Genome-Wide Identification of MYB Gene Family in Peach and Identification of MYBs Involved in Carotenoid Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Protein Characterization of MYB Family Members in the Peach Genome
2.2. Chromosomal Localization and Collinearity Analysis of MYB Family Members
2.3. Ka and Ks Calculations
2.4. Gene Structure, Conserved Motifs, and Structural Domain Analysis of the PpMYB Family
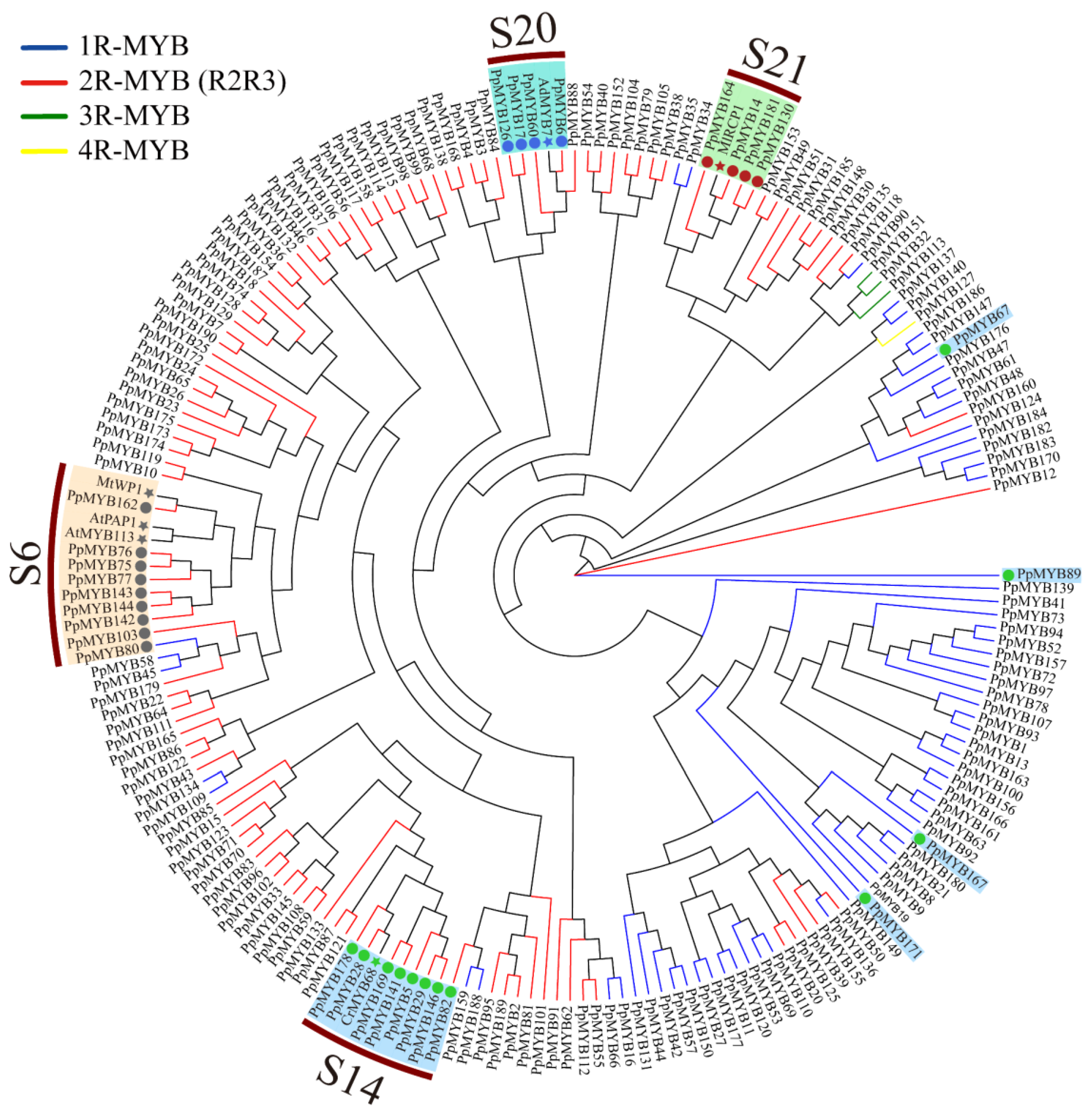
2.5. Phylogenetic Tree Analysis of MYB Family Genes in Peach
2.6. Analysis of Cis-Acting Elements of PpMYB Family Genes
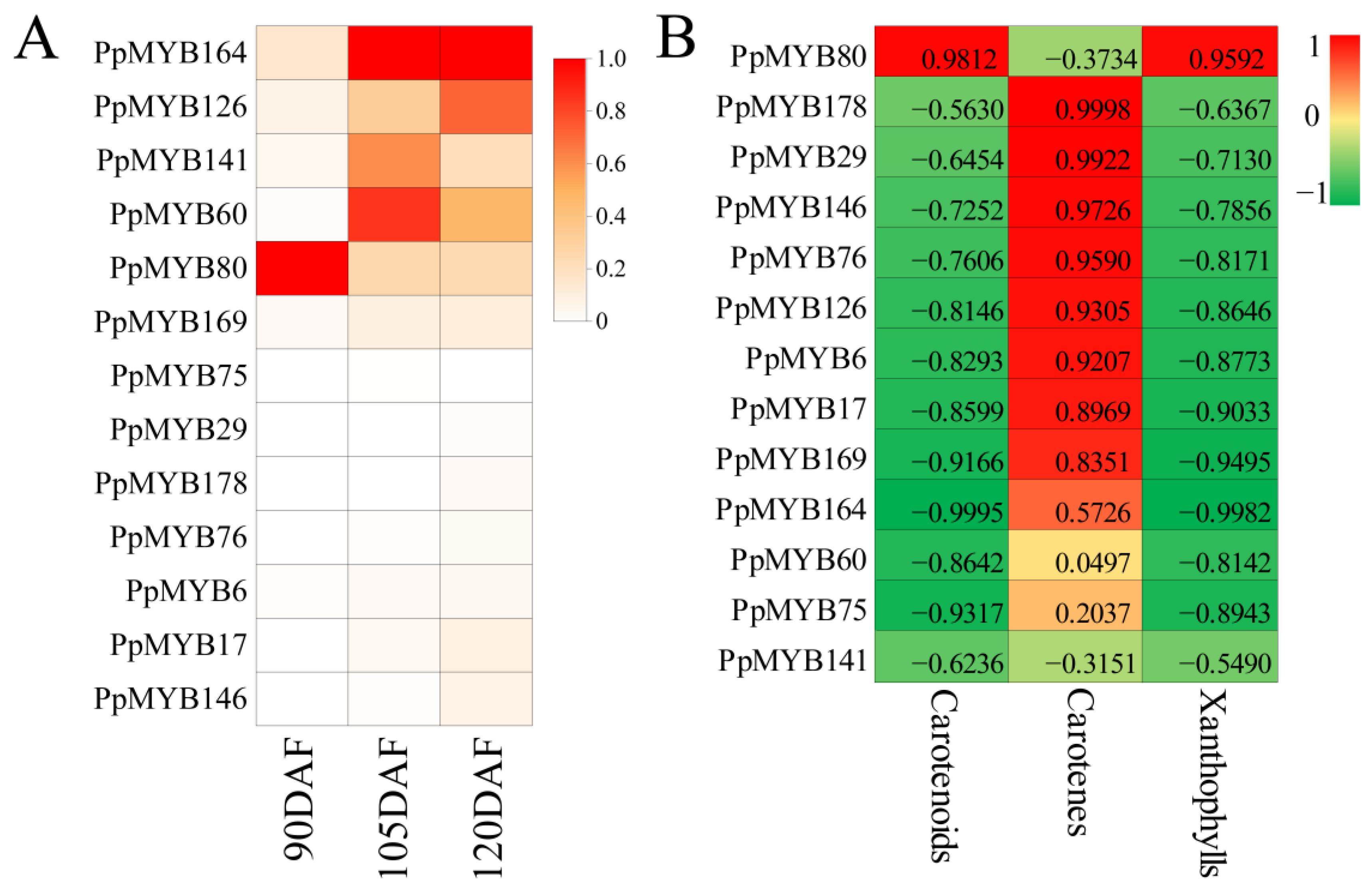
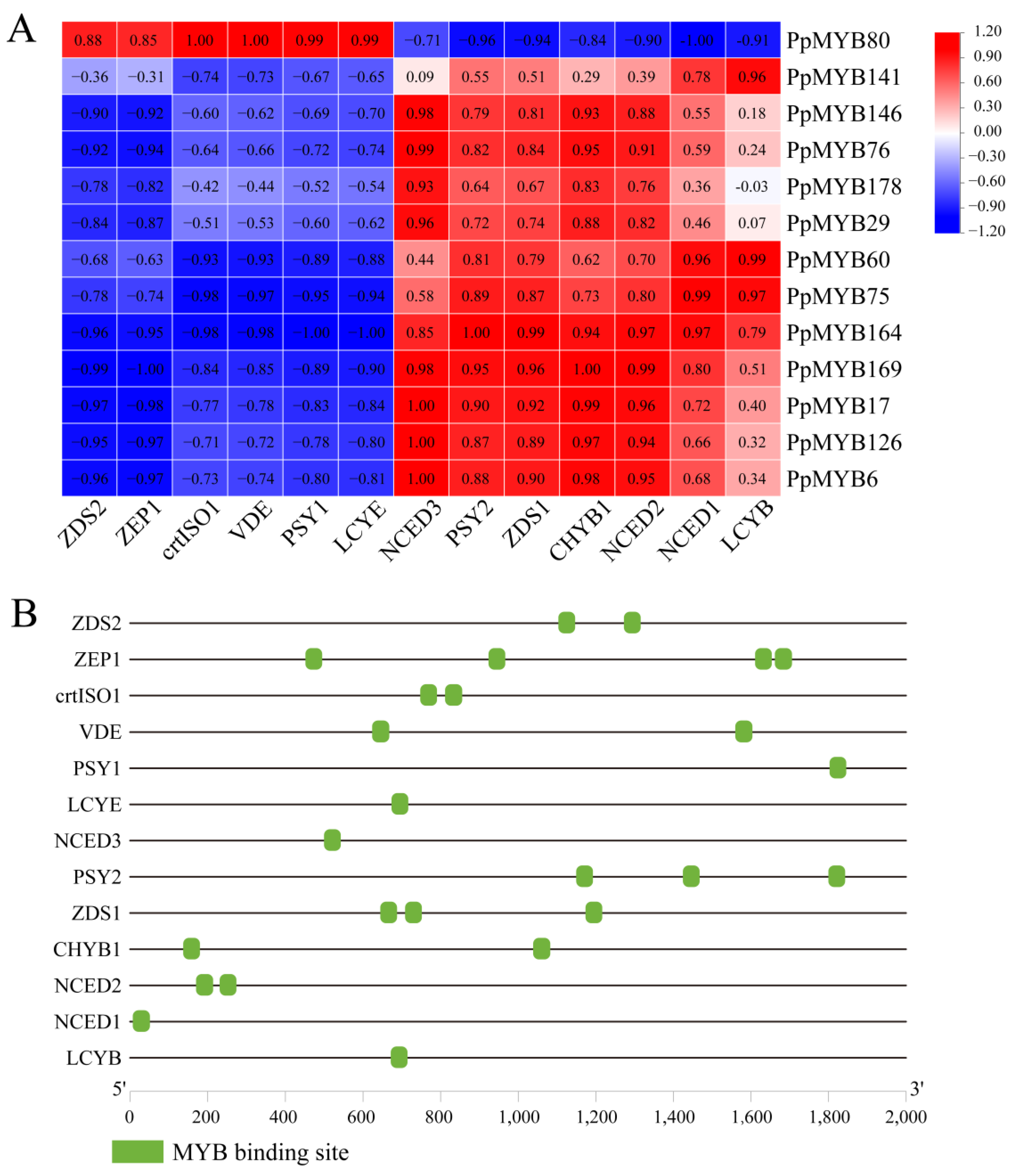
2.7. Heat Mapping and Correlation Analysis
3. Results
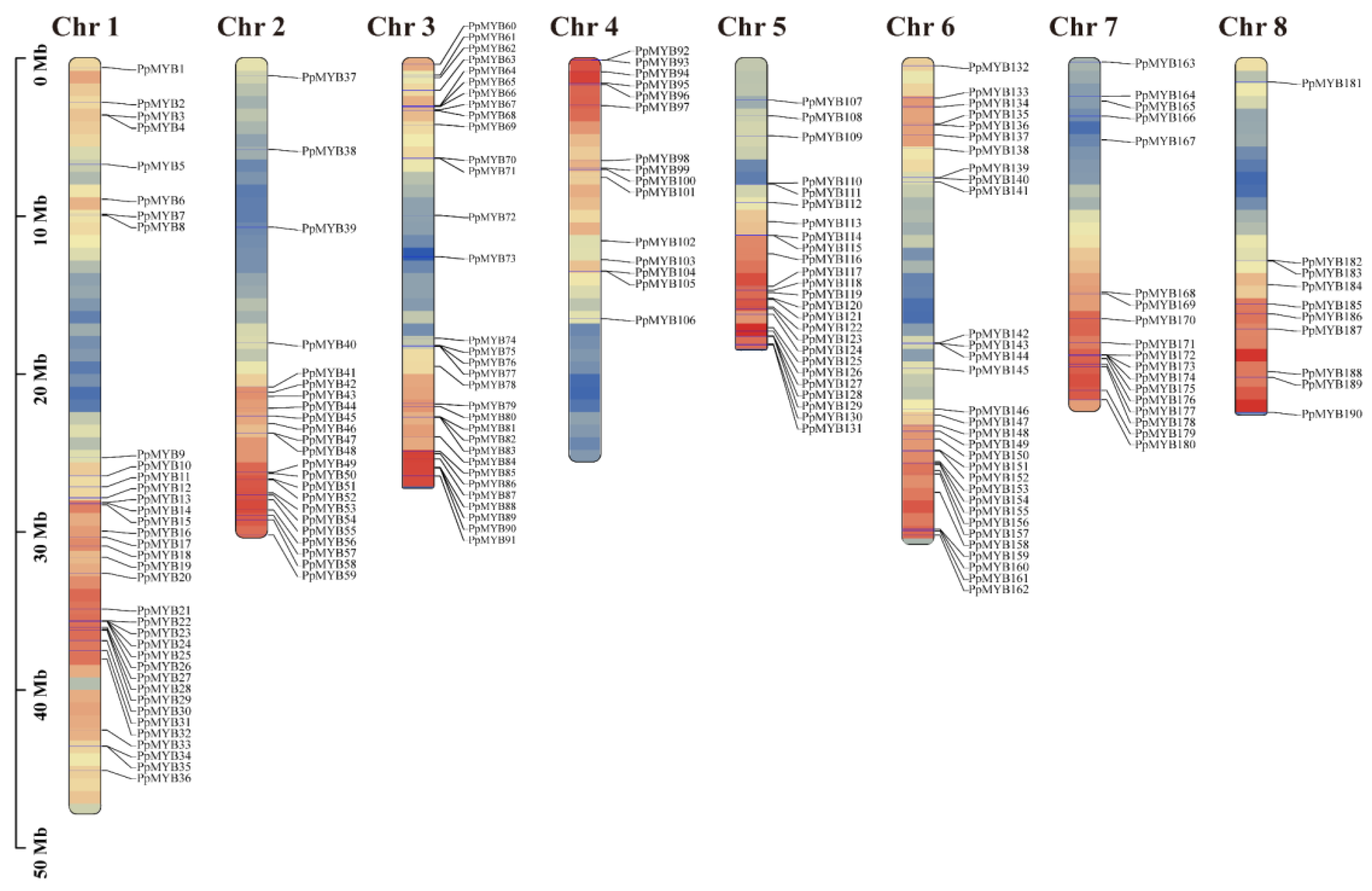
3.1. Identification of PpMYB Family Members and Analysis of Chromosomal Localization
3.2. Analysis of Physicochemical Properties of PpMYB Family Members
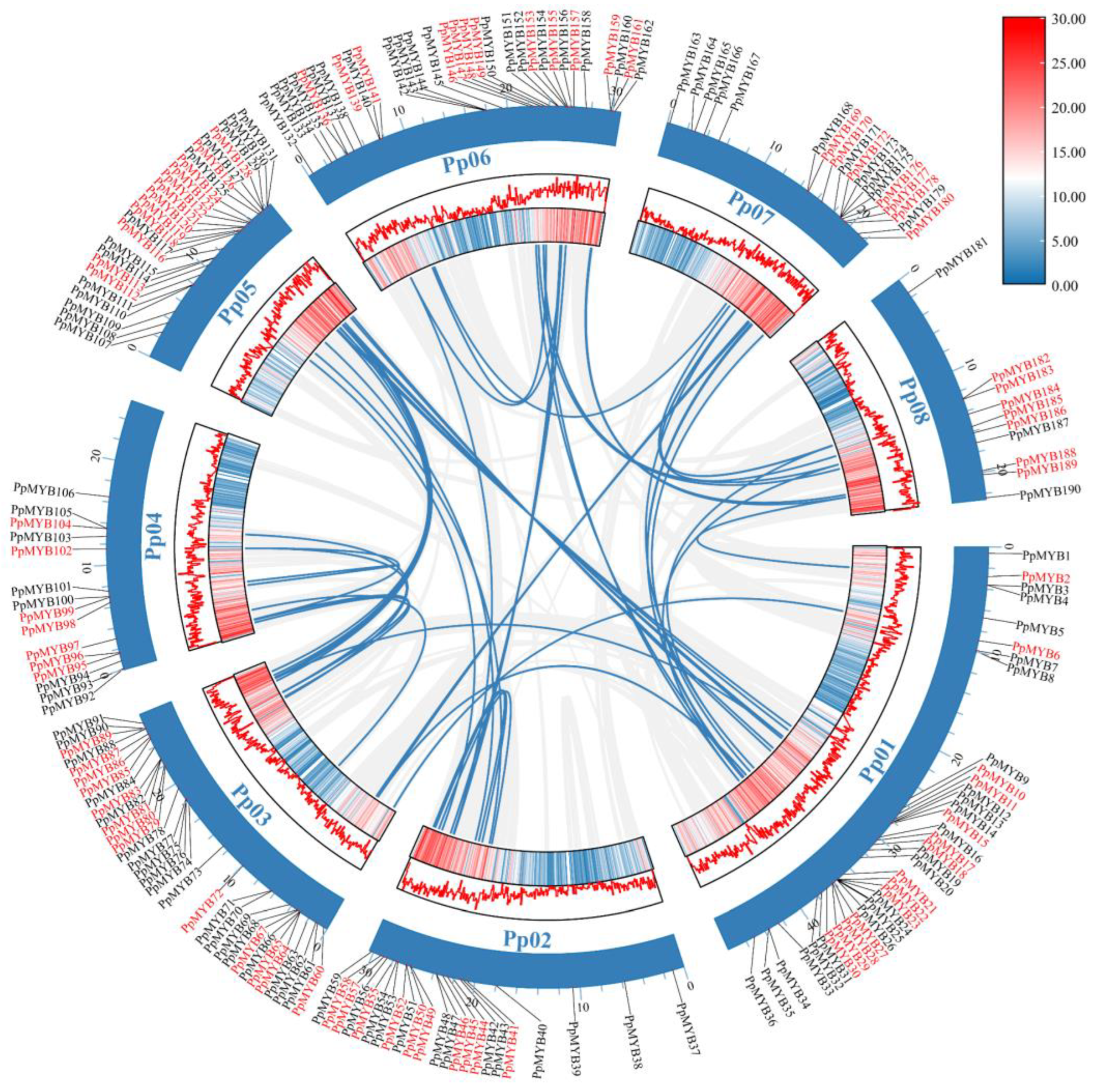
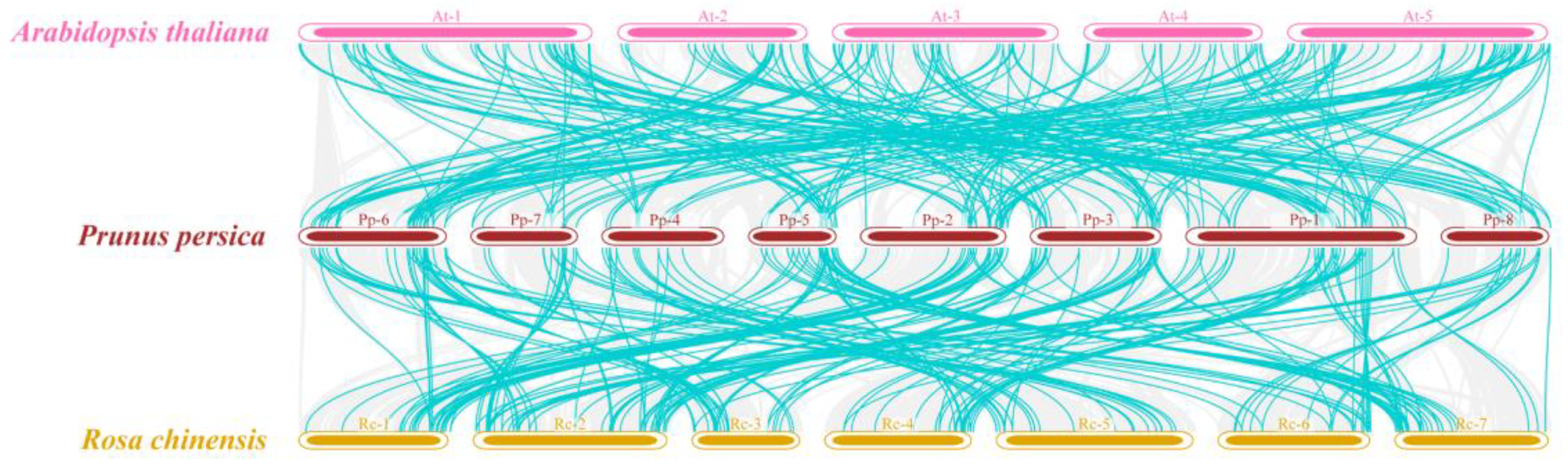
3.3. Intraspecific and Interspecific Collinearity Analysis of PpMYB Family Genes
3.4. Phylogenetic Relationship between Peach and Arabidopsis MYB Proteins
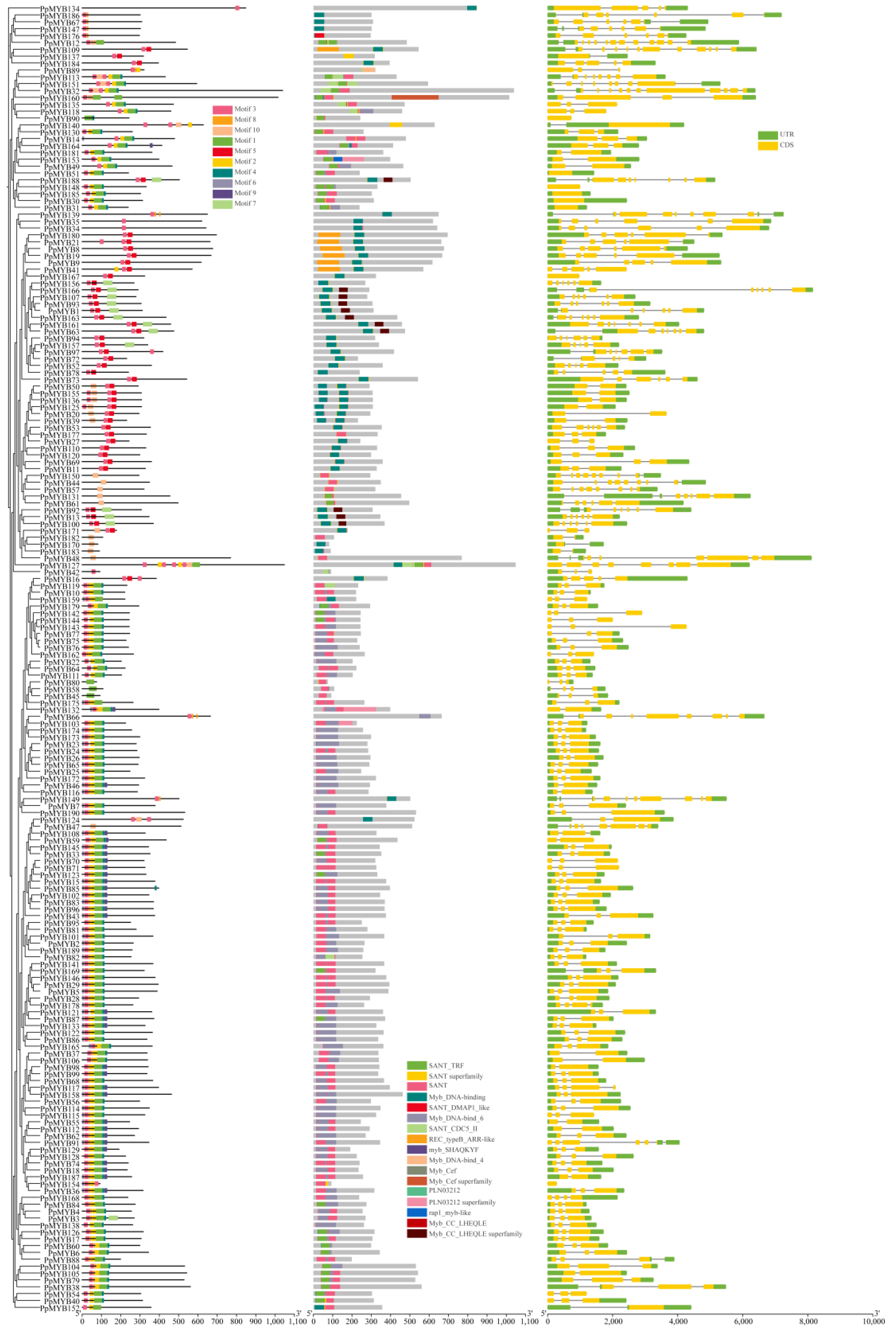
3.5. Motif Analysis, Domain, and Gene Structure Analysis of PpMYB Family Genes
3.6. Analysis of Cis-Acting Elements in the Promoters of MYB Family Genes
3.7. Identification of PpMYB Involved in Carotenoid Biosynthesis
3.8. PpMYB Regulates the Expression of Key Genes for Carotenoid Biosynthesis
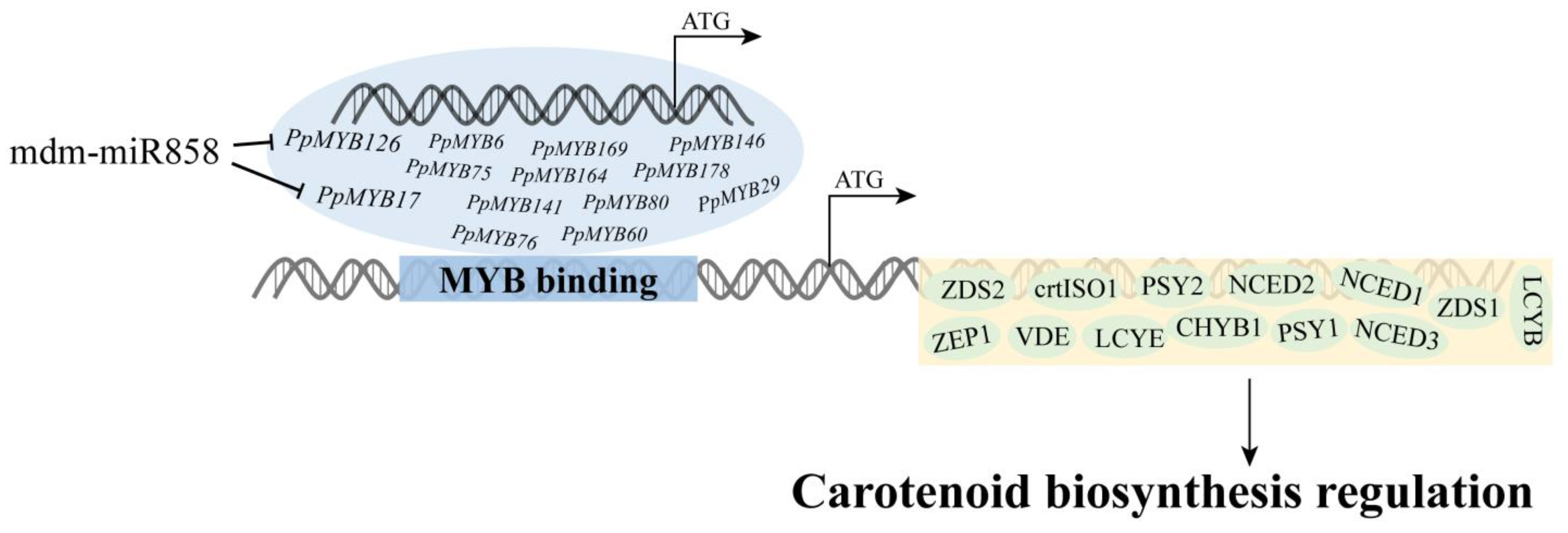
3.9. miRNA Targeting PpMYB Involved in Carotenoid Biosynthesis
4. Discussion
4.1. Bioinformatics Analysis of the Peach MYB Family
4.2. PpMYB Is Involved in Carotenoid Biosynthesis
4.3. miRNA Targets PpMYB Involved in the Regulation of Carotenoid Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Q.; Yang, S.; Zhang, R.; Liu, S.; Zhang, C.; Li, Y.; Li, J. Characterization of honey peach (Prunus persica (L.) Batsch) aroma variation and unraveling the potential aroma metabolism mechanism through proteomics analysis under abiotic stress. Food Chem. 2022, 386, 132720. [Google Scholar] [CrossRef] [PubMed]
- Serra, S.; Anthony, B.; Masia, A.; Giovannini, D.; Musacchi, S. Determination of Biochemical Composition in Peach (Prunus persica L. Batsch) Accessions Characterized by Different Flesh Color and Textural Typologies. Foods 2020, 9, 1452. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Fan, J.; Li, Y.; Cao, K.; Chen, C.; Wang, X.; Fang, W.; Zhu, G.; Wang, L. Characterizing of carotenoid diversity in peach fruits affected by the maturation and varieties. J. Food Compos. Anal. 2022, 113, 104711. [Google Scholar] [CrossRef]
- Kim, J.; Kwon, S.; Jang, M.; Lee, J.; Kim, G.; Kang, H.; Hwang, I. Analysis of Ascorbic Acid, Anthocyanin and Carotenoid Contents of Parts from Selected Peach Cultivars. J. Korean Soc. Food Sci. Nutr. 2021, 50, 962–970. [Google Scholar] [CrossRef]
- Zheng, J.; Yang, X.; Ye, J.; Su, D.; Wang, L.; Liao, Y.; Zhang, W.; Wang, Q.; Chen, Q.; Xu, F. Multiomics analysis provides new insights into the regulatory mechanism of carotenoid biosynthesis in yellow peach peel. Mol. Hortic. 2023, 3, 23. [Google Scholar] [CrossRef]
- Fraser, P.; Bramley, P. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Jiang, J.G.; Chen, Q. Progress on molecular breeding and metabolic engineering of biosynthesis pathways of C(30), C(35), C(40), C(45), C(50) carotenoids. Biotechnol. Adv. 2007, 25, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Yabuzaki, J. Carotenoids Database: Structures, chemical fingerprints and distribution among organisms. Database 2017, 2017, bax004. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, E.; Uppal, S.; Rogozin, I.; Gentleman, S.; Redmond, T. Evolutionary aspects and enzymology of metazoan carotenoid cleavage oxygenases. Biochim. Biophys. Acta 2020, 1865, 158665. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T. Recent progress in structural studies of carotenoids in animals and plants. Arch. Biochem. Biophys. 2009, 483, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Domonkos, I.; Kis, M.; Gombos, Z.; Ughy, B. Carotenoids, versatile components of oxygenic photosynthesis. Prog. Lipid Res. 2013, 52, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Zhang, J.; Nageswaran, D.; Li, L. Carotenoid metabolism and regulation in horticultural crops. Hortic. Res. 2015, 2, 15036. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Uragami, C.; Cogdell, R. Carotenoids and Photosynthesis. Sub-Cell. Biochem. 2016, 79, 111–139. [Google Scholar]
- Howitt, C.; Pogson, B. Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ. 2006, 29, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Hannoufa, A.; Soroka, J.; Xu, N.; Li, X.; Zebarjadi, A.; Gruber, M. Enhanced β-ionone emission in Arabidopsis over-expressing AtCCD1 reduces feeding damage in vivo by the crucifer flea beetle. Environ. Entomol. 2011, 40, 1622–1630. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.; Pogson, B. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Cazzonelli, C. Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 2011, 38, 833–847. [Google Scholar] [CrossRef] [PubMed]
- Eggersdorfer, M.; Wyss, A. Carotenoids in human nutrition and health. Arch. Biochem. Biophys. 2018, 652, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Li, L. Carotenoid metabolism: Biosynthesis, regulation, and beyond. J. Integr. Plant Biol. 2008, 50, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Jahns, P.; Holzwarth, A. The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim. Biophys. Acta 2012, 1817, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Perreau, F.; Frey, A.; Effroy-Cuzzi, D.; Savane, P.; Berger, A.; Gissot, L.; Marion-Poll, A. ABSCISIC ACID-DEFICIENT4 Has an Essential Function in Both cis-Violaxanthin and cis-Neoxanthin Synthesis. Plant Physiol. 2020, 184, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Tadmor, Y.; Li, L. Pathways for Carotenoid Biosynthesis, Degradation, and Storage. Methods Mol. Biol. 2020, 2083, 3–23. [Google Scholar] [PubMed]
- Stanley, L.; Yuan, Y. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed]
- Hermanns, A.; Zhou, X.; Xu, Q.; Tadmor, Y.; Li, L. Carotenoid pigment accumulation in horticultural plants. Hortic. Plant J. 2020, 6, 343–360. [Google Scholar] [CrossRef]
- Sun, T.; Li, L. Toward the ‘golden’ era: The status in uncovering the regulatory control of carotenoid accumulation in plants. Plant Sci. 2020, 290, 110331. [Google Scholar] [CrossRef] [PubMed]
- Meng, N.; Wei, Y.; Gao, Y.; Yu, K.; Cheng, J.; Li, X.; Duan, C.; Pan, Q. Characterization of Transcriptional Expression and Regulation of Carotenoid Cleavage Dioxygenase 4b in Grapes. Front. Plant Sci. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Zeng, Y.; Meng, Q.; Guan, Y.; Li, C.; Yang, H.; Zhang, Y.; Ampomah-Dwamena, C.; Liu, P.; Chen, C.; et al. Red light-induced kumquat fruit coloration is attributable to increased carotenoid metabolism regulated by FcrNAC22. J. Exp. Bot. 2021, 72, 6274–6290. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A fruit ripening-associated transcription factor CsMADS5 positively regulates carotenoid biosynthesis in citrus. J. Exp. Bot. 2021, 72, 3028–3043. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A Citrus Phosphate Starvation Response Factor CsPHL3 Negatively Regulates Carotenoid Metabolism. Plant Cell Physiol. 2021, 62, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The Citrus Transcription Factor CsMADS6 Modulates Carotenoid Metabolism by Directly Regulating Carotenogenic Genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shen, Y.; Zhou, P.; Fatima, M.; Lin, J.; Yue, J.; Zhang, X.; Chen, L.; Ming, R. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res. 2019, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 16861. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Luo, T.; Liu, C.; Wang, Y.; Yang, H.; Yang, W.; Zheng, L.; Xiao, X.; Zhang, M.; Xu, R.; et al. An R2R3-MYB transcription factor represses the transformation of α- and β-branch carotenoids by negatively regulating expression of CrBCH2 and CrNCED5 in flavedo of Citrus reticulate. New Phytol. 2017, 216, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, J.; Stanley, L.; LaFountain, A.; Frank, H.; Liu, C.; Yuan, Y. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Thrimawithana, A.; Dejnoprat, S.; Lewis, D.; Espley, R.; Allan, A. A kiwifruit (Actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 2019, 221, 309–325. [Google Scholar] [CrossRef] [PubMed]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genom. 2017, 18, 225. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Yates, A.D.; Allen, J.; Amode, R.M.; Azov, A.G.; Barba, M.; Becerra, A.; Bhai, J.; Campbell, L.I.; Carbajo Martinez, M.; Chakiachvili, M.; et al. Ensembl Genomes 2022: An expanding genome resource for non-vertebrates. Nucleic Acids Res. 2022, 50, D996–D1003. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- He, Z.; Zhang, H.; Gao, S.; Lercher, M.J.; Chen, W.H.; Hu, S. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Fan, C.; Wei, Y.; Meng, J.; Li, Z.; Zhong, C. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guo, T.; Li, J.; Chen, Z.; Guo, B.; An, X. Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int. J. Biol. Macromol. 2021, 191, 359–376. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhang, S.; Wang, R.; Zhang, R.; Hao, Y. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 2013, 8, e69955. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Dong, Q.; Wu, J.; Li, H.; Luan, H.; Jia, P.; Zhang, X.; Guo, S.; Yang, M.; Qi, G. Genome-wide analysis of the R2R3-MYB transcription factor gene family expressed in Juglans regia under abiotic and biotic stresses. Ind. Crops Prod. 2023, 198, 116709. [Google Scholar] [CrossRef]
- Wilkins, O.; Nahal, H.; Foong, J.; Provart, N.; Campbell, M. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 2009, 149, 981–993. [Google Scholar] [CrossRef]
- Li, P.; Xia, E.; Fu, J.; Xu, Y.; Zhao, X.; Tong, W.; Tang, Q.; Tadege, M.; Fernie, A.R.; Zhao, J. Diverse roles of MYB transcription factors in regulating secondary metabolite biosynthesis, shoot development, and stress responses in tea plants (Camellia sinensis). Plant J. 2022, 110, 1144–1165. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Wang, Z.; Wang, Y.; Wang, C.; Zhu, B.; Liu, H.; Ji, W.; Wen, J.; Chu, C.; Tadege, M.; et al. The MYB Activator WHITE PETAL1 Associates with MtTT8 and MtWD40-1 to Regulate Carotenoid-Derived Flower Pigmentation in Medicago truncatula. Plant Cell 2019, 31, 2751–2767. [Google Scholar] [CrossRef]
- Wu, G. Plant microRNAs and development. J. Genet. Genom. 2013, 40, 217–230. [Google Scholar] [CrossRef]
- Xu, Q.; Liu, Y.; Zhu, A.; Wu, X.; Ye, J.; Yu, K.; Guo, W.; Deng, X. Discovery and comparative profiling of microRNAs in a sweet orange red-flesh mutant and its wild type. BMC Genom. 2010, 11, 246. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Ju, Z.; Cao, D.; Zhai, B.; Qin, G.; Zhu, H.; Fu, D.; Luo, Y.; Zhu, B. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 2015, 13, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Koul, A.; Yogindran, S.; Sharma, D.; Kaul, S.; Rajam, M.; Dhar, M. Carotenoid profiling, in silico analysis and transcript profiling of miRNAs targeting carotenoid biosynthetic pathway genes in different developmental tissues of tomato. Plant Physiol. Biochem. 2016, 108, 412–421. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, W.; Chen, Q.; Zhang, S.; Mei, Z.; Yu, L.; Wang, C.; Mao, Z.; Chen, Z.; Chen, X.; et al. Mdm-miR858 targets MdMYB9 and MdMYBPA1 to participate anthocyanin biosynthesis in red-fleshed apple. Plant J. 2023, 113, 1295–1309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Yang, H.J.; Qu, D.; Zhu, Z.Z.; Yang, Y.Z.; Zhao, Z.Y. The MdBBX22-miR858-MdMYB9/11/12 module regulates proanthocyanidin biosynthesis in apple peel. Plant Biotechnol. J. 2022, 20, 1683–1700. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Badola, P.K.; Bhatia, C.; Sharma, D.; Trivedi, P.K. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 2020, 6, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Q.; Liu, X.F.; Zhu, Y.J.; Zhu, J.Z.; Liu, C.; Wang, Z.Y.; Shen, X.X.; Allan, A.C.; Yin, X.R. Identification of miRNA858 long-loop precursors in seed plants. Plant Cell 2023, 36, 1637–1654. [Google Scholar] [CrossRef] [PubMed]
- Premachandran, Y. Triggered in distress: A miRNA-controlled switch for drought-induced ABA biosynthesis in rice. Plant Physiol. 2022, 189, 447–449. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Zheng, J.; Yi, Y.; Yang, X.; Jiang, L.; Ye, J.; Zhang, W.; Xu, F. Genome-Wide Identification of MYB Gene Family in Peach and Identification of MYBs Involved in Carotenoid Biosynthesis. Forests 2024, 15, 1119. https://doi.org/10.3390/f15071119
Liu F, Zheng J, Yi Y, Yang X, Jiang L, Ye J, Zhang W, Xu F. Genome-Wide Identification of MYB Gene Family in Peach and Identification of MYBs Involved in Carotenoid Biosynthesis. Forests. 2024; 15(7):1119. https://doi.org/10.3390/f15071119
Chicago/Turabian StyleLiu, Fengyi, Jiarui Zheng, Yuwei Yi, Xiaoyan Yang, Leiyu Jiang, Jiabao Ye, Weiwei Zhang, and Feng Xu. 2024. "Genome-Wide Identification of MYB Gene Family in Peach and Identification of MYBs Involved in Carotenoid Biosynthesis" Forests 15, no. 7: 1119. https://doi.org/10.3390/f15071119
APA StyleLiu, F., Zheng, J., Yi, Y., Yang, X., Jiang, L., Ye, J., Zhang, W., & Xu, F. (2024). Genome-Wide Identification of MYB Gene Family in Peach and Identification of MYBs Involved in Carotenoid Biosynthesis. Forests, 15(7), 1119. https://doi.org/10.3390/f15071119





