Response of Rhododendron simsii and Rhododendron delavayi Superoxide Dismutase Family Genes to High-Temperature Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of R. simsii and R. delavayi SOD Gene Family Members
2.2. Bioinformatics Analysis
2.3. Experimental Materials and High-Temperature Stress Treatment
2.4. Total RNA Extraction and cDNA First Strand Synthesis
2.5. Quantitative Real-Time Fluorescence Polymerase Chain Reaction
3. Results
3.1. Identification of SOD Proteins in R. simsii and R. delavayi
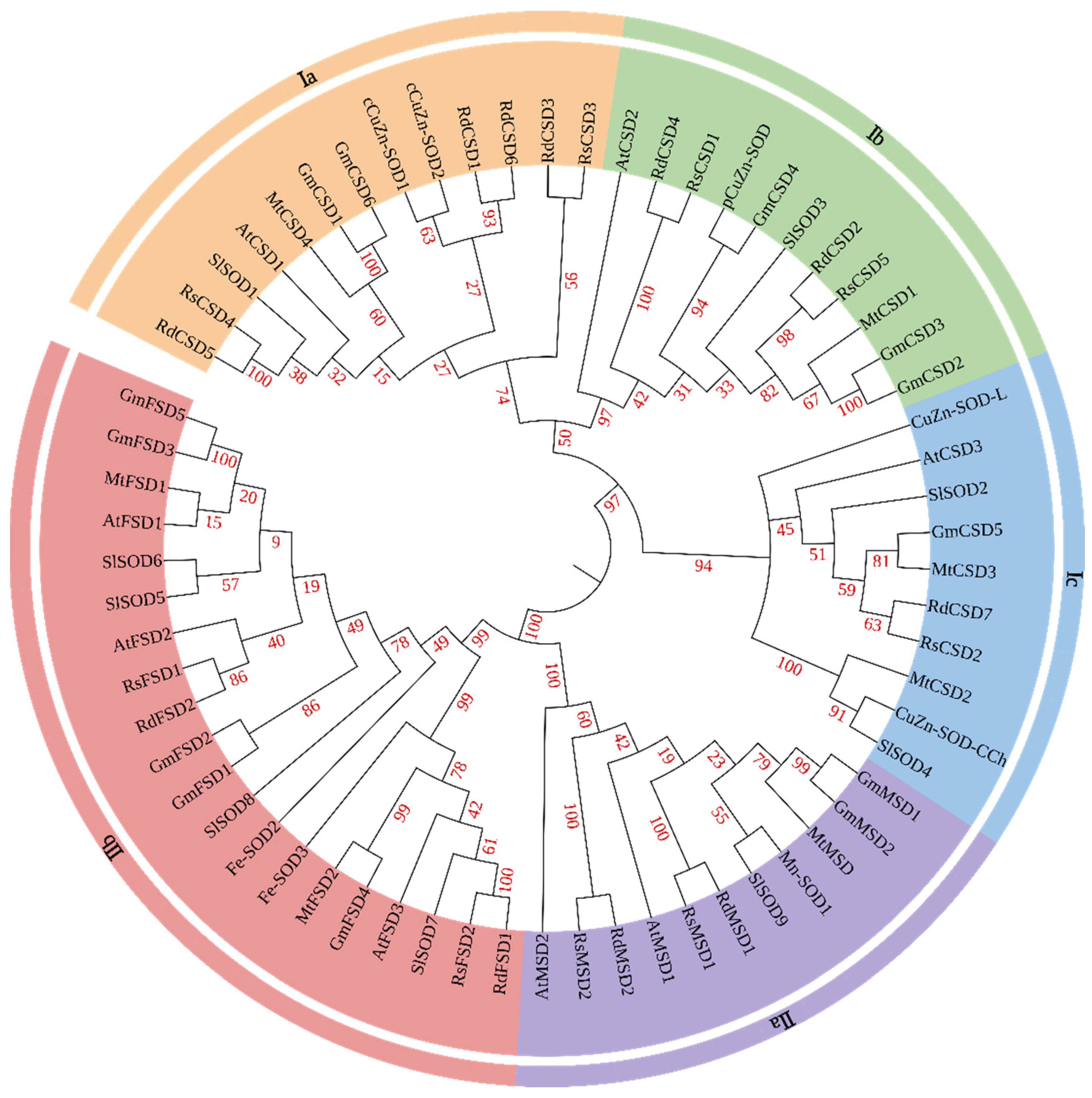
3.2. Phylogenetic Relationships
3.3. Gene Structure and Motif Composition of the R. simsii and R. delavayi SOD Gene Families
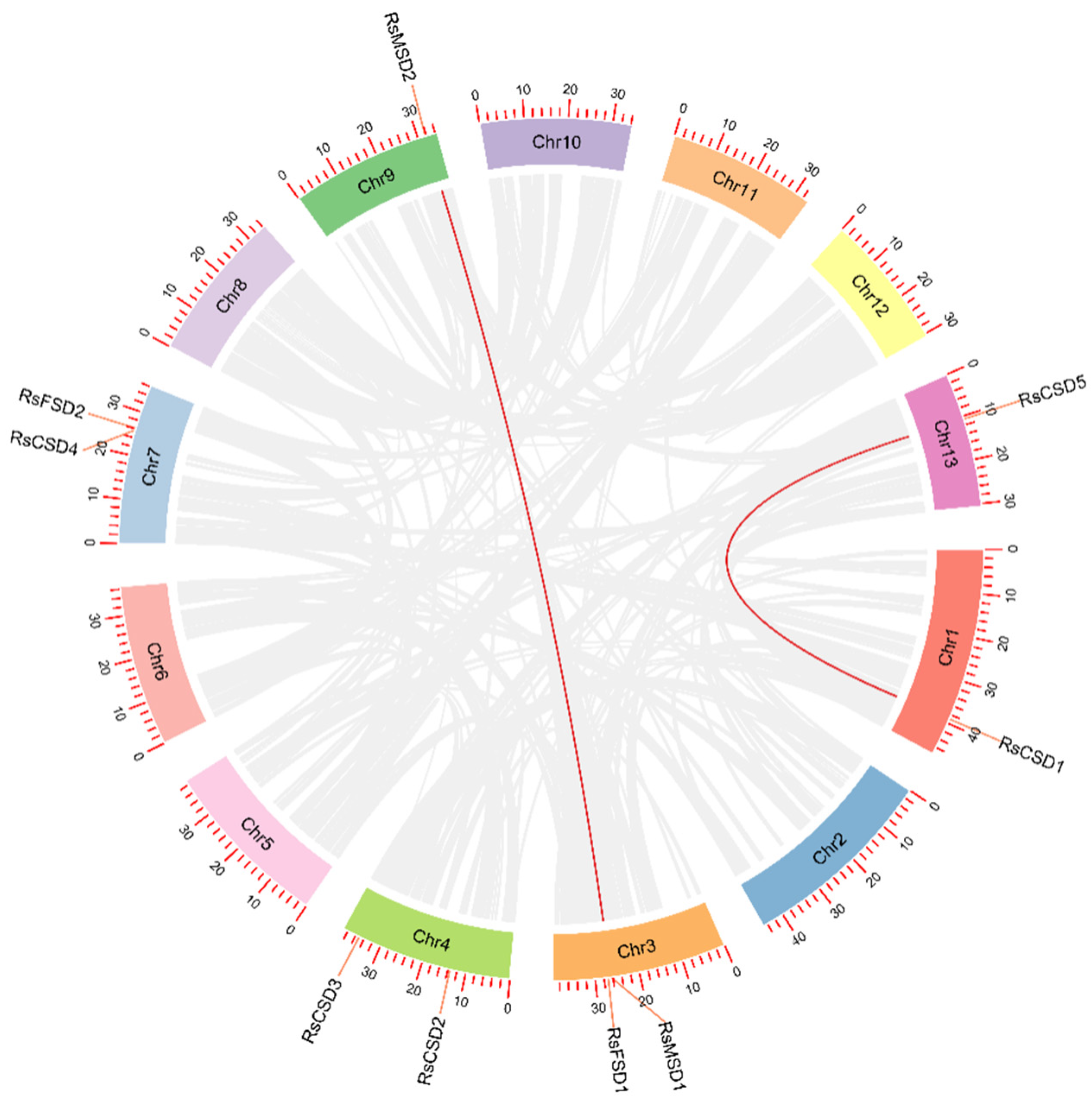
3.4. Chromosomal Distribution and Synthesis Analysis of RsSOD Genes
3.5. Examination of Cis-Elements in the Promoters of RsSOD and RdSOD Genes
3.6. RsSOD and RdSOD Family Protein Interaction Networks
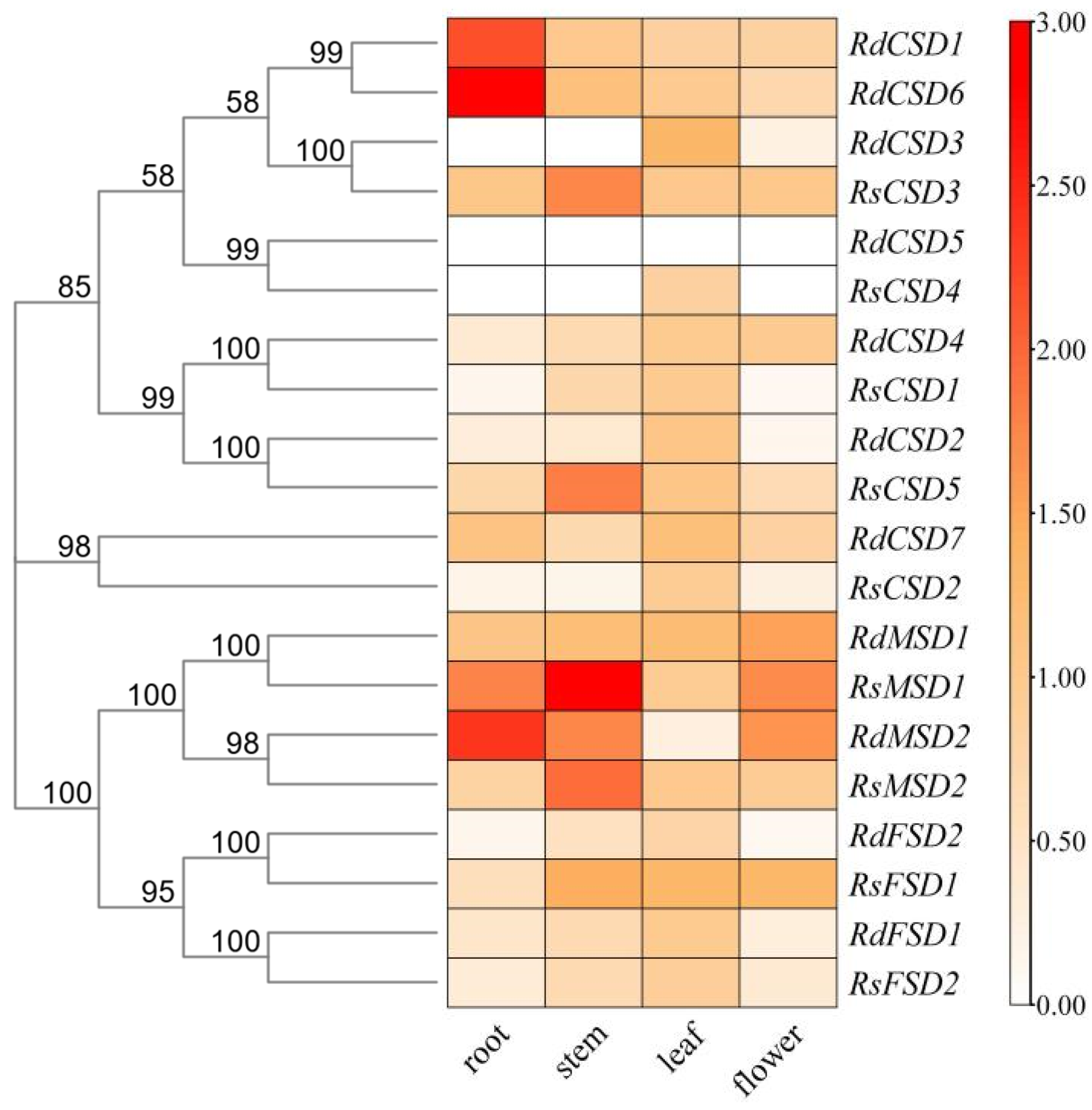
3.7. Tissue-Specific Expression Profiles of RsSODs and RdSODs
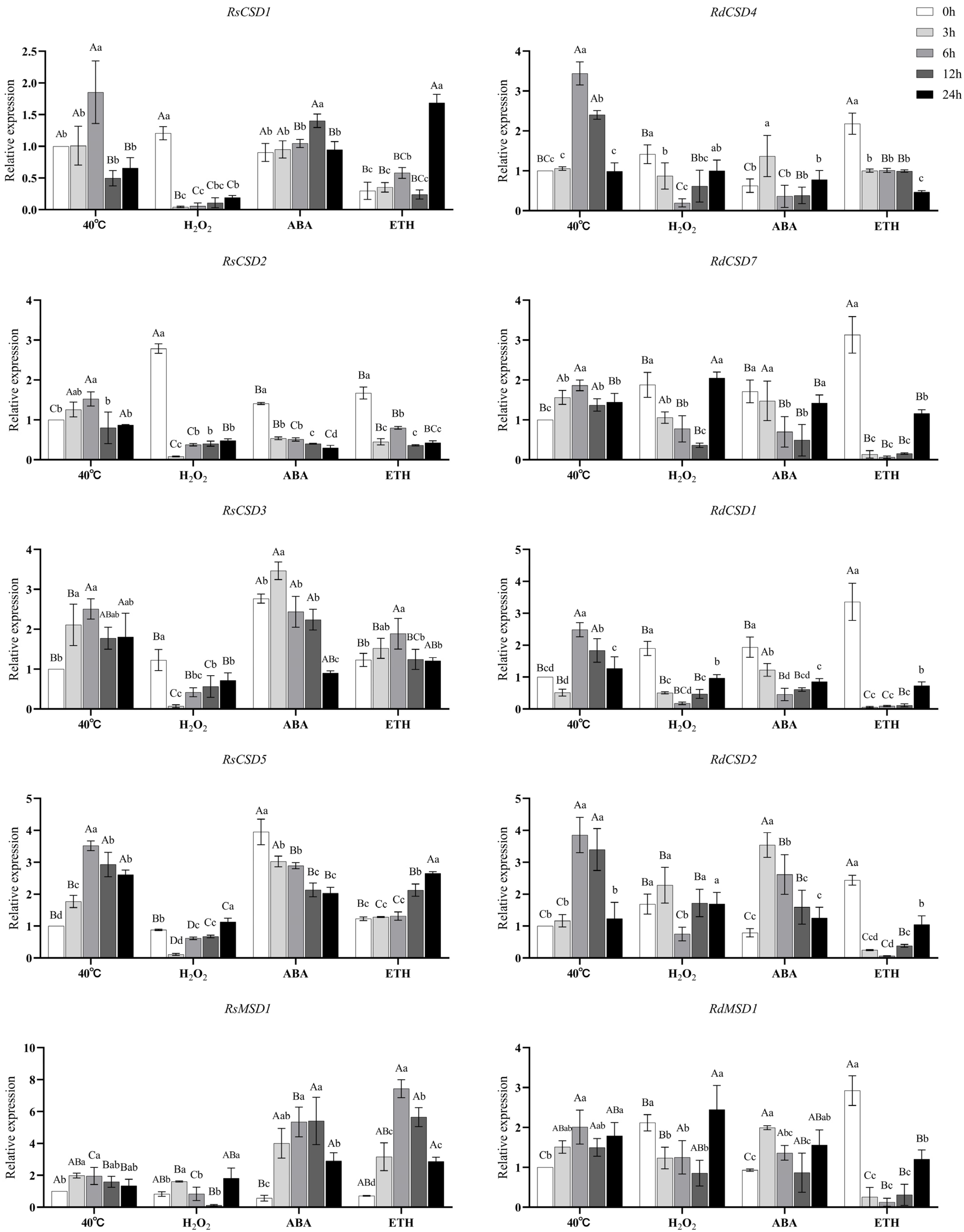
3.8. Expression of RsSODs and RdSODs under Exogenous Reagents and High-Temperature Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Perez, I.B.; Brown, P.J. The Role of ROS Signaling in Cross-Tolerance: From Model to Crop. Front. Plant Sci. 2014, 5, 754. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive Oxygen Species (ROS) and Response of Antioxidants as ROS-Scavengers during Environmental Stress in Plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Fink, R.C.; Scandalios, J.G. Molecular Evolution and Structure--Function Relationships of the Superoxide Dismutase Gene Families in Angiosperms and Their Relationship to Other Eukaryotic and Prokaryotic Superoxide Dismutases. Arch. Biochem. Biophys. 2002, 399, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Bao, X.; Zhi, Y.; Wu, Q.; Guo, Y.; Yin, X.; Zeng, L.; Li, J.; Zhang, J.; He, W.; et al. Overexpression of a MYB Family Gene, OsMYB6, Increases Drought and Salinity Stress Tolerance in Transgenic Rice. Front. Plant Sci. 2019, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.S.; Bang, S.G.; Ahn, M.-A.; Kim, G.; Kim, E.; Eom, S.H.; Hyun, T.K. Molecular Cloning and Functional Characterization of Heat Stress-Responsive Superoxide Dismutases in Garlic (Allium sativum L.). Antioxidants 2021, 10, 815. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Wang, Y.; Ding, C.; Lei, L.; Qian, W.; Zeng, J.; Yang, Y.; et al. Transcriptional and Physiological Analyses Reveal the Association of ROS Metabolism with Cold Tolerance in Tea Plant. Environ. Exp. Bot. 2019, 160, 45–58. [Google Scholar] [CrossRef]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-Wide Identification and Characterization of the Superoxide Dismutase Gene Family in Musa acuminata Cv. Tianbaojiao (AAA Group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef]
- Alscher, R.G. Role of Superoxide Dismutases (SODs) in Controlling Oxidative Stress in Plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Tan, M.; Lu, J.; Zhang, A.; Hu, B.; Zhu, X.; Li, W. The distribution and cooperation of antioxidant (iso) enzymes and antioxidants in different subcellular compartments in maize leaves during water stress. J. Plant Growth Regul. 2011, 30, 255–271. [Google Scholar] [CrossRef]
- Li, X.; Cai, J.; Liu, F.; Dai, T.; Cao, W.; Jiang, D. Cold Priming Drives the Sub-Cellular Antioxidant Systems to Protect Photosynthetic Electron Transport against Subsequent Low Temperature Stress in Winter Wheat. Plant Physiol. Biochem. 2014, 82, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Duanmu, H.; Qiao, Y.; Jin, X.; Yu, Y.; Yu, L.; Chen, C. Genome-Wide Identification and Characterization of the Soybean SOD Family during Alkaline Stress. PeerJ 2020, 8, e8457. [Google Scholar] [CrossRef]
- Feng, K.; Yu, J.; Cheng, Y.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Li, Z.; Yao, Z.; Yang, Y.; et al. The SOD Gene Family in Tomato: Identification, Phylogenetic Relationships, and Expression Patterns. Front. Plant Sci. 2016, 7, 1279. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, L.; Shang, J.; Hu, X.; Yu, H.; Wu, H.; Lv, W.; Zhao, Y. Genome-Wide Analysis of the Maize Superoxide Dismutase (SOD) Gene Family Reveals Important Roles in Drought and Salt Responses. Genet. Mol. Biol. 2021, 44, e20210035. [Google Scholar] [CrossRef]
- Han, L.-M.; Hua, W.-P.; Cao, X.-Y.; Yan, J.-A.; Chen, C.; Wang, Z.-Z. Genome-Wide identification and Expression Analysis of the Superoxide dismutase (SOD) Gene Family in Salvia miltiorrhiza. Gene 2020, 742, 144603. [Google Scholar] [CrossRef]
- Zhou, Y.; Hu, L.; Wu, H.; Jiang, L.; Liu, S. Genome-Wide Identification and Transcriptional Expression Analysis of Cucumber Superoxide Dismutase (SOD) Family in Response to Various Abiotic Stresses. Int. J. Genom. 2017, 2017, 7243973. [Google Scholar] [CrossRef]
- Zhou, C.; Zhu, C.; Fu, H.; Li, X.; Chen, L.; Lin, Y.; Lai, Z.; Guo, Y. Genome-Wide Investigation of Superoxide Dismutase (SOD) Gene Family and Their Regulatory miRNAs Reveal the Involvement in Abiotic Stress and Hormone Response in Tea Plant (Camellia sinensis). PLoS ONE 2019, 14, e0223609. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Genome-Wide Analysis of Superoxide Dismutase Gene Family in Gossypium raimondii and G. arboreum. Plant Gene 2016, 6, 18–29. [Google Scholar] [CrossRef]
- Yu, Q.; Osborne, L.D.; Rengel, Z. Increased tolerance to Mn deficiency in transgenic tobacco overproducing superoxide dismutase. Ann. Bot. 1999, 84, 543–547. [Google Scholar] [CrossRef][Green Version]
- Tang, L.; Kwon, S.-Y.; Kim, S.-H.; Kim, J.-S.; Choi, J.S.; Cho, K.Y.; Sung, C.K.; Kwak, S.-S.; Lee, H.-S. Enhanced Tolerance of Transgenic Potato Plants Expressing Both Superoxide Dismutase and Ascorbate Peroxidase in Chloroplasts against Oxidative Stress and High Temperature. Plant Cell Rep. 2006, 25, 1380–1386. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Raven, P.H.; Hong, D.Y. Flora of China. Vol. 14 (Apiaceae through Ericaceae); Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2005; pp. 242–440. [Google Scholar]
- Geng, X.M.; Liu, P.; Li, Z.F.; Xiao, L.Y. Improving heat tolerance of Rhododendron by H2O2 pretreatment. J. Anhui Agric. Univ. 2019, 46, 167–172, (In Chinese with English Abstract). [Google Scholar]
- Shen, H.F.; Zhao, B.; Xu, J.J.; Liang, W.; Huang, W.M.; Li, H.H. Effects of Heat Stress on Changes in Physiology and Anatomy in Two Cultivars of Rhododendron. S. Afr. J. Bot. 2017, 112, 338–345. [Google Scholar] [CrossRef]
- Zhao, H.; Geng, X.M.; Wang, L.L.; Xu, S.D. Research on the effect of ethylene in heat resistance mechanism of Rhododendron. Acta Hortic. Sin. 2022, 49, 561–570, (In Chinese with English Abstract). [Google Scholar]
- Geng, X.M.; Xiao, L.Y.; Zhao, H.; Liu, P. Sub-cellular Localization of ROS-scavenging System in Rhododendron Leaves under Heat Stress and H2O2 Pretreatment. Northwest Bot. 2019, 39, 791–800, (In Chinese with English Abstract). [Google Scholar]
- Zhang, L.; Xu, P.; Cai, Y.; Ma, L.; Li, S.; Li, S.; Xie, W.; Song, J.; Peng, L.; Yan, H.; et al. The Draft Genome Assembly of Rhododendron delavayi Franch. var. delavayi. Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Yang, F.-S.; Nie, S.; Liu, H.; Shi, T.-L.; Tian, X.-C.; Zhou, S.-S.; Bao, Y.-T.; Jia, K.-H.; Guo, J.-F.; Zhao, W.; et al. Chromosome-Level Genome Assembly of a Parent Species of Widely Cultivated Azaleas. Nat. Commun. 2020, 11, 5269. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the Functional Annotation of Proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide Dismutase in Arabidopsis: An Eclectic Enzyme Family with Disparate Regulation and Protein Localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef]
- Nath, K.; Kumar, S.; Poudyal, R.S.; Yang, Y.N.; Timilsina, R.; Park, Y.S.; Nath, J.; Chauhan, P.S.; Pant, B.; Lee, C.-H. Developmental Stage-Dependent Differential Gene Expression of Superoxide Dismutase Isoenzymes and Their Localization and Physical Interaction Network in Rice (Oryza sativa L.). Genes. Genom. 2014, 36, 45–55. [Google Scholar] [CrossRef]
- Song, J.; Zeng, L.; Chen, R.; Wang, Y.; Zhou, Y. In Silico Identification and Expression Analysis of Superoxide Dismutase (SOD) Gene Family in Medicago Truncatula. 3 Biotech 2018, 8, 348. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Amend, A.; Borkowski, J.; DeMarco, R.; Bailey, W.; Liu, Y.; Xie, G.; Blevins, R. MULTICLUSTAL: A Systematic Method for Surveying Clustal W Alignment Parameters. Bioinformatics 1999, 15, 862–863. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; Yin, H.; Li, L.; Wang, R.; Wu, J.; Wu, J.; Zhang, S. Different Modes of Gene Duplication Show Divergent Evolutionary Patterns and Contribute Differently to the Expansion of Gene Families Involved in Important Fruit Traits in Pear (Pyrus bretschneideri). Front. Plant Sci. 2018, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a Plant Cis-Acting Regulatory Element Database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Li, X.L.; Hua, Z.R.; Zhang, F.; Wang, X.J. Evaluation of Heat Tolerance of Different Rhododendron Varieties. Acta Agric. Jiangxi 2022, 34, 82–86, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Fernández-Ocaña, A.; Chaki, M.; Luque, F.; Gómez-Rodríguez, M.V.; Carreras, A.; Valderrama, R.; Begara-Morales, J.C.; Hernández, L.E.; Corpas, F.J.; Barroso, J.B. Functional Analysis of Superoxide Dismutases (SODs) in Sunflower under Biotic and Abiotic Stress Conditions. Identification of Two New Genes of Mitochondrial Mn-SOD. J. Plant Physiol. 2011, 168, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Song, H.; Zhang, B.; Lu, Q.; Liu, Z.; Zhang, S.; Guo, R.; Wang, C.; Zhao, Z.; Liu, J.; et al. Genome-Wide Identification, Characterization, and Expression Analysis of Superoxide Dismutase (SOD) Genes in Foxtail Millet (Setaria italica L.). 3Biotech 2018, 8, 486. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xia, M.; Chen, J.; Deng, F.; Yuan, R.; Zhang, X.; Shen, F. Data Set for Phylogenetic Tree and RAMPAGE Ramachandran Plot Analysis of SODs in Gossypium raimondii and G. arboreum. Data Brief 2016, 9, 345–348. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. A Modern Circadian Clock in the Common Angiosperm Ancestor of Monocots and Eudicots. BMC Biol. 2010, 8, 55. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of Duplicate Genes in Exon-Intron Structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Feng, Z.; Bian, L.; Xie, H.; Liang, J. miR398 Regulation in Rice of the Responses to Abiotic and Biotic Stresses Depends on CSD1 and CSD2 Expression. Funct. Plant Biol. 2010, 38, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Raja, V.; Majeed, U.; Kang, H.; Andrabi, K.I.; John, R. Abiotic Stress: Interplay between ROS, Hormones and MAPKs. Environ. Exp. Bot. 2017, 137, 142–157. [Google Scholar] [CrossRef]
- Prerostova, S.; Dobrev, P.I.; Kramna, B.; Gaudinova, A.; Knirsch, V.; Spichal, L.; Zatloukal, M.; Vankova, R. Heat Acclimation and Inhibition of Cytokinin Degradation Positively Affect Heat Stress Tolerance of Arabidopsis. Front. Plant Sci. 2020, 11, 87. [Google Scholar] [CrossRef] [PubMed]
- Larkindale, J.; Huang, B. Thermotolerance and Antioxidant Systems in Agrostis stolonifera: Involvement of Salicylic Acid, Abscisic Acid, Calcium, Hydrogen Peroxide, and Ethylene. J. Plant Physiol. 2004, 161, 405–413. [Google Scholar] [CrossRef]
- Hsieh, E.-J.; Cheng, M.-C.; Lin, T.-P. Functional Characterization of an Abiotic Stress-Inducible Transcription Factor AtERF53 in Arabidopsis Thaliana. Plant Mol. Biol. 2013, 82, 223–237. [Google Scholar] [CrossRef]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 Transcriptionally Activates CaWRKY40, Regulates Ralstonia Solanacearum Resistance, and Confers High-Temperature and High-Humidity Tolerance in Pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Zhen, Z.; Peng, J.; Chang, L.; Gong, Q.; Wang, N.N. Loss of ACS7 Confers Abiotic Stress Tolerance by Modulating ABA Sensitivity and Accumulation in Arabidopsis. J. Exp. Bot. 2011, 62, 4875–4887. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geng, X.; Hua, L.; Gong, J.; Yi, Y.; Tang, M.; Ceng, F. Response of Rhododendron simsii and Rhododendron delavayi Superoxide Dismutase Family Genes to High-Temperature Stress. Forests 2024, 15, 931. https://doi.org/10.3390/f15060931
Geng X, Hua L, Gong J, Yi Y, Tang M, Ceng F. Response of Rhododendron simsii and Rhododendron delavayi Superoxide Dismutase Family Genes to High-Temperature Stress. Forests. 2024; 15(6):931. https://doi.org/10.3390/f15060931
Chicago/Turabian StyleGeng, Xingmin, Li Hua, Jiyi Gong, Yin Yi, Ming Tang, and Fanyu Ceng. 2024. "Response of Rhododendron simsii and Rhododendron delavayi Superoxide Dismutase Family Genes to High-Temperature Stress" Forests 15, no. 6: 931. https://doi.org/10.3390/f15060931
APA StyleGeng, X., Hua, L., Gong, J., Yi, Y., Tang, M., & Ceng, F. (2024). Response of Rhododendron simsii and Rhododendron delavayi Superoxide Dismutase Family Genes to High-Temperature Stress. Forests, 15(6), 931. https://doi.org/10.3390/f15060931





