Transcriptome Analysis Reveals the Genetic Basis of Phenotypic Traits of Vaccinium uliginosum L. at Different Elevations in the Changbai Mountains
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Morphological Traits of V. uliginosum at Different Elevations
2.3. RNA Extraction, cDNA Library Construction, and Sequencing
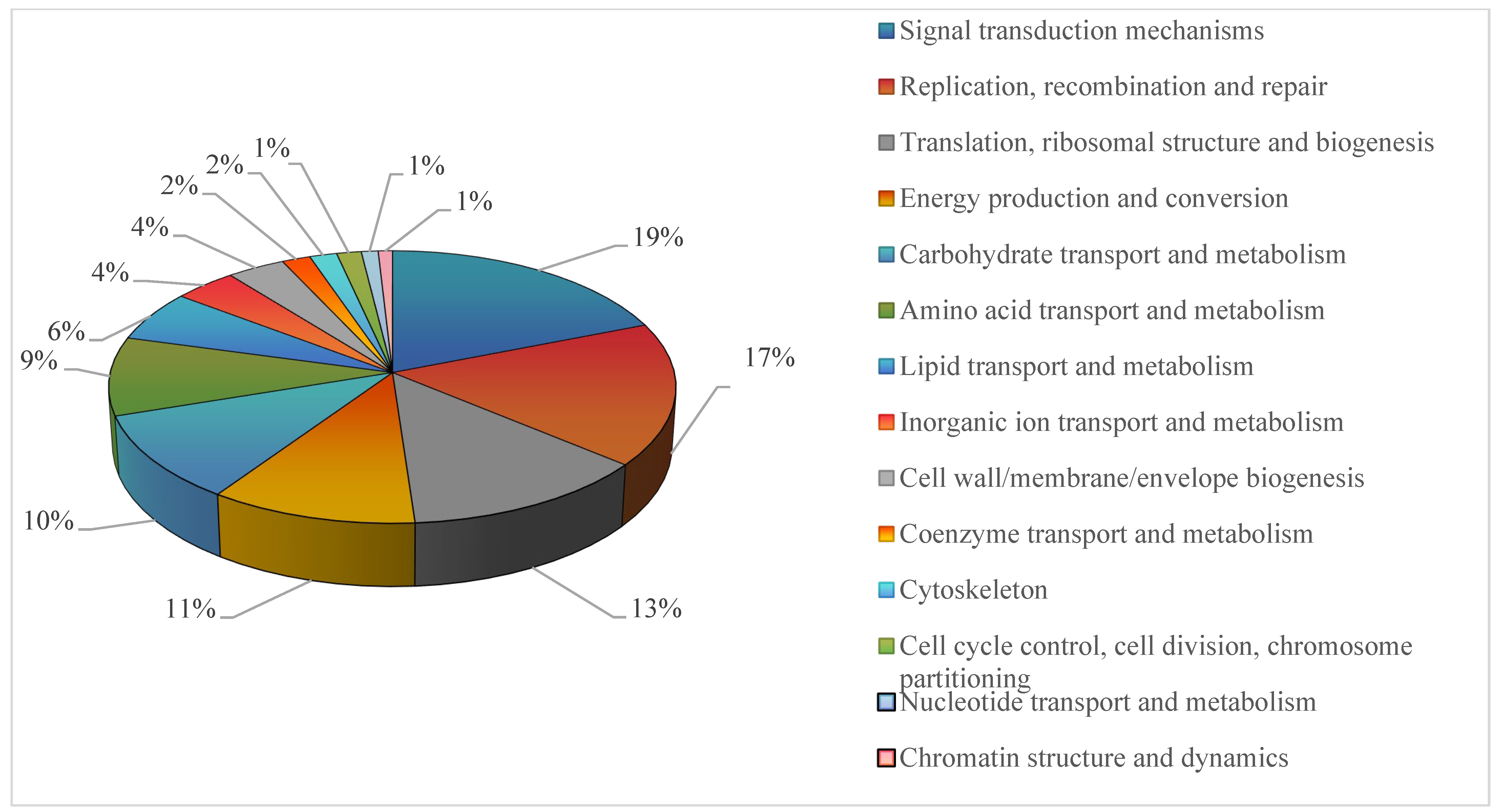
2.4. Transcriptomic Assembly and Functional Annotation of Genes
2.5. Analysis of Differentially Expressed Genes (DEGs)
2.6. Data Analysis
2.7. Real-Time Quantitative PCR Validation
3. Results
3.1. Evaluation of Effects of Elevation on Morphological Traits in Populations of V. uliginosum Individuals Sampled
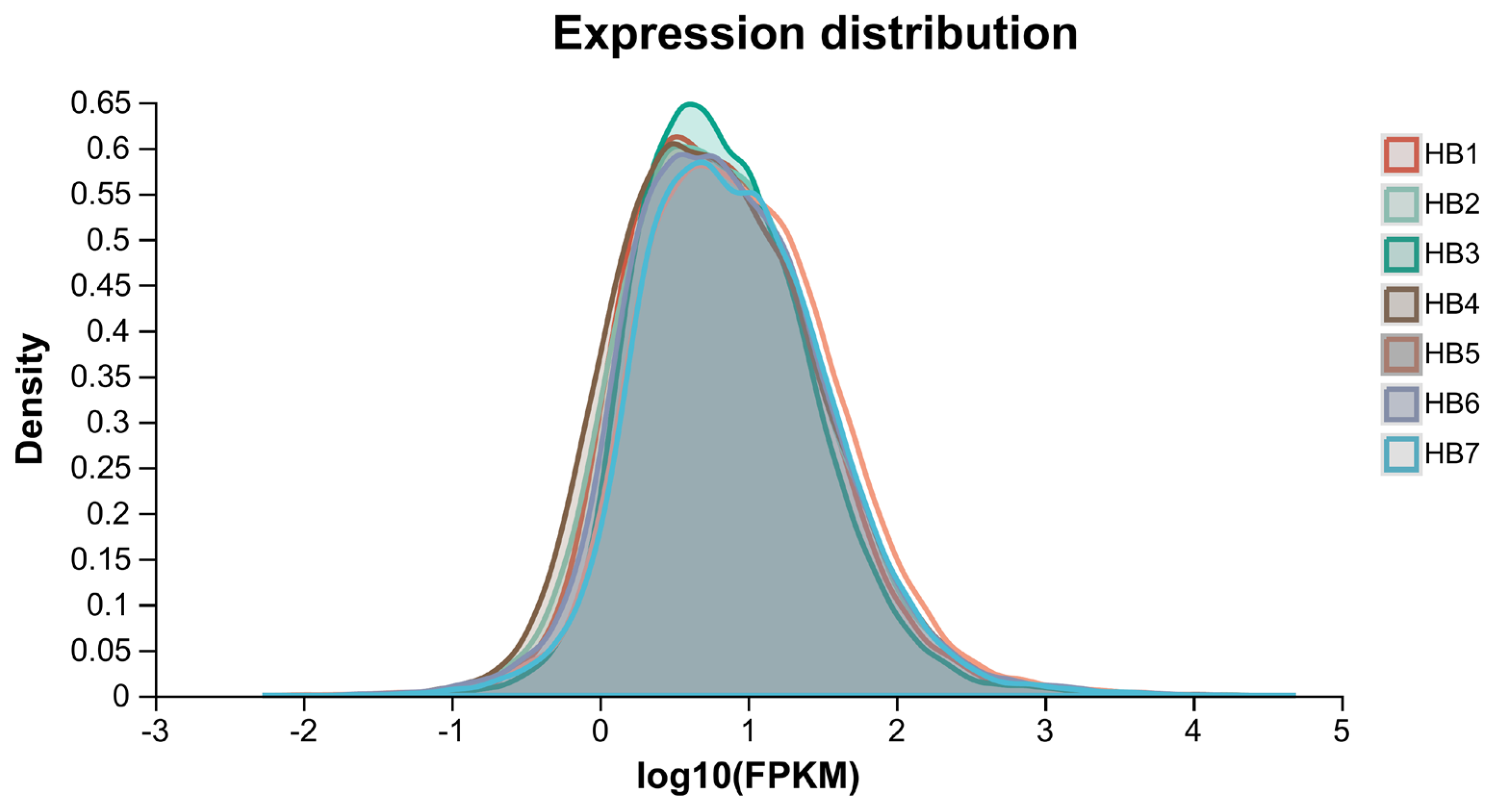
3.2. Overview of Transcriptome Data
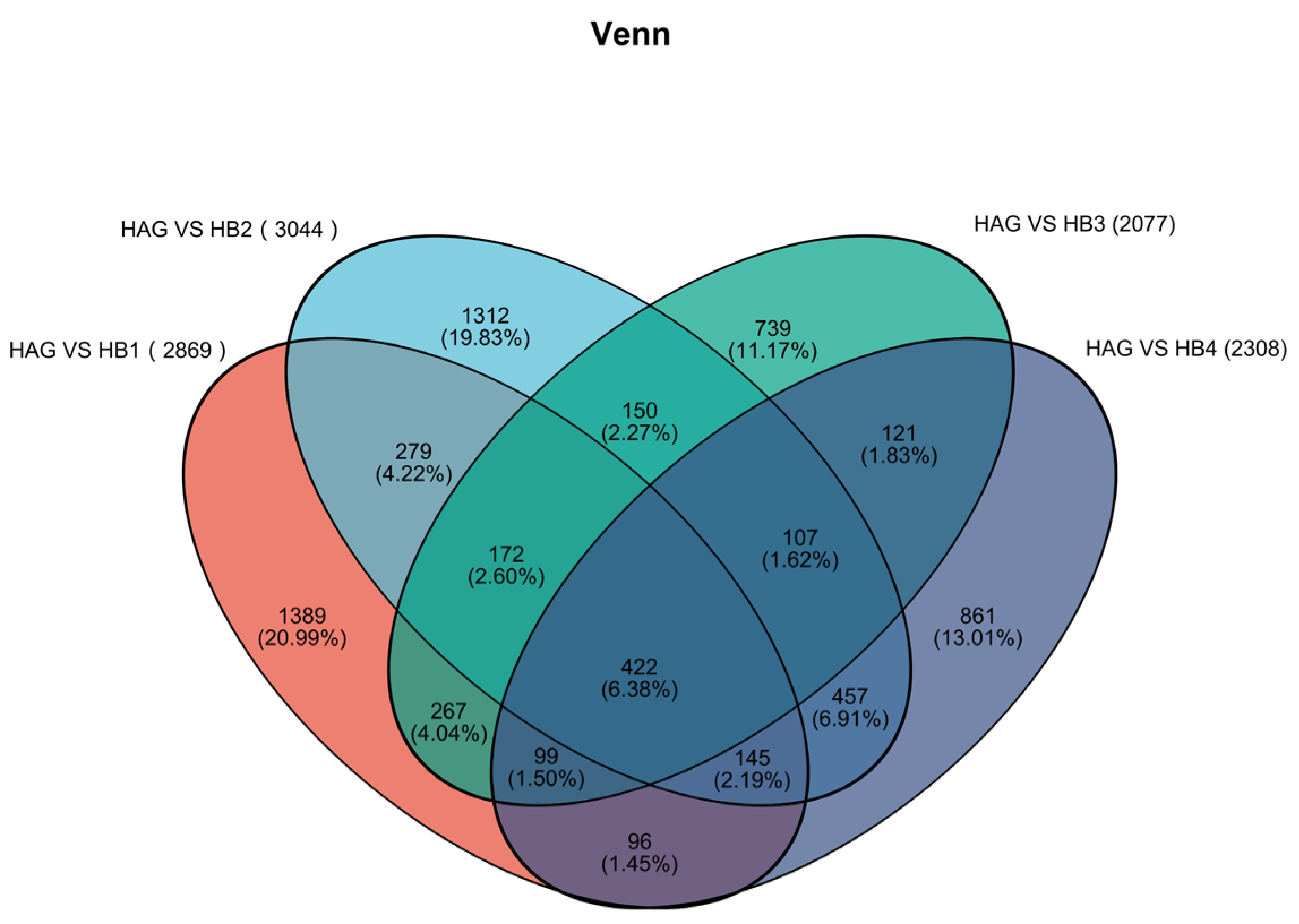
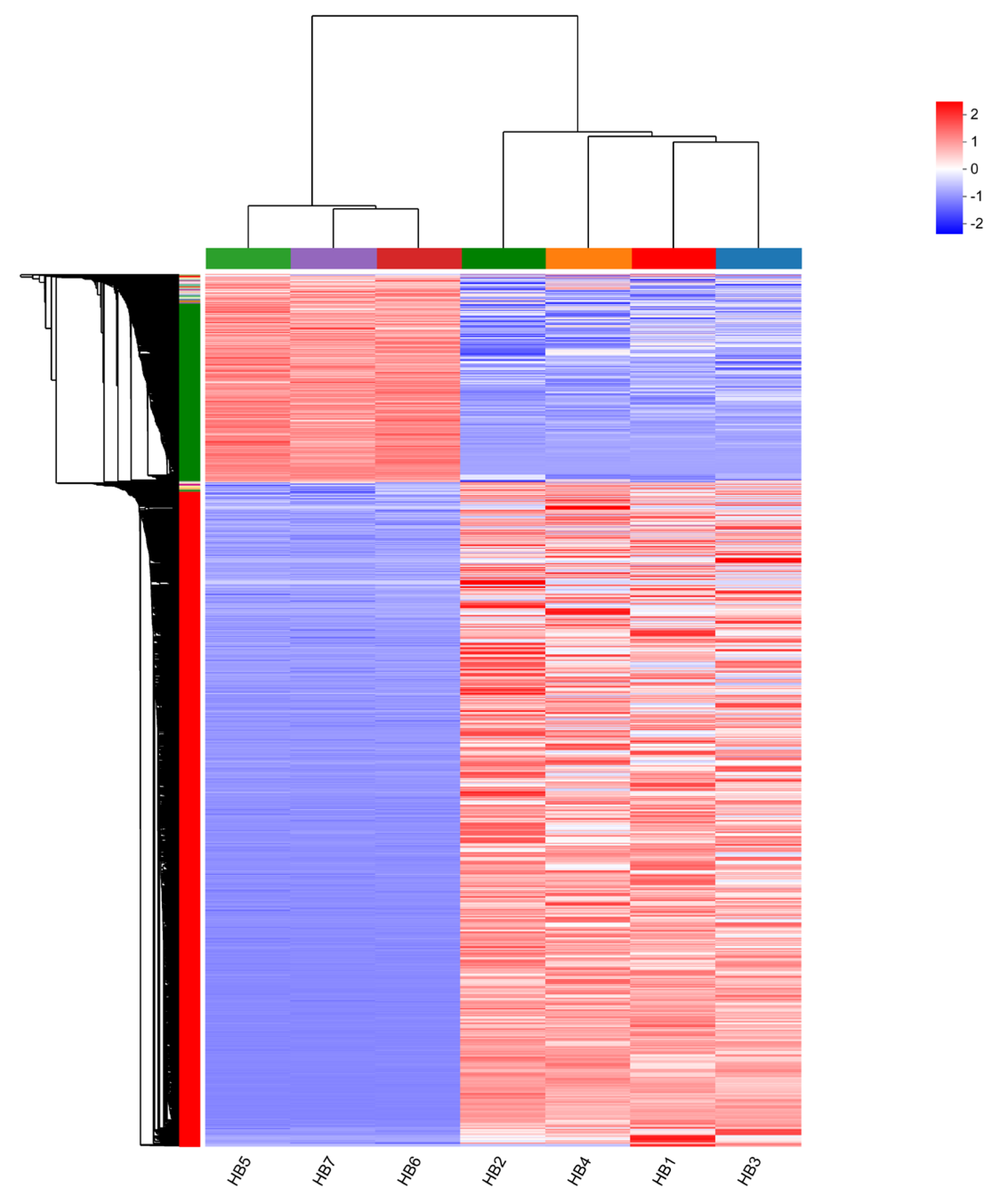
3.3. Analysis of Differentially Expressed Genes (DEGs)
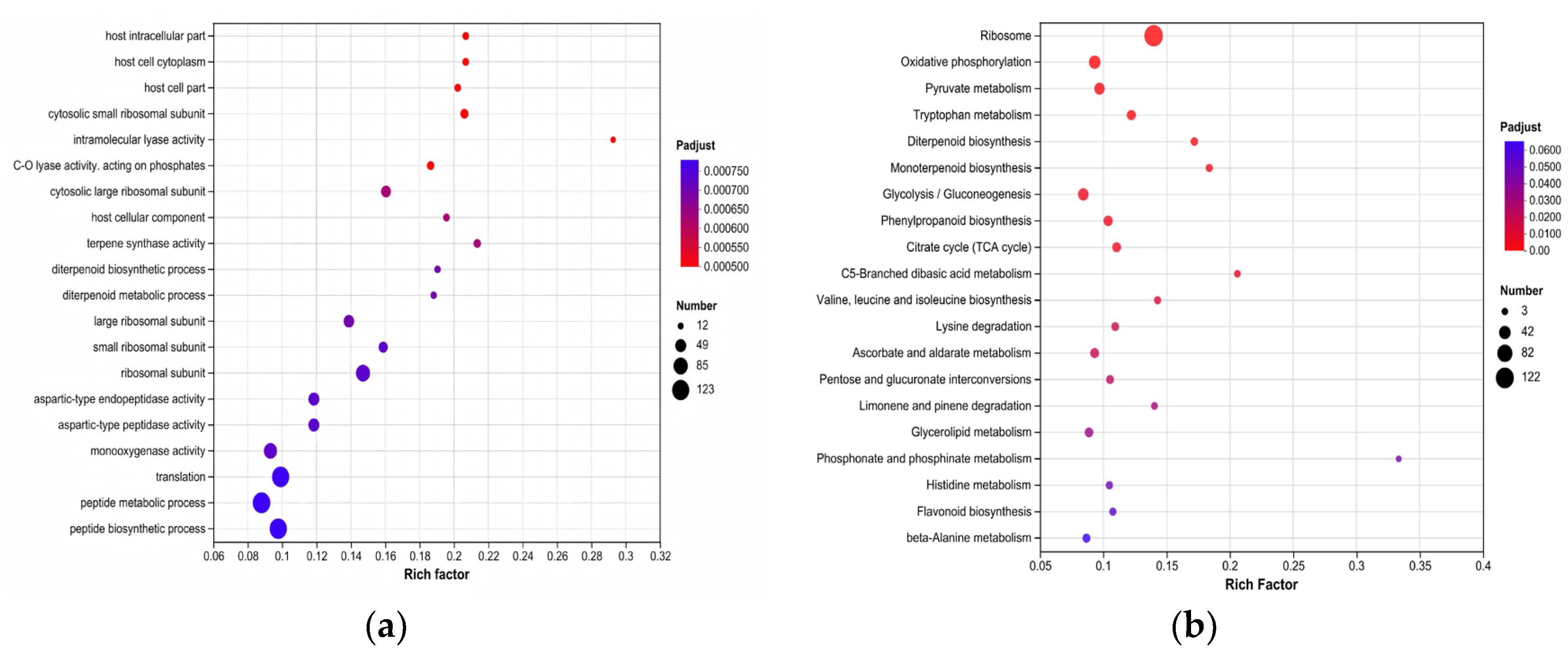
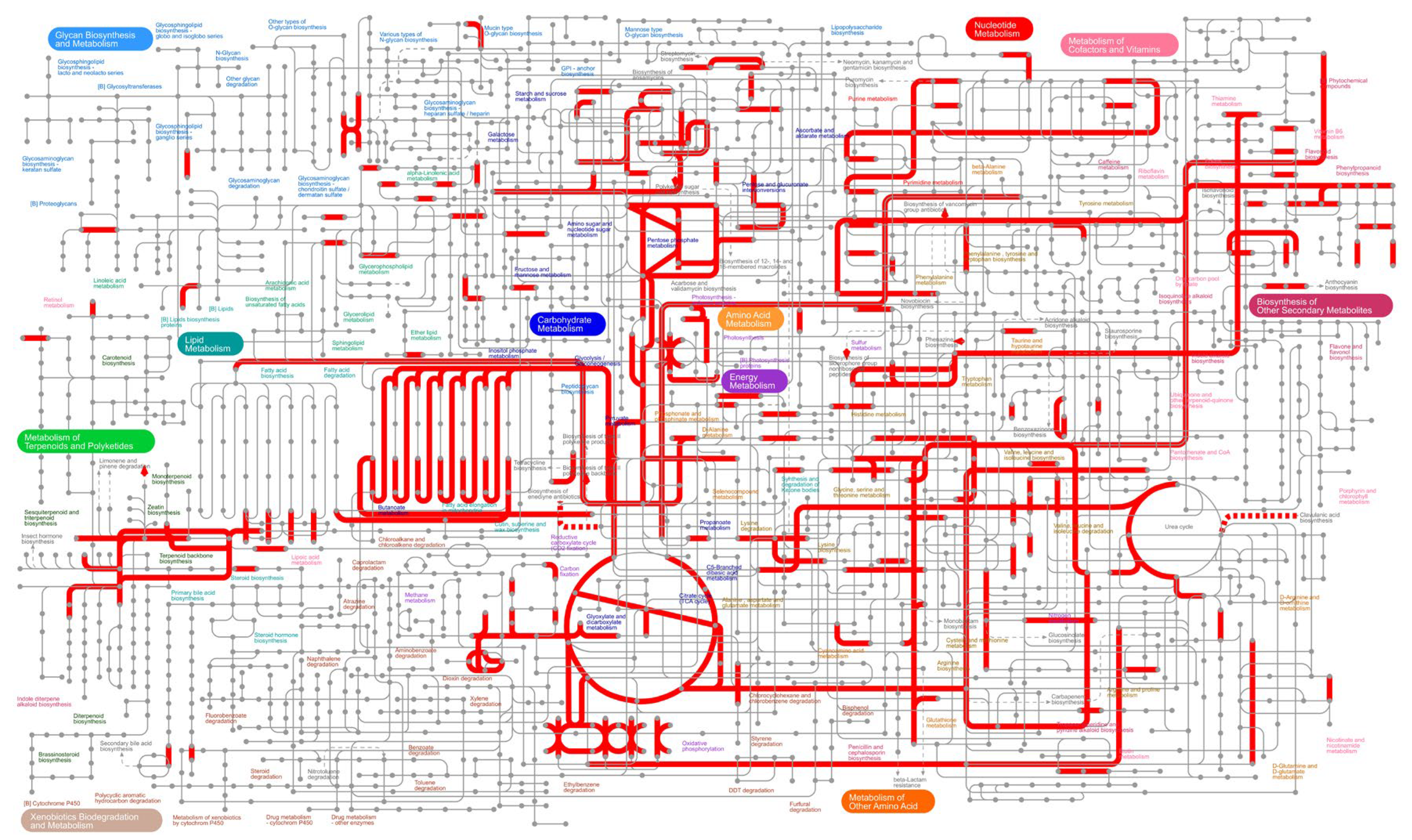
3.4. GO and KEGG Enrichment Analysis of Differentially Expressed Genes (DEGs)
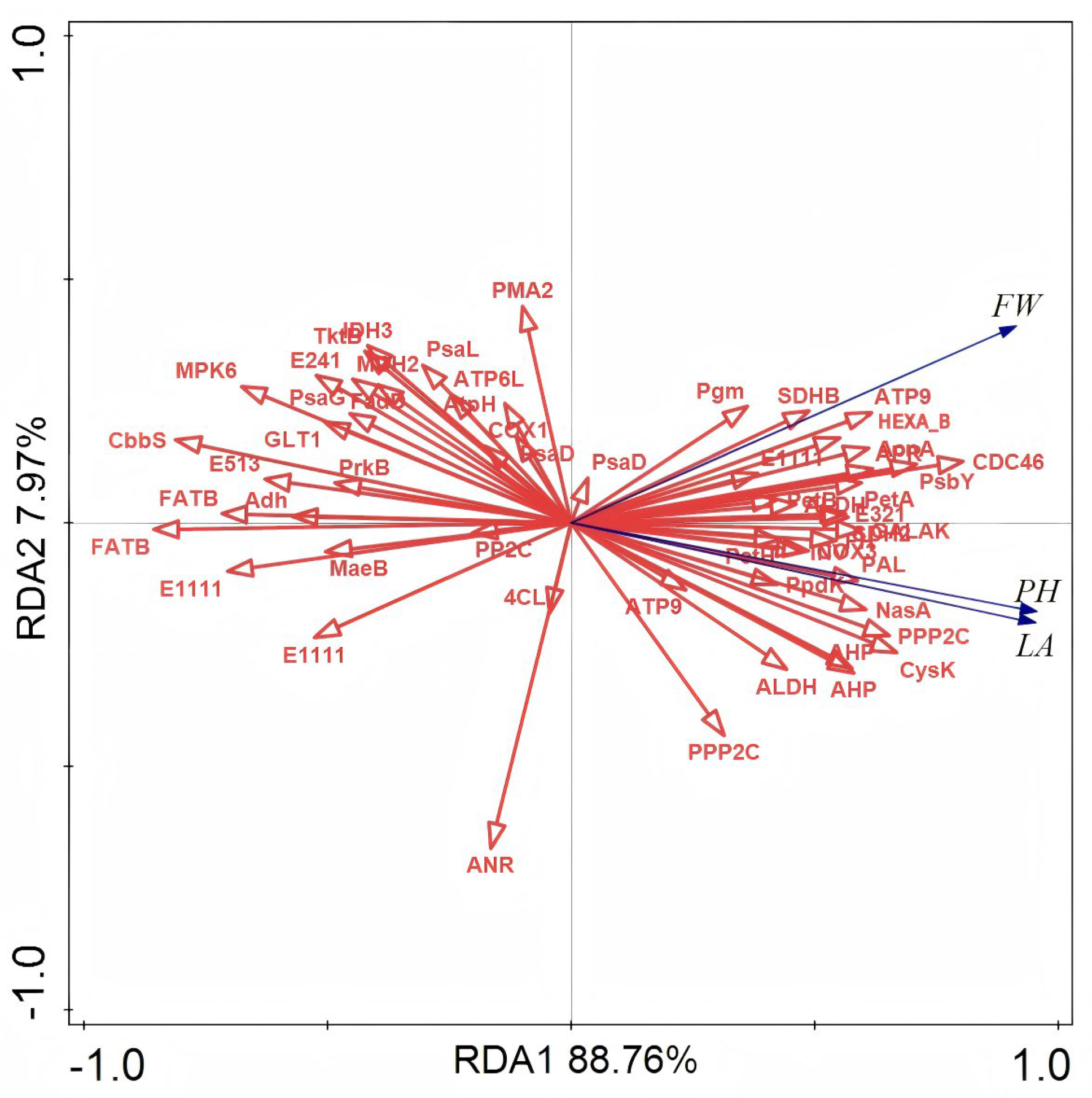
3.5. Screening of Differentially Expressed Genes (DEGs) Correlated with Morphological Features
3.6. Enrichment of DEGs Associated with Morphological Traits
3.7. Analysis of DEGs Associated with Photosynthesis
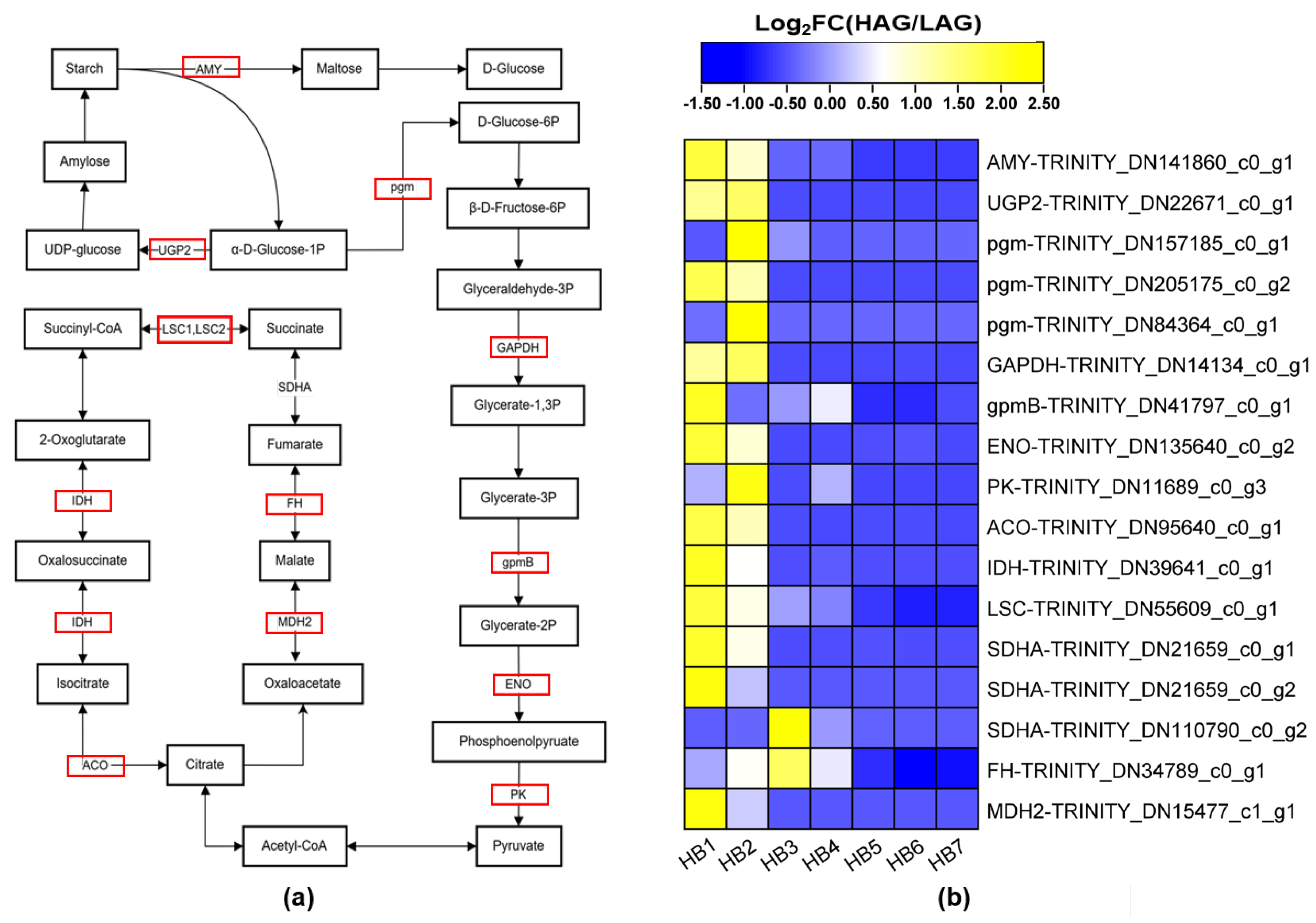
3.8. Analysis of DEGs Associated with Carbohydrate Metabolism
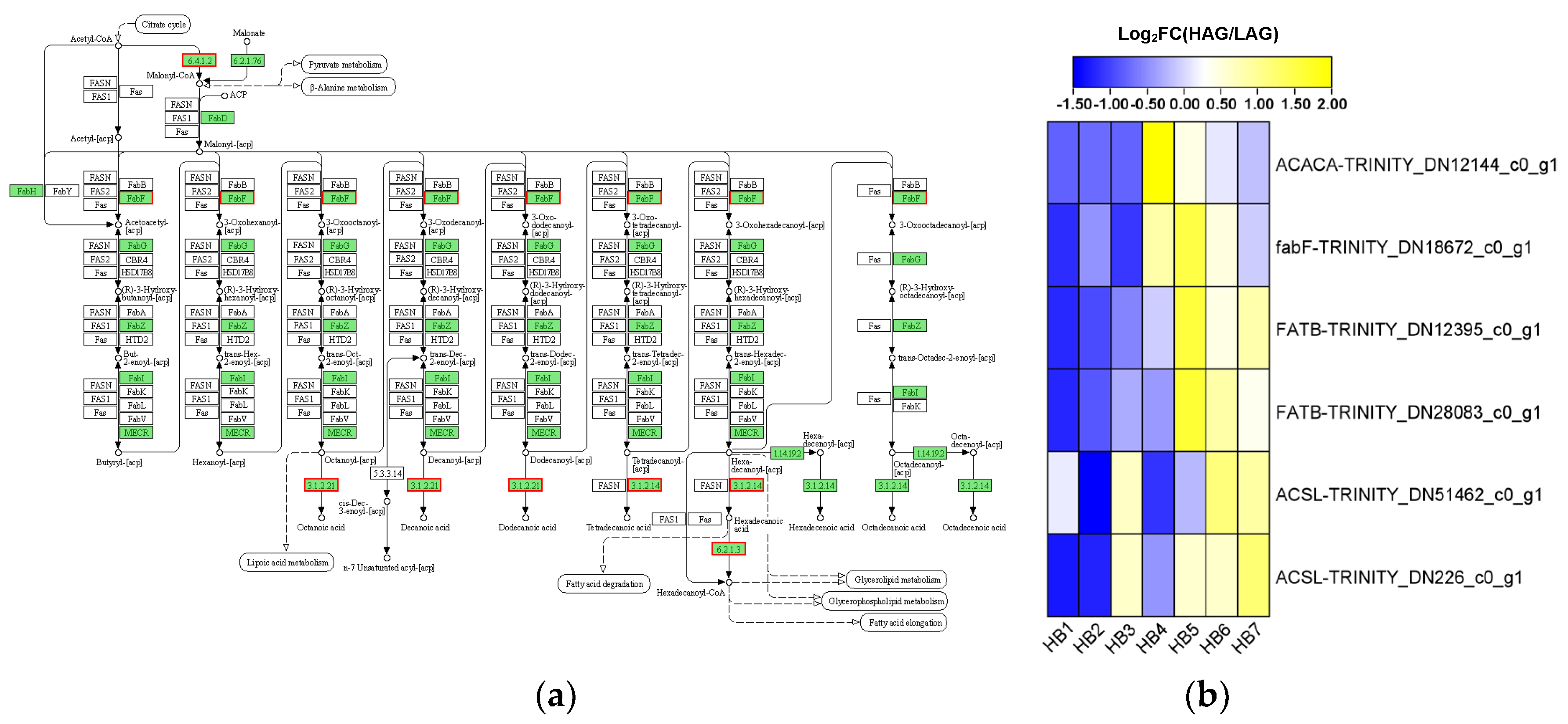
3.9. Analysis of DEGs Associated with Lipid Metabolism
3.10. Analysis of DEGs Related to Plant Hormone Signal Transduction
3.11. Validation of RNA-Seq with qRT-PCR
4. Discussion
4.1. Morphological Adaptations of Plants at High Elevations
4.2. The Gene Expression and Metabolic Processes in Plants Are Altered Under High-Altitude Conditions
4.3. The Morphogenesis Mechanism of the Adaptation of V. uliginosum to High-Altitude Environments
4.3.1. Lipid Metabolism Pathway
4.3.2. Carbohydrate Metabolism Pathway
4.3.3. Photosynthesis Pathway
4.3.4. The Modulation of Phytohormones in High-Altitude Environments
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RNA-Seq | Transcriptome sequencing technology |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| log2FC | Log2-fold change |
| FATB | Fatty acyl-ACP thioesterase B |
| cbbS | Ribulose-bisphosphate carboxylase small subunit |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| RDA | Redundancy analysis |
| LAG | Low-elevation samples |
| HAG | High-elevation samples |
| HEXA_B | Hexosaminidase gene |
| ANR | Anthocyanidin reductase gene |
| PPP2C | Serine/threonine-protein phosphatase 2A catalytic subunit |
| SDHA | Succinate dehydrogenase (ubiquinone) flavoprotein subunit gene |
| UGP2 | UTP-glucose-1-phosphate uridylyltransferase gene |
| AMY | Alpha-amylase gene |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase gene |
| gpmB | 2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase gene |
| FH | Fumarate hydratase gene |
| ENO | Enolase 1/2/3 gene |
| PK | Pyruvate kinase gene |
| ACO | Aconitate hydratase gene |
| LSC | Succinyl-CoA synthetase alpha subunit gene |
| ACSL | Long-chain acyl-CoA synthetase gene |
| fabF | 3-Oxoacyl-[acyl-carrier-protein] gene |
| ACACA | Acetyl-CoA carboxylase/biotin carboxylase 1 gene |
| AUX/IAA | Auxin/Indole-3-Acetic Acid gene |
| GLS | Glucosinolate |
| GA | Gibberellin |
| ABA | Abscisic acid |
| BAK1 | Brassinosteroid-insensitive 1 (BRI1)-Associated Kinase 1 |
| BRs | Brassinosteroids |
| XTH | Xyloglucan endotransglucosylase/hydrolase |
References
- Yang, X.; Xu, M. Biodiversity conservation in Changbai Mountain Biosphere Reserve, northeastern China: Status, problem, and strategy. Biodivers. Conserv. 2003, 12, 883–903. [Google Scholar] [CrossRef]
- He, H.S.; Hao, Z.; Mladenoff, D.J.; Shao, G.; Hu, Y.; Chang, Y. Simulating forest ecosystem response to climate warming incorporating spatial effects in north-eastern China. J. Biogeogr. 2005, 32, 2043–2056. [Google Scholar] [CrossRef]
- Abbott, R.J.; Brennan, A.C. Altitudinal gradients, plant hybrid zones and evolutionary novelty. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130346. [Google Scholar] [CrossRef]
- Wei, J.; Wu, G.; Deng, H. Vegetation biomass distribution characteristics of alpine tundra ecosystem in Changbai Mountains. Ying Yong Sheng Tai Xue Bao 2004, 15, 1999–2004. [Google Scholar]
- Wang, L.; Wang, W.J.; Wu, Z.; Du, H.; Zong, S.; Ma, S. Potential distribution shifts of plant species under climate change in Changbai Mountains, China. Forests 2019, 10, 498. [Google Scholar] [CrossRef]
- Fan, C.; Guo, Z.; Zheng, J. Study on Morphological Traits of Natural Populations of Vaccinium uliginosum at Different Altitudinal Gradients on Changbai Mountain. Horticulturae 2024, 10, 224. [Google Scholar] [CrossRef]
- Zheng, D.; Liu, Q. Effects of environmental factors on forest community distribution in Changbai Mountain Nature Reserve of northeastern China. J. Beijing For. Univ. 2023, 45, 57–64. [Google Scholar] [CrossRef]
- Wheeler, J.A.; Hoch, G.; Cortes, A.J.; Sedlacek, J.; Wipf, S.; Rixen, C. Increased spring freezing vulnerability for alpine shrubs under early snowmelt. Oecologia 2014, 175, 219–229. [Google Scholar] [CrossRef]
- Rathore, N.; Kumar, P.; Mehta, N.; Swarnkar, M.K.; Shankar, R.; Chawla, A. Time-series RNA-Seq transcriptome profiling reveals novel insights about cold acclimation and de-acclimation processes in an evergreen shrub of high altitude. Sci. Rep. 2022, 12, 15553. [Google Scholar] [CrossRef]
- Nong, M.L.; Luo, X.H.; Zhu, L.X.; Zhang, Y.N.; Dun, X.Y.; Huang, L. Insights into the Adaptation to High Altitudes from Transcriptome Profiling: A Case Study of an Endangered Species, Kingdonia uniflora. Genes 2023, 14, 1291. [Google Scholar] [CrossRef]
- Jinqiu, Y.; Bing, L.; Tingting, S.; Jinglei, H.; Zelai, K.; Lu, L.; Wenhua, H.; Tao, H.; Xinyu, H.; Zengqing, L.; et al. Integrated Physiological and Transcriptomic Analyses Responses to Altitude Stress in Oat (Avena sativa L.). Front. Genet. 2021, 12, 638683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.L.; Leng, Y.N.; Hao, R.R.; Zhang, W.Y.; Li, H.F.; Chen, M.X.; Zhu, F.Y. Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques. Int. J. Mol. Sci. 2024, 25, 12666. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Yang, J.; He, H.; Huangfu, S.; Wang, J.; Li, H.; Zhang, B.; Wang, X.; Zhang, X.; et al. Reprogramming of Metabolome and Transcriptome Shaped the Elevational Adaptation of Quercus variabilis by Regulating Leaf Functional Traits. Plant Cell Environ. 2025, 48, 6189–6208. [Google Scholar] [CrossRef]
- Albert, K.R.; Mikkelsen, T.N.; Ro-Poulsen, H. Ambient UV-B radiation decreases photosynthesis in high arctic Vaccinium uliginosum. Physiol. Plant 2008, 133, 199–210. [Google Scholar] [CrossRef]
- Boesgaard, K.S.; Albert, K.R.; Ro-Poulsen, H.; Michelsen, A.; Mikkelsen, T.N.; Schmidt, N.M. Long-term structural canopy changes sustain net photosynthesis per ground area in high arctic Vaccinium uliginosum exposed to changes in near-ambient UV-B levels. Physiol. Plant 2012, 145, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Li, H.; Meng, J.; Zhnag, Y.; Zhang, B. Study on cloning of VuLhca4 gene from Vaccinium uliginosum L. and functional analysis under abiotic stress. J. Northeast Agric. Univ. 2023, 54, 34–44. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, F. Cloning and functional analysis of bog bilberry E3 ligase VuARI2 gene in cold resistance. Acta Bot. Boreali-Occident. Sin. 2024, 44, 1420–1432. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Smith-Unna, R.; Boursnell, C.; Patro, R.; Hibberd, J.M.; Kelly, S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016, 26, 1134–1144. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Klopfenstein, D.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Warwick Vesztrocy, A.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef] [PubMed]
- Petrucelli, M.F.; Martins-Santana, L.; Sanches, P.R.; Oliveira, V.M.; Rossi, A.; Martinez-Rossi, N.M. The Transcription Factor StuA Regulates the Glyoxylate Cycle in the Dermatophyte Trichophyton rubrum under Carbon Starvation. Int. J. Mol. Sci. 2023, 25, 405. [Google Scholar] [CrossRef]
- Deng, Y.; Li, Y.; Sun, H. Selection of reference genes for RT-qPCR normalization in blueberry (Vaccinium corymbosum × angustifolium) under various abiotic stresses. FEBS Open Bio 2020, 10, 1418–1435. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Skinner, J.; Bennett, J.E. Evaluation of reference genes for real-time quantitative PCR studies in Candida glabrata following azole treatment. BMC Mol. Biol. 2012, 13, 22. [Google Scholar] [CrossRef]
- Darzi, Y.; Letunic, I.; Bork, P.; Yamada, T. iPath3.0: Interactive pathways explorer v3. Nucleic Acids Res. 2018, 46, W510–W513. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Chen, J.-Z.; Tian, X.; Zheng, Y.-H.; Hao, J.; Xue, Y.-J.; Ding, S.-Y.; Zong, C.-W. The variation of total flavonoids, anthocyanins and total phenols in Vaccinium uliginosum fruits in Changbai Mountain of China is closely related to spatial distribution. J. Berry Res. 2022, 12, 463–481. [Google Scholar] [CrossRef]
- Yang, H.; Chen, J.; Wang, Y.; Tian, X.; Zheng, Y.; Jin, Z.; Hao, J.; Xue, Y.; Ding, S.; Zong, C. Determination of the genetic diversity and population structure of Vaccinium uliginosum in Northeast China based on the chloroplast mat K gene and EST-SSRseq molecular markers. Genet. Resour. Crop Evol. 2024, 71, 1035–1052. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, Z.; Zhang, C.; Zhang, J.; Ran, A. Ecophysiological responses of three tree species to a high-altitude environment in the southeastern tibetan plateau. Forests 2018, 9, 48. [Google Scholar] [CrossRef]
- Halbritter, A.H.; Fior, S.; Keller, I.; Billeter, R.; Edwards, P.J.; Holderegger, R.; Karrenberg, S.; Pluess, A.R.; Widmer, A.; Alexander, J.M. Trait differentiation and adaptation of plants along elevation gradients. J. Evol. Biol. 2018, 31, 784–800. [Google Scholar] [CrossRef]
- Zhang, X.; Kuang, T.; Dong, W.; Qian, Z.; Zhang, H.; Landis, J.B.; Feng, T.; Li, L.; Sun, Y.; Huang, J. Genomic convergence underlying high-altitude adaptation in alpine plants. J. Integr. Plant Biol. 2023, 65, 1620–1635. [Google Scholar] [CrossRef]
- Rosli, K.A.; Misran, A.; Yazan, L.S.; Wahab, P.E.M. Light-nutrient interaction orchestrates leaf dynamics, nitrogen assimilation, and cellular energetics in Agastache rugosa (Fisch. & CA Mey.) Kuntze. Environ. Exp. Bot. 2024, 229, 106044. [Google Scholar] [CrossRef]
- Kortstee, A.; Appeldoorn, N.; Oortwijn, M.; Visser, R. Differences in regulation of carbohydrate metabolism during early fruit development between domesticated tomato and two wild relatives. Planta 2007, 226, 929–939. [Google Scholar] [CrossRef]
- Zheng, Y.; Sun, J.; Luo, Z.; Li, Y.; Huang, Y. Emerging mechanisms of lipid peroxidation in regulated cell death and its physiological implications. Cell Death Dis. 2024, 15, 859. [Google Scholar] [CrossRef] [PubMed]
- Karamat, U.; Sun, X.; Li, N.; Zhao, J. Genetic regulators of leaf size in Brassica crops. Hortic. Res. 2021, 8, 91. [Google Scholar] [CrossRef]
- Xuan, T.D.; Minh, T.N.; Anh, L.H.; Khanh, T.D. Allelopathic momilactones A and B are implied in rice drought and salinity tolerance, not weed resistance. Agron. Sustain. Dev. 2016, 36, 52. [Google Scholar] [CrossRef]
- Anh, L.H.; Khanh, T.D.; Xuan, T.D. Biological roles of momilactones: Achievements, challenges, and promising approaches to exploit their beneficial properties. Front. Nat. Prod. 2023, 2, 1245869. [Google Scholar] [CrossRef]
- Henschel, J.M.; Andrade, A.N.d.; dos Santos, J.B.L.; da Silva, R.R.; da Mata, D.A.; Souza, T.; Batista, D.S. Lipidomics in Plants Under Abiotic Stress Conditions: An Overview. Agronomy 2024, 14, 1670. [Google Scholar] [CrossRef]
- Okazaki, Y.; Saito, K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014, 79, 584–596. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, L.; Qi, D. Variation in leaf traits at different altitudes reflects the adaptive strategy of plants to environmental changes. Ecol. Evol. 2020, 10, 8166–8175. [Google Scholar] [CrossRef]
- Hu, J.; Li, S.; Zhang, Y.; Du, D.; Zhu, X. Potential Regulatory Effects of Arbuscular Mycorrhizal Fungi on Lipid Metabolism of Maize in Response to Low-Temperature Stress. J. Agric. Food Chem. 2024, 72, 22574–22587. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Xu, Y.; Wang, J.; Singer, S.D.; Chen, G. The role of triacylglycerol in plant stress response. Plants 2020, 9, 472. [Google Scholar] [CrossRef]
- Mashek, D.G.; Li, L.O.; Coleman, R.A. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2007, 2, 465. [Google Scholar] [CrossRef]
- Martínez-Vilalta, J.; Sala, A.; Asensio, D.; Galiano, L.; Hoch, G.; Palacio, S.; Piper, F.I.; Lloret, F. Dynamics of non-structural carbohydrates in terrestrial plants: A global synthesis. Ecol. Monogr. 2016, 86, 495–516. [Google Scholar] [CrossRef]
- Liu, J.-F.; Deng, Y.-P.; Wang, X.-F.; Ni, Y.-Y.; Wang, Q.; Xiao, W.-F.; Lei, J.-P.; Jiang, Z.-P.; Li, M.-H. The concentration of non-structural carbohydrates, N, and P in Quercus variabilis does not decline toward its northernmost distribution range along a 1500 km transect in China. Front. Plant Sci. 2018, 9, 1444. [Google Scholar] [CrossRef] [PubMed]
- Atkin, O.; Millar, A.; Gardeström, P.; Day, D. Photosynthesis, carbohydrate metabolism and respiration in leaves of higher plants. Photosynth. Physiol. Metab. 2000, 9, 153–175. [Google Scholar] [CrossRef]
- Ma, W.; Wei, X.; Su, M.; Luo, Q.; Zhao, Y. Responses of non-structural carbohydrate metabolism of Medicago sativa seedlings to nitric oxide under drought stress. Acta Ecol. Sin. 2019, 39, 8068–8077. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, H.P.; Batish, D.R.; Kaur, S.; Kohli, R.K. EMF radiations (1800 MHz)-inhibited early seedling growth of maize (Zea mays) involves alterations in starch and sucrose metabolism. Protoplasma 2016, 253, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Zhai, B.; Hu, Z.; Sun, S.; Tang, Z.; Wang, G. Characteristics of photosynthetic rates in different vegetation types at high-altitude in mountainous regions. Sci. Total Environ. 2024, 907, 168071. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; JI, Y.; Liu, Y.; Wu, X.; Chen, F.; Liu, X. Study on the adaptive mechanisms of five plants to high-altitude light based on transcriptome sequencing in Maidica wetland of Tibet. Plant Sci. J. 2021, 39, 632–642. [Google Scholar] [CrossRef]
- Didaran, F.; Kordrostami, M.; Ghasemi-Soloklui, A.A.; Pashkovskiy, P.; Kreslavski, V.; Kuznetsov, V.; Allakhverdiev, S.I. The mechanisms of photoinhibition and repair in plants under high light conditions and interplay with abiotic stressors. J. Photochem. Photobiol. B Biol. 2024, 259, 113004. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Divi, U.K.; Rahman, T.; Krishna, P. Brassinosteroid-mediated stress tolerance in Arabidopsis shows interactions with abscisic acid, ethylene and salicylic acid pathways. BMC Plant Biol. 2010, 10, 151. [Google Scholar] [CrossRef]
- Ghorbel, M.; Brini, F. Hormone mediated cell signaling in plants under changing environmental stress. Plant Gene 2021, 28, 100335. [Google Scholar] [CrossRef]
- Yu, Z.; Duan, X.; Luo, L.; Dai, S.; Ding, Z.; Xia, G. How plant hormones mediate salt stress responses. Trends Plant Sci. 2020, 25, 1117–1130. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Krishna, P. Brassinosteroid-mediated stress responses. J. Plant Growth Regul. 2003, 22, 289–297. [Google Scholar] [CrossRef]
- Popko, J.; Hänsch, R.; Mendel, R.R.; Polle, A.; Teichmann, T. The role of abscisic acid and auxin in the response of poplar to abiotic stress. Plant Biol. 2010, 12, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 2005, 17, 1979–1993. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Sun, T.-p.; Gubler, F. Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 2004, 55, 197–223. [Google Scholar] [CrossRef]
- Du, R.; Niu, S.; Liu, Y.; Sun, X.; Porth, I.; El-Kassaby, Y.A.; Li, W. The gibberellin GID1-DELLA signalling module exists in evolutionarily ancient conifers. Sci. Rep. 2017, 7, 16637. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Qi, K.; Gao, X.; Guo, L.; Cao, P.; Li, Q.; Qiao, X.; Gu, C.; Zhang, S. Genome-wide identification and comparative analysis of the PYL gene family in eight Rosaceae species and expression analysis of seeds germination in pear. BMC Genom. 2022, 23, 233. [Google Scholar] [CrossRef]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Jiang, Z.; Kim, J.-M.; To, T.K.; Li, W.; Zhang, X. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef]
- Zhang, C.; He, M.; Jiang, Z.; Liu, L.; Pu, J.; Zhang, W.; Wang, S.; Xu, F. The xyloglucan endotransglucosylase/hydrolase gene XTH22/TCH4 regulates plant growth by disrupting the cell wall homeostasis in Arabidopsis under boron deficiency. Int. J. Mol. Sci. 2022, 23, 1250. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Nucleic Acids Research. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2024. Nucleic Acids Res. 2024, 52, D18–D32. [Google Scholar] [CrossRef] [PubMed]




| Locality | Elevation (m) | Longitude | Latitude |
|---|---|---|---|
| Xinglong Forest Farm, Block 24, Subplot 13 | 706 | 128°18′39.19″ | 42°25′5.35″ |
| Xinglong Forest Farm, Block 91, Subplot 9 | 939 | 128°21′8.06″ | 42°18′0.56″ |
| Dongfanghong Forest Farm, Block 80, Subplot 11 | 1226 | 128°23′12.11″ | 42°6′59.27″ |
| Dongfanghong Forest Farm, Block 112, Subplot 15 | 1384 | 128°17′34.67″ | 42°3′24.39″ |
| Changbai Mountain Nature Reserve | 1985 | 128°4′3.69″ | 42°3′27.06″ |
| Changbai Mountain Nature Reserve | 2190 | 128°3′59.72″ | 42°2′44.66″ |
| Changbai Mountain Nature Reserve | 2366 | 128°4′3.05″ | 42°2′14.72″ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, J.; Zhao, L.; Mu, K.; Wang, R.; Zhang, Q. Transcriptome Analysis Reveals the Genetic Basis of Phenotypic Traits of Vaccinium uliginosum L. at Different Elevations in the Changbai Mountains. Forests 2025, 16, 1571. https://doi.org/10.3390/f16101571
Wang Y, Li J, Zhao L, Mu K, Wang R, Zhang Q. Transcriptome Analysis Reveals the Genetic Basis of Phenotypic Traits of Vaccinium uliginosum L. at Different Elevations in the Changbai Mountains. Forests. 2025; 16(10):1571. https://doi.org/10.3390/f16101571
Chicago/Turabian StyleWang, Yue, Jun Li, Luying Zhao, Kai Mu, Ruijian Wang, and Qichang Zhang. 2025. "Transcriptome Analysis Reveals the Genetic Basis of Phenotypic Traits of Vaccinium uliginosum L. at Different Elevations in the Changbai Mountains" Forests 16, no. 10: 1571. https://doi.org/10.3390/f16101571
APA StyleWang, Y., Li, J., Zhao, L., Mu, K., Wang, R., & Zhang, Q. (2025). Transcriptome Analysis Reveals the Genetic Basis of Phenotypic Traits of Vaccinium uliginosum L. at Different Elevations in the Changbai Mountains. Forests, 16(10), 1571. https://doi.org/10.3390/f16101571






