; Hammerbacher, A.; Krokene, P.; Niu, X.M.; Gershenzon, J.; Schneider, B. Localization of phenolics in phloem parenchyma cells of Norway spruce (Picea abies). ChemBioChem 2012, 13, 2707–2713. [Google Scholar] [CrossRef]
- Briones-Roblero, C.I.; Rodríguez-Díaz, R.; Santiago-Cruz, J.A.; Zúñiga, G.; Rivera-Orduña, F.N. Degradation capacities of bacteria and yeasts isolated from the gut of Dendroctonus rhizophagus (Curculionidae: Scolytinae). Folia Microbiol. 2017, 62, 1–9. [Google Scholar] [CrossRef]
- Adams, A.S.; Boone, C.K.; Bohlmann, J.; Raffa, K.F. Responses of bark beetle-associated bacteria to host monoterpenes and their relationship to insect life histories. J. Chem. Ecol. 2011, 37, 808–817. [Google Scholar] [CrossRef]
- Bracewell, R.R.; Six, D.L. Experimental evidence of bark beetle adaptation to a fungal symbiont. Ecol. Evol. 2015, 5, 5109–5119. [Google Scholar] [CrossRef]
- Guevara-Rozo, S.; Hussain, A.; Cale, J.A.; Klutsch, J.G.; Rajabzadeh, R.; Erbilgin, N. Nitrogen and ergosterol concentrations varied in live jack pine phloem following inoculations with fungal associates of mountain pine beetle. Front. Microbiol. 2020, 11, 1703. [Google Scholar] [CrossRef]
- Scott, J.J.; Oh, D.C.; Yuceer, M.C.; Klepzig, K.D.; Clardy, J.; Currie, C.R. Bacterial protection of beetle-fungus mutualism. Science 2008, 322, 63. [Google Scholar] [CrossRef]
- Kandasamy, D.; Gershenzon, J.; Andersson, M.N.; Hammerbacher, A. Volatile organic compounds influence the interaction of the Eurasian spruce bark beetle (Ips typographus) with its fungal symbionts. ISME J. 2019, 13, 1788–1800. [Google Scholar] [CrossRef]
- Ceja-Navarro, J.A.; Vega, F.E.; Karaoz, U.; Hao, Z.; Jenkins, S.; Lim, H.C.; Kosina, P.; Infante, F.; Northen, T.R.; Brodie, E.L. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat. Commun. 2015, 6, 7618. [Google Scholar] [CrossRef]
- Ge, Y.; Jing, Z.; Diao, Q.; He, J.Z.; Liu, Y.J. Host species and geography differentiate honeybee gut bacterial communities by changing the relative contribution of community assembly processes. mBio 2021, 12, e0075-21. [Google Scholar] [CrossRef]

| Sample Type | Sample ID | Host Tree | Latitude Longitude | Elevation/m | Location |
|---|---|---|---|---|---|
| Gut | DAC1 | Pinus armandii | 33.6891° N, 107.8966° E | 1701.70 ± 4.62 | Foping, Hanzhong City in Shanxi Province, China |
| DAC2 | |||||
| DAC3 | |||||
| Cuticle | DA1 | ||||
| DA2 | |||||
| DA3 |
| Sample Name | Bioproject ID | Biosample ID | Sequence Read Archive (SRA) |
|---|---|---|---|
| DAC1 | PRJNA1251992 | SAMN48028048 | SRR33208850 |
| DAC2 | SAMN48028049 | SRR33208849 | |
| DAC3 | SAMN48028050 | SRR33208848 | |
| DA1 | SAMN48028045 | SRR33208853 | |
| DA2 | SAMN48028046 | SRR33208852 | |
| DA3 | SAMN48028047 | SRR33208851 |
| Sample Type | Fungi Unclassified | Erysiphe | Rhizophagus | Enterospora | Rhizopus | Ophiostomatoid Fungi |
|---|---|---|---|---|---|---|
| Gut | 2.50 | 0.62 | 6.15 | 0.00 | −0.10 | NA |
| Cuticle | 0.00 | −0.47 | 1.54 | 0.00 | −0.19 | 0.11 |
| Sample Type | Bacteria Unclassified | Thiobacillus | Enterobacter | Escherichia | Wolbachia |
|---|---|---|---|---|---|
| Gut | 0.70 | NA | 16.14 | NA | 9.86 |
| Cuticle | −0.31 | 0.00 | 0.21 | 2.05 | −0.21 |
| Sample Type | Prevotella | Mircobacterium | Bacteroides | Clostridium | Ruminococcus |
| Gut | 0.00 | NA | NA | NA | 0.00 |
| Cuticle | 0.54 | 0.11 | 0.62 | 0.52 | 0.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liang, L.; Wang, H.; Wang, Z.; Lu, Q. Specific Function and Assembly of Crucial Microbes for Dendroctonus armandi Tsai et Li. Forests 2025, 16, 1584. https://doi.org/10.3390/f16101584
Liu C, Liang L, Wang H, Wang Z, Lu Q. Specific Function and Assembly of Crucial Microbes for Dendroctonus armandi Tsai et Li. Forests. 2025; 16(10):1584. https://doi.org/10.3390/f16101584
Chicago/Turabian StyleLiu, Caixia, Lingyu Liang, Huimin Wang, Zheng Wang, and Quan Lu. 2025. "Specific Function and Assembly of Crucial Microbes for Dendroctonus armandi Tsai et Li" Forests 16, no. 10: 1584. https://doi.org/10.3390/f16101584
APA StyleLiu, C., Liang, L., Wang, H., Wang, Z., & Lu, Q. (2025). Specific Function and Assembly of Crucial Microbes for Dendroctonus armandi Tsai et Li. Forests, 16(10), 1584. https://doi.org/10.3390/f16101584






