1. Introduction
Vitamin B6 (VB6) is an essential water-soluble micronutrient required by all living organisms [
1]. In plants, its active form, pyridoxal 5′-phosphate (PLP) [
2], functions as a ubiquitous enzymatic cofactor involved in amino acid metabolism [
3], reactive oxygen species detoxification [
4], and the synthesis of diverse secondary metabolites. VB6 levels strongly influence plant development, stress tolerance, and nutritional quality [
4]. De novo biosynthesis of VB6 occurs through a DXP-independent pathway catalyzed by pyridoxal 5′-phosphate synthase (PDX1) and the glutaminase PDX2 [
5,
6,
7], with PDX2 initiating the pathway by deaminating glutamine to generate ammonia, which is subsequently utilized by PDX1 to form PLP [
7,
8]. Although
PDX genes have been characterized in several model angiosperms such as
Arabidopsis thaliana and
Oryza sativa [
9,
10,
11], the functional roles of
PDX2 remain poorly understood in evolutionarily ancient lineages.
G. biloba, the only extant member of Ginkgoaceae, represents a “living fossil” with more than 270 million years of evolutionary history [
12]. Beyond its phylogenetic importance, ginkgo kernels are widely consumed as both medicine and food, in part due to their relatively high VB6 content, which has been associated with beneficial effects on cognition and antioxidant capacity [
13,
14,
15,
16,
17]. Previous analyses have demonstrated that VB6 levels in ginkgo kernels surpass those in many common nuts and seeds, suggesting the presence of unique genetic or regulatory mechanisms [
18,
19,
20]. However, the molecular regulation of VB6 biosynthesis in ginkgo remains largely unexplored, and no systematic investigation of
PDX2 genes in gymnosperms has been reported. This gap limits our understanding of whether VB6 metabolism in ginkgo is conserved with angiosperms or follows distinct strategies.
Therefore, the present study was designed with three primary objectives: (i) to perform a genome-wide identification and characterization of PDX2 genes in G. biloba; (ii) to investigate their molecular properties, evolutionary relationships, and expression patterns during kernel development; and (iii) to validate their functional roles in VB6 biosynthesis through heterologous expression in N. benthamiana. Collectively, this work provides the first functional insights into PDX2 genes in ginkgo, enriches current understanding of vitamin metabolism in gymnosperms, and offers potential molecular targets for nutritional enhancement through metabolic engineering.
3. Results
3.1. Identification of PDX2 Gene Family Members and Analysis of Protein Physicochemical Properties
This study combined genome-wide identification of
PDX2 homologous genes in
G. biloba with functional validation of
Gb_34755 through heterologous overexpression and biochemical assays, HMM (Hidden Markov Model) profiling was employed to validate results from the NCBI Batch Conserved Domain Database (CDD) and Pfam. The analysis identified two
PDX2 gene family members.
Table 2 presents the physicochemical characteristics and predicted subcellular localization of the two PDX2 proteins,
Gb_34755 and
Gb_34990.
Physicochemical analysis revealed that the protein encoded by Gb_34755 consists of 254 amino acids, with a molecular weight of 27.66 kDa and a theoretical isoelectric point (pI) of 7.82, suggesting a slightly basic nature under physiological conditions. The instability index of this protein is 45.86, which exceeds the threshold of 40, indicating it may be an unstable protein. Its grand average of hydropathicity (GRAVY) score is –0.13, with a hydropathy index of 91.42, suggesting that this protein is generally hydrophilic and may function in aqueous cellular environments.
In contrast, Gb_34990 encodes a shorter protein of 213 amino acids, with a molecular weight of 22.60 kDa and a pI of 6.59, indicating a mildly acidic profile. The instability index of 35.62 classifies it as a stable protein. Its GRAVY score is slightly higher (0.006), yet still indicative of a predominantly hydrophilic protein.
Subcellular localization predictions suggest that Gb_34755 is localized in the cytoplasm, whereas Gb_34990 is predicted to localize in both the chloroplast and cytoplasm. This divergence indicates that members of the PDX2 gene family may participate in distinct metabolic pathways of VB6-related metabolic pathways through different subcellular compartments. For instance, the chloroplast-localized Gb_34990 may be directly involved in coenzyme synthesis related to photosynthesis, while the cytoplasmic Gb_34755 may play a role in primary metabolic processes.
In summary, the variation in physicochemical properties and subcellular localization among the PDX2 gene family members in G. biloba reflects their potential functional differentiation. These findings provide important clues for further exploration of the molecular mechanisms underlying their roles in different physiological contexts.
3.2. Conserved Motif and Gene Structure Analysis of the PDX2 Gene Family
In the genome-wide identification and functional analysis of the
PDX2 gene family in
G. biloba, the identification of conserved motifs and domains provides critical insights into the functional divergence and evolutionary relationships within this gene family (
Figure 1). Sequence alignment of
Gb_34755 and
Gb_34990 revealed that both genes contain multiple conserved motifs (
Figure 1A,C). Although the biological functions of some motifs remain to be experimentally validated, their conservation among
PDX2 family members suggests that these regions are essential for protein function.
PDX2 proteins are known to function as part of the VB6 biosynthetic complex, typically interacting with PDX1 proteins to carry out PLP biosynthesis. Using the Conserved Domain Database (CDD) from NCBI, conserved domain prediction was conducted for PDX2 protein sequences. The results indicated that PDX2 proteins belong to the GAT-1 superfamily and are annotated as type I glutamine amidotransferases (
Figure 1B). Specifically, the region spanning amino acids 1–244 of the PDX2 protein contains the PLN02832 domain (glutamine amidotransferase subunit of the pyridoxal 5′-phosphate synthase complex), which is a characteristic domain of the trimeric glutamine amidotransferase family and plays a crucial role in the biosynthesis pathway of VB6.
In this study, Gb_34755 and Gb_34990 exhibited similar distributions of conserved motifs, although sequence variations in certain motifs may reflect functional specialization. For instance, variations observed in some motifs of Gb_34990 may be associated with its subcellular localization in chloroplasts and its adaptation to photosynthesis-related metabolic pathways. The conservation and divergence of these motifs suggest that the PDX2 gene family has undergone functional differentiation during evolution. The differences in motif composition between Gb_34755 and Gb_34990 may be related to their regulatory roles in distinct subcellular environments, providing a basis for further investigation into their expression patterns across different tissues or under various stress conditions.
In summary, the analysis of conserved motifs not only supports the functional conservation of the PDX2 gene family but also lays a molecular foundation for elucidating the VB6-related metabolic pathways in G. biloba. Future studies may validate the functions of key motifs through site-directed mutagenesis or transgenic approaches.
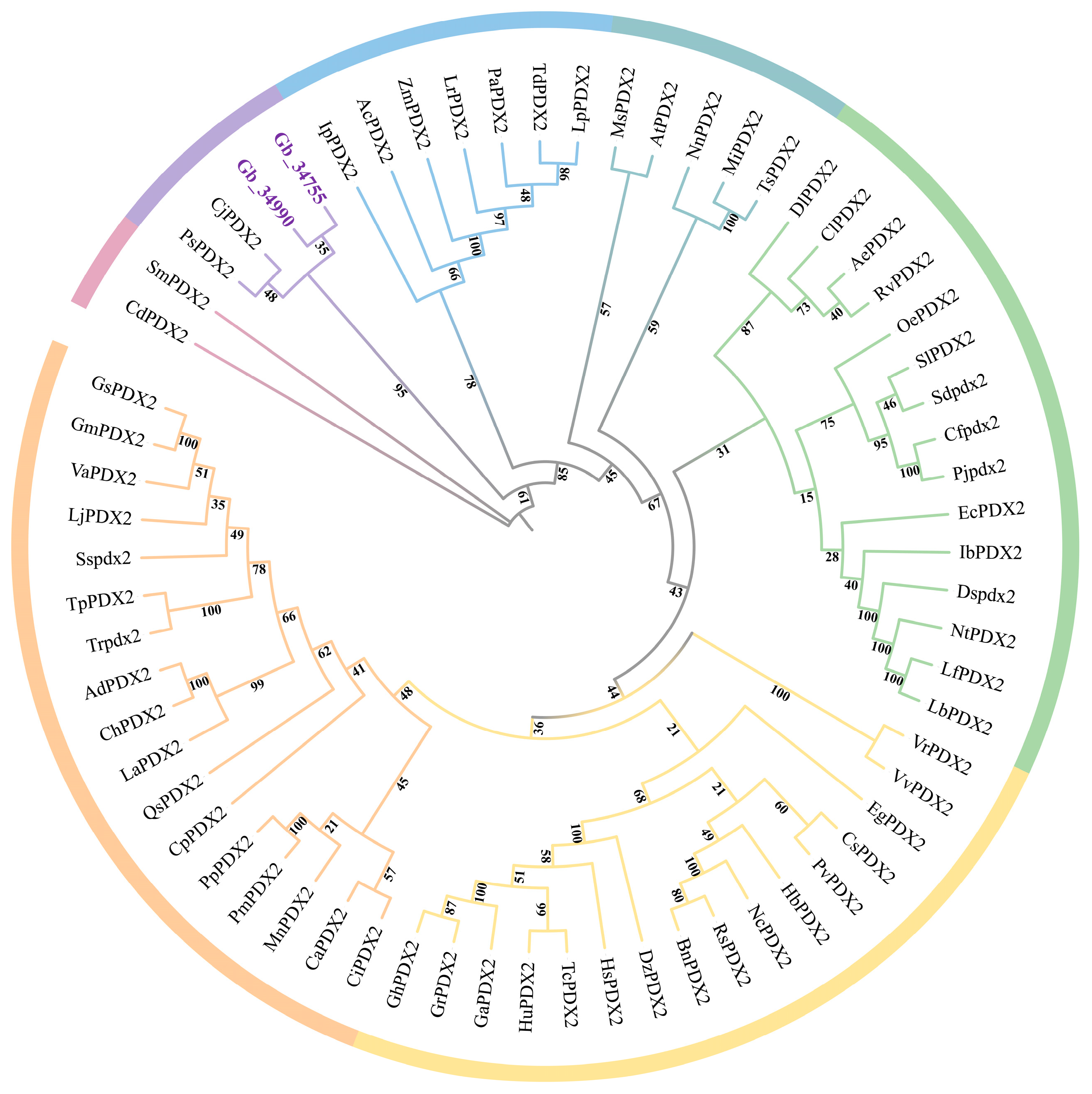
3.3. Phylogenetic Analysis of the PDX2 Gene Family
Phylogenetic tree analysis revealed that the
PDX2 gene family in
G. biloba (
GbPDX2) comprises two members:
Gb_34755 and
Gb_34990 (
Figure 2). Together with
PDX2 genes from other species, these genes form several evolutionary clades, reflecting both the evolutionary conservation and functional divergence of the gene family. The clustering of Ginkgo
PDX2 genes with those of
Selaginella moellendorffii (
SmPDX2) and
Ceratopteris richardii (
CdPDX2) indicates a high degree of conservation of
PDX2 genes between Ginkgo and early land plants such as ferns. In addition, the grouping of
GbPDX2 genes with
PDX2 genes from gymnosperms, such as
Picea sitchensis (
PsPDX2) and
Cryptomeria japonica (
CjPDX2), suggests that Ginkgo, as one of the most ancient extant gymnosperms, possesses unique evolutionary features within the gymnosperm lineage. This pattern reflects adaptive modifications of the VB6–related metabolism in Ginkgo during evolution to meet distinct physiological requirements or environmental stresses.
Notably, the GbPDX2 genes of Ginkgo do not cluster directly with the PDX2 genes of most angiosperms, such as A. thaliana (AtPDX2), O. sativa (OsPDX2), or Zea mays (ZmPDX2), indicating that the PDX2 genes in Ginkgo diverged from those of angiosperms at an early stage of evolution. Angiosperm PDX2 genes mainly group into several independent clades (e.g., monocots and dicots), while GbPDX2 genes are more closely related to those of gymnosperms and ferns, which is consistent with the phylogenetic position of Ginkgo. This study provides important clues for understanding the evolutionary history of PDX2 genes in Ginkgo and their roles in VB6-related metabolic pathways. Future research involving gene expression and functional experiments will be essential to further verify their biological functions.
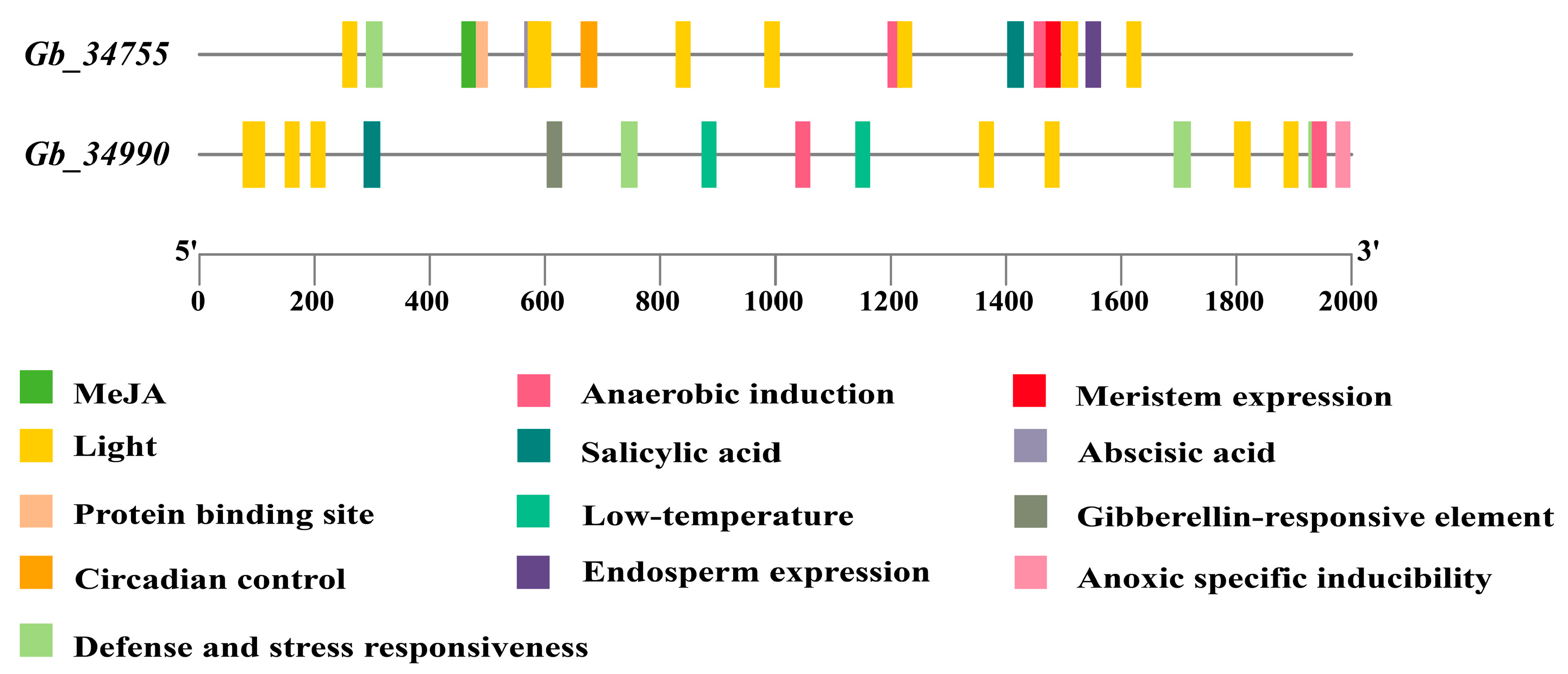
3.4. Analysis of Cis-Acting Elements in the Promoters of PDX2 Gene Family Members
Promoters are regulatory regions of genes that contain various cis-acting elements capable of responding to environmental stimuli and modulating gene expression. Based on the analysis of cis-acting elements in the promoter regions of
G. biloba PDX2 gene family members (
Figure 3), we systematically identified the distribution patterns of multiple functional cis-elements in the promoters of two genes (
Gb_34755 and
Gb_34990). The results revealed that light-responsive elements are highly enriched in the promoter regions of both genes, suggesting that light signaling may play a significant role in regulating the expression of Ginkgo
PDX2 genes.
In addition, elements associated with defense and stress responsiveness, as well as methyl jasmonate (MeJA) responsive elements, are widely present, indicating the potential involvement of these genes in plant stress response mechanisms. Notably, the promoter of Gb_34990 is enriched in low-temperature responsive elements and salicylic acid-responsive elements, implying that this gene may function in cold stress responses and immune defense signaling pathways.
In contrast, the promoter of Gb_34755 contains a higher abundance of meristem expression elements and endosperm expression elements, suggesting its specific regulatory role during plant growth and developmental stages. Furthermore, both promoters harbor multiple elements related to anaerobic induction and anoxic-specific inducibility, indicating that Ginkgo PDX2 genes may be activated under hypoxic stress conditions. The presence of abscisic acid (ABA)-responsive elements further supports the potential involvement of these genes in environmental adaptation and endogenous hormone-regulated pathways.
In summary, the enrichment of diverse cis-acting elements in the promoters of Ginkgo PDX2 gene family members suggests that their expression is regulated by a variety of internal and external environmental factors. These include light signals and hormonal responses, as well as multiple stress conditions, reflecting their important roles in Ginkgo’s adaptation to complex environments and in the regulation of growth and development. These findings provide a theoretical basis for subsequent functional validation and regulatory mechanism studies.
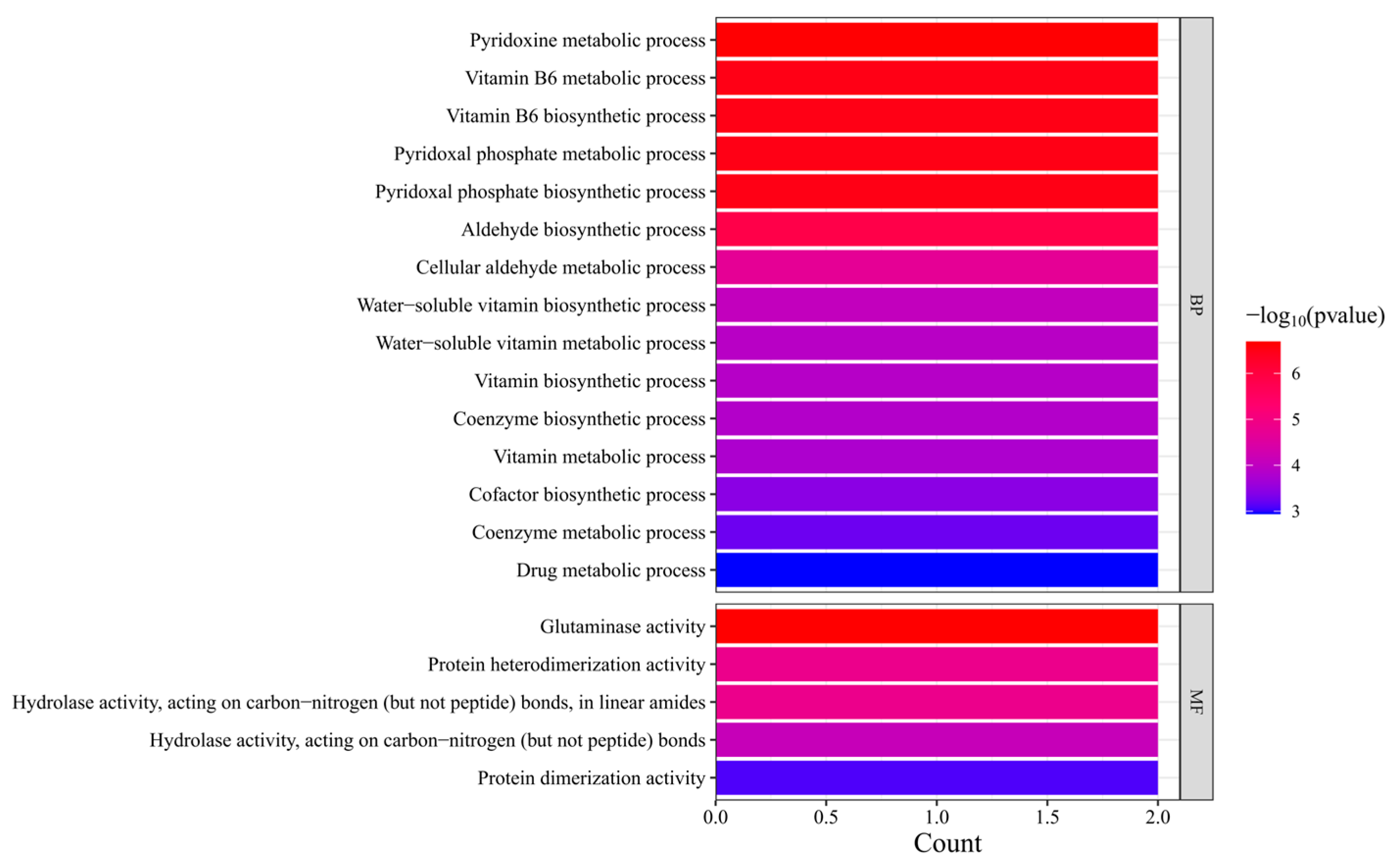
3.5. Function Annotation Display of the PDX2 Family Genes in G. biloba
A comprehensive functional characterization of the
G. biloba PDX2 gene family was conducted through systematic Gene Ontology (GO) function annotation display. According to the function annotation results, members of the
PDX2 gene family exhibit distinct functional features at both the Biological Process and Molecular Function levels. The analysis showed that Ginkgo
PDX2 genes are significantly enriched in pathways related to VB6 metabolism (
Figure 4). Among these, the pyridoxine metabolic process and VB6 biosynthetic process displayed the highest enrichment levels (represented by red bars), indicating that this gene family plays a central role in the VB6-related metabolic pathways.
In addition, these genes are notably involved in the pyridoxal phosphate metabolic process, water-soluble vitamin biosynthetic process, and coenzyme biosynthetic process. Interestingly, the cellular aldehyde metabolic process also showed significant enrichment, suggesting that the PDX2 gene family may participate in the regulation of secondary metabolic pathways that are specific to Ginkgo.
At the Molecular Function level, the PDX2 gene family is primarily associated with the following activities: Glutaminase activity: directly related to the catalytic function of the VB6 synthase complex; Protein heterodimerization activity: forming the molecular basis for functional complexes with PDX1 proteins; Hydrolase activity acting on C–N bonds: involved in the hydrolysis of amide bonds during VB6-related metabolic pathways.
Of particular interest is the significant enrichment of protein dimerization activity, which aligns well with the known molecular mechanism by which PDX2 proteins function through homo- or heterodimer formation.
Comparative analysis revealed that the enrichment pattern of Ginkgo PDX2 genes in the VB6 synthesis pathway is conserved with that observed in angiosperms such as A. thaliana. However, the specific enrichment in aldehyde metabolic processes may reflect a unique adaptation of secondary metabolism in gymnosperms. Moreover, the pronounced enrichment of glutaminase activity (highlighted in red bars) supports a potential novel role of PDX2 genes in nitrogen metabolism in Ginkgo.
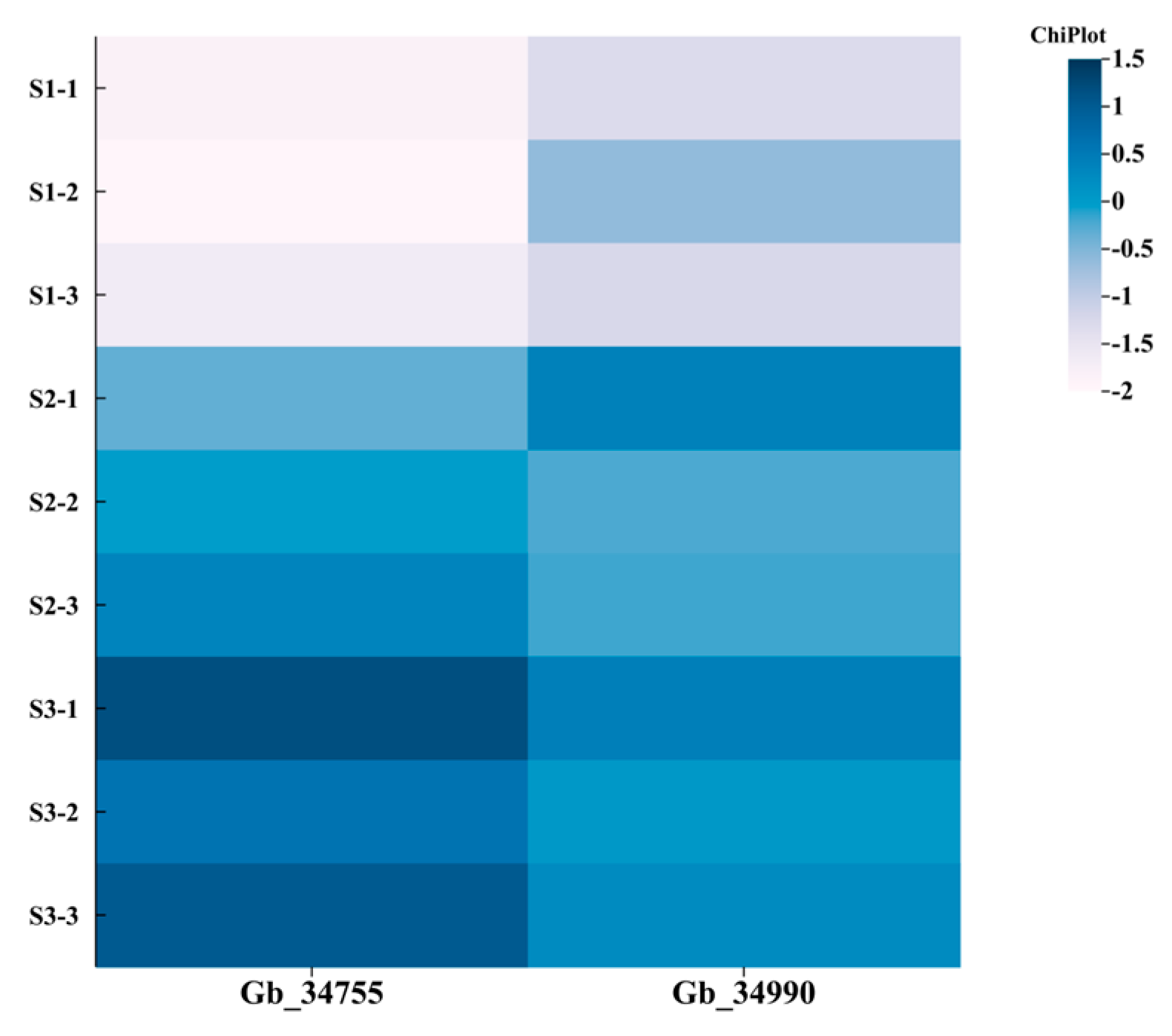
3.6. Expression Analysis of the PDX2 Gene Family
All kernel samples were collected from a single healthy and mature
G. biloba tree (cultivar ‘Foshou’) at three different developmental stages (June, July, and August). At each stage, seeds were harvested from multiple fruits and pooled to form a biological sample representing that timepoint. Based on the heatmap analysis results (
Figure 5), the
G. biloba PDX2 gene family members (
Gb_34755 and
Gb_34990) exhibit distinct expression patterns across three developmental stages of the kernel (S1—early developmental stage (June), S2—mid-transition stage (July), and S3—late/mature stage (August)). Early developmental stage (June–S1): Both genes show the lowest expression levels across all biological replicates (S1-1 to S1-3). Mid-transition stage (July–S2): Expression levels approach the baseline. Maturation and accumulation stage (August–S3): Expression levels increase significantly, showing high expression in this phase.
A comparative analysis across the three stages revealed a monotonic upward trend in expression from S1 to S3, with Gb_34755 and Gb_34990 displaying highly synchronized expression patterns. This expression dynamic likely reflects the period of high VB6 accumulation in Ginkgo seed kernels during August, suggesting that PDX2 genes may be involved in vitamin synthesis during the maturation stage. The co-expression of these two genes implies potential functional complementarity within the metabolic pathway.
Using a threshold of |log2FC| ≥ 1 to identify candidate genes, Gb_34755 was ultimately selected as the key candidate gene.
3.7. Construction of the PDX2 Gene Overexpression Vector
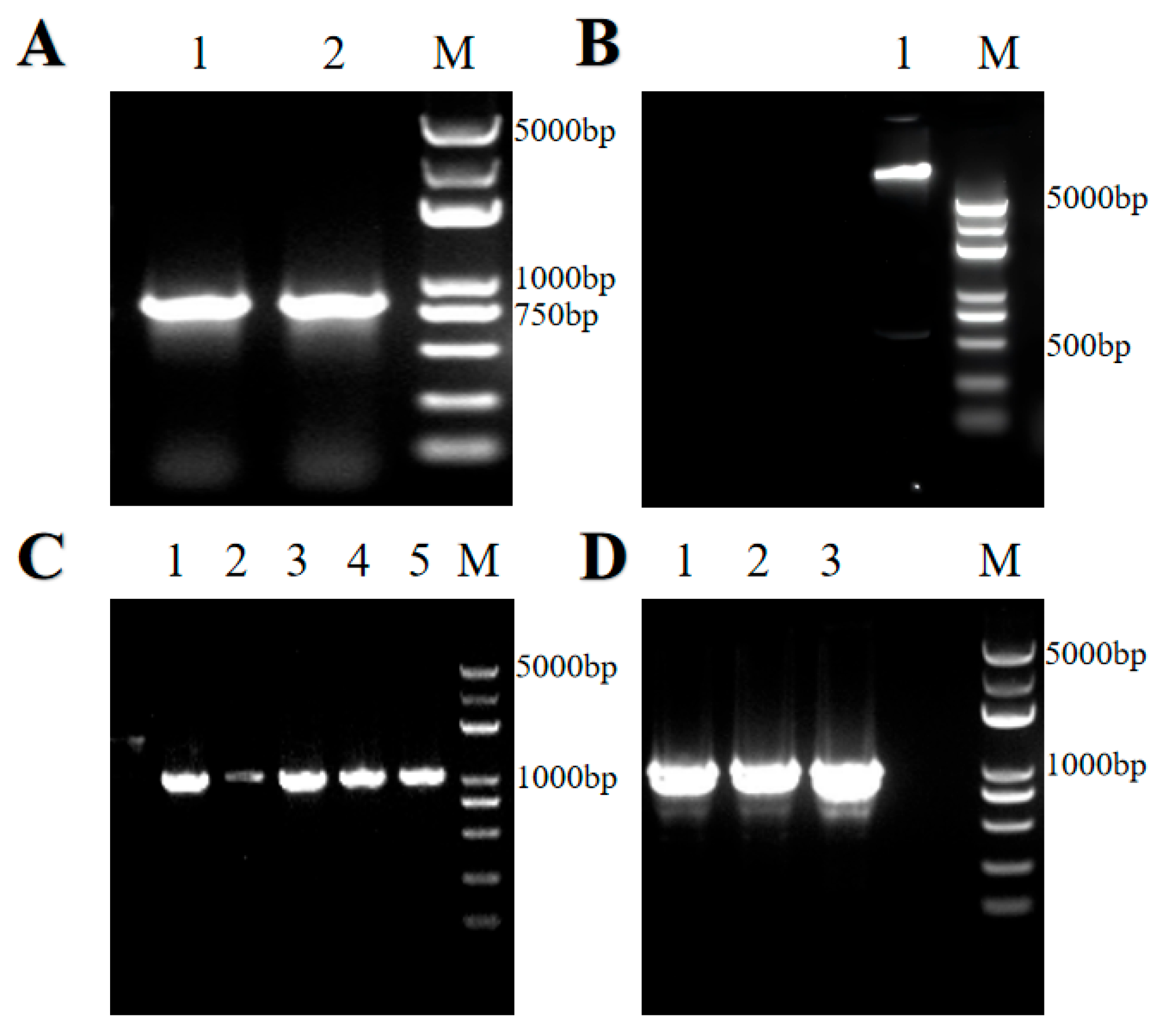
The length of the PCR amplified product was 805 bp, which corresponds to the sum of the target gene size obtained from genomic sequencing (765 bp) and the homologous arm sequences (40 bp) (
Figure 6A). The vector was linearized by restriction enzyme digestion, producing a faint fragment of approximately 550 bp, indicating successful vector digestion (
Figure 6B).
After positive clones were identified, plasmids were extracted from expanded cultures and sequenced. Agarose gel electrophoresis of the PCR product showed a final band length of 1063 bp, confirming the successful construction of the overexpression vector containing the target gene (
Figure 6C).
The validated recombinant plasmid was then introduced into
Agrobacterium tumefaciens. Single colonies were randomly selected from LB plates containing the appropriate antibiotics and cultured for 4–6 h. PCR was performed on
Agrobacterium tumefaciens bacterial suspensions to identify positive clones. Gel electrophoresis revealed bands of the expected length, consistent with the size of the target gene, indicating successful transformation (
Figure 6D).
The Agrobacterium cultures identified as positive by electrophoresis were further amplified and mixed with an appropriate amount of glycerol, then stored at ultra-low temperatures for subsequent genetic transformation experiments.
3.8. Transient Overexpression Functional Validation in N. benthamiana Leaves
In this study, a
N. benthamiana transformation system based on the ruby reporter gene was successfully established (
Figure 7), serving as a heterologous expression platform for functional studies of
G. biloba PDX2 genes. Experimental results showed that wild-type control (WT) leaves exhibited the typical green phenotype (
Figure 7A), while the overexpression transgenic lines (OE), due to stable transformation of the ruby gene (containing the
PDX2 candidate gene), displayed a distinct red coloration in the leaves (
Figure 7B).
This system confirmed the visual detectability of the ruby reporter gene in N. benthamiana and provides a reliable platform for subsequent functional validation of Ginkgo PDX2 genes.
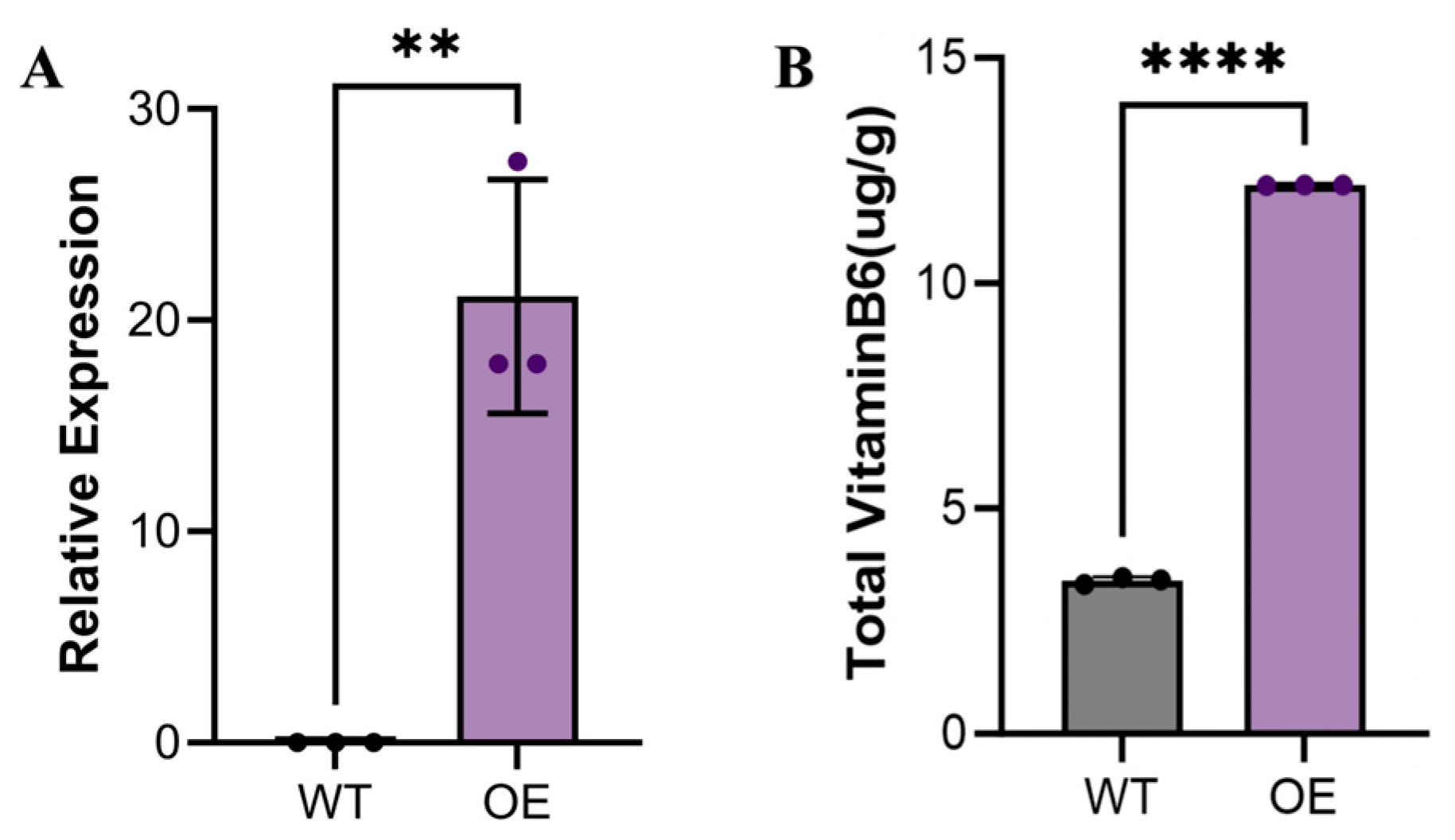
Using Agrobacterium-mediated transient transformation, we successfully achieved overexpression of the
G. biloba PDX2 gene in
N. benthamiana leaves. qRT-PCR analysis (
Figure 8A) revealed that the mRNA expression level of the
PDX2 gene in overexpression lines (OE) was significantly upregulated compared to the wild-type (WT) (
p < 0.05), indicating that the exogenous gene was successfully integrated and efficiently expressed. This result validates the reliability of the experimental system and establishes a foundation for subsequent functional studies.
To investigate the biological function of the
PDX2 gene, we measured the VB6 content in both WT and OE
N. benthamiana leaves (
Figure 8B). The results demonstrated that VB6 levels in the OE lines were significantly higher than those in the WT (
p < 0.05). Specifically, the VB6 content in WT leaves was 3.38 ± 0.08 μg/g, while in OE lines it increased significantly to 12.172 ± 0.006 μg/g—representing an approximately four-fold increase (
p < 0.05). This substantial enhancement strongly supports the positive regulatory role of the Ginkgo
PDX2 gene in VB6-related metabolic pathways.
This finding is consistent with the known function of PDX2 genes in the VB6 metabolic pathway and further underscores their critical role in coenzyme biosynthesis and metabolic regulation in plants. All data are presented as the mean ± standard deviation (SD) of three independent biological replicates, and statistical significance was confirmed (p < 0.05), ensuring the reliability and reproducibility of the results.
The increased VB6 content in OE lines not only confirms the functional role of the PDX2 gene but also highlights its potential application in enhancing VB6 levels in plants. Through this transient overexpression system, we preliminarily elucidated the role of the G. biloba PDX2 gene in VB6-related metabolic pathways and demonstrated its capacity to significantly enhance VB6 accumulation in N. benthamiana. These findings provide important evidence for further elucidating the molecular mechanisms and application potential of Ginkgo PDX2 genes.
4. Discussion
This study presents the first systematic identification and functional analysis of
PDX2 genes in
G. biloba, an evolutionarily ancient gymnosperm species. Two homologs,
Gb_34755 and
Gb_34990, were identified, both containing the conserved glutaminase domain (PLN02832), and their expression patterns were shown to differ across developmental stages of kernels, suggesting functional divergence under spatial and temporal regulation. Notably, transient overexpression of these genes in
N. benthamiana significantly enhanced VB6 (pyridoxal 5′-phosphate, PLP) content, providing supporting evidence for their active role in VB6 biosynthesis. These results extend current understanding of plant vitamin metabolism, which has so far primarily focused on angiosperms such as
Arabidopsis thaliana and
Oryza sativa [
9,
27], by introducing gymnosperms into the comparative framework.
Mechanistically, the functional divergence of the two
PDX2 homologs can be linked to differences in predicted subcellular localization and cis-regulatory architectures.
Gb_34755 is localized to the cytoplasm, whereas
Gb_34990 is predicted to be dual-localized in both the cytoplasm and the chloroplast. The presence of chloroplast localization is unusual for PDX2 proteins, which are predominantly cytosolic in angiosperms [
6], and may reflect evolutionary adaptations in Ginkgo that compartmentalize VB6 biosynthesis for more efficient metabolic flux or plastid-specific functions. This is consistent with studies showing that the subcellular localization of vitamin biosynthetic enzymes can impact metabolic regulation and cofactor availability [
28].
Promoter analysis revealed that
Gb_34990 is enriched with stress- and hormone-responsive elements, including those responsive to salicylic acid, abscisic acid, and low temperature, indicating possible induction under abiotic stress. In contrast,
Gb_34755 harbors more elements associated with growth and developmental processes, particularly auxin and gibberellin responsiveness, suggesting a role in basal metabolic function during seed development. This partitioning supports a model of subfunctionalization following gene duplication, where duplicated genes acquire distinct regulatory contexts to diversify function [
29]. Similar differential regulation and specialization of
PDX family members have also been reported in rice and tomato under abiotic stress [
5]. Moreover, the comparatively high protein instability index predicted for
Gb_34755 may suggest transient or fine-tuned expression during specific stages, whereas
Gb_34990 appears to maintain more stable expression over developmental stages, reflected by its expression peak in late kernel development.
Phylogenetic analysis placed the Ginkgo
PDX2 genes closer to those in ferns and gymnosperms than to angiosperm counterparts, which aligns with Ginkgo’s long-diverged evolutionary history [
30]. The evolutionary conservation of the GAT-1 domain through different plant lineages suggests that this core enzymatic function has been maintained since the early land plant ancestors. However, the regulatory divergence observed in our study may represent an early evolutionary shift in functional specialization within gymnosperms [
31], potentially shaped by adaptation to unique photoperiods, longevity, and stress resistance characteristics of woody perennials [
32].
While the current study provides functional insights, several limitations remain. Firstly, subcellular localization was predicted computationally and not validated experimentally; future work involving fluorescent tagging, yeast two-hybrid assays, or co-immunoprecipitation is warranted. Secondly, although transient expression in N. benthamiana offers a rapid tool for initial functional assessment, it does not fully recapitulate Ginkgo’s native physiological context. Differences in metabolic pathways and expression networks between the two species may affect the interpretation of heterologous overexpression results. Finally, detailed analysis of VB6 biosynthesis flux, expression under different stress conditions, and gene regulation in diverse tissues will be necessary to further delineate the roles of PDX2 genes in vivo.
In conclusion, this research expands the current understanding of vitamin B6 metabolism by demonstrating for the first time the expression, localization, and functional characteristics of PDX2 homologs in a gymnosperm species. The novel insights into subcellular compartmentalization, promoter responsiveness, and evolutionary divergence underscore the biological complexity and specialization of VB6 biosynthetic regulation in ancient plant lineages. These findings pave the way for future research into metabolic engineering of VB6 pathways in woody plants and may contribute to the development of micronutrient-fortified crops with enhanced stress tolerance and health-promoting properties.
5. Conclusions
This study provides the first comprehensive characterization of the PDX2 gene family in G. biloba, revealing their potential roles in vitamin B6 (VB6) biosynthesis. Two members, Gb_34755 and Gb_34990, were identified and shown to contain conserved glutaminase domains characteristic of key enzymes in VB6 metabolism. Their predicted differential subcellular localization suggests functional divergence between developmental and stress-responsive pathways. Expression profiling demonstrated strong upregulation of both genes during late seed development, coinciding with VB6 accumulation, and heterologous overexpression in N. benthamiana confirmed that Gb_34755 enhances VB6 content. Promoter analyses revealed distinct cis-regulatory elements, indicating specialized regulatory roles in growth and environmental adaptation.
While this work expands our understanding of VB6 metabolic pathways in gymnosperms, several limitations should be acknowledged. Functional interpretations were partly based on computational predictions that require in planta validation, and the lack of stable genetic transformation systems in ginkgo currently restricts direct functional testing. Future efforts should therefore focus on developing transformation techniques for gymnosperms, performing tissue- and organ-specific expression analyses, and dissecting upstream transcriptional regulation and protein–protein interactions within the PDX complex. The application of advanced tools such as CRISPR/Cas genome editing and multi-omics integration will further enable precise functional dissection and metabolic engineering of VB6 pathways.
Taken together, this study not only provides the first insights into PDX2 gene function in an ancient gymnosperm but also highlights the novelty and evolutionary significance of ginkgo as a model for studying metabolic adaptations. The resources and knowledge generated here offer a foundation for future functional genomics investigations and provide promising targets for improving nutritional quality and stress resilience in woody crops through metabolic engineering.