1. Introduction
Adventitious roots are produced from non-middle column sheath tissues such as stems or leaves of plants [
1]. The occurrence of adventitious roots is a critical factor for successful asexual propagation of plants. The difficulty of rooting in histocultured seedlings of woody plants hinders the construction of a regeneration system of tissue culture plants and the rapid multiplication technology. The formation and development of adventitious roots are regulated by multiple genes and transcription factors, which mainly include
WOX,
PIN,
ARF,
ARL,
CRL, and
SHR [
2]. The WUSCHEL-related homeobox (WOX) is a family of plant-specific transcription factors, and the prototypical members of the plant-specific WUSCHEL-related homeobox (WOX) protein family, all of which contain the conserved structural domains of the homology box (HB) and are part of the HB superfamily [
3]. The structural domains of the
WOX gene family have a helix-loop-helix-turn-helix (HTH) structure, which contains 60–66 amino acids essential for specific functions in plants [
4]. Based on the phylogenetic analysis of WOX proteins, the WOX family is classified into three clades: the ancient clade, the intermediate clade, and the modern/WUS clade [
4]. Previous studies have demonstrated that
WOX genes play an important role in plant growth and development. The
WOX gene family is involved in developmental processes such as plant embryonic development, maintenance of stem cells, and formation of various organs, as well as in the formation and maintenance of healing tissue [
5,
6], particularly during the critical rooting period, when this gene family plays an important regulatory role. In
Rosa canina L.,
RcWOX1 play a pivotal role in auxin-induced rhizoid formation [
7].
AtWOX4 is also a
MC-WOX gene and primarily regulates stem cell maintenance of CAM through the
CLE41/
CLE44-
PXY-
WOX4/
WOX14 pathway [
8].
AtWOX5 is specifically expressed at the initiation of root primordia in
Arabidopsis thaliana and is a marker gene for the quiescent center (QC) [
1].
AtWOX13 is a key regulator of healing tissue formation in
Arabidopsis and promotes the initiation of primordial and lateral roots [
9,
10,
11]. In woody plants, the
JsWOX1 gene plays a role in root primordia initiation in
Jasminum sambac, and the overexpression of this gene increases the number of healing tissues and healing tissue rooting [
12]. The
PtoWOX5a gene is specifically expressed in
Populus tomentosa at the tip of adventitious roots and lateral roots. This gene can regulate the development of adventitious roots in poplar plants. Phenotypic complementation experiments have demonstrated that
PtoWOX5a can functionally complement
AtWOX5 in QC cells [
13]. The genes
PeWOX11a and
PeWOX11b in
P. tomentosa play important roles in adventitious root genesis and the morphogenesis process [
14]. The transgenesis of
JrWOX11 from
Juglans sp. into poplar plants increased root hair length and the number of adventitious roots. Therefore, it was hypothesized that the
JrWOX11 gene may regulate root development and growth [
15].
Eucalyptus is a dicotyledonous plant of Myrtaceae, which is the most commonly planted artificial forest tree species in the world. It is characterized by strong adaptability, rapid growth, short rotation period, various species, and strong disease resistance [
16]. Tissue culture technology is an important means of forest genetic breeding that has been widely used in the expansion, popularization, and molecular breeding of eucalyptus varieties. Following the completion of whole-genome sequencing of different plants and the accumulation of many DNA and protein sequences in databases such as NCBI and Phytozome, the completion of whole-genome sequencing of the
E. grandis genome in 2011 greatly promoted the development of molecular biology of eucalyptus [
17]. On the other hand, eucalyptus is a perennial woody plant, and the hybrid progeny generated after artificial heterogametic pollination is likely to segregate and have a certain degree of variability. Rooting is critical to establish a rapid multiplication system in histoculture. The results of rooting vary with the differences in the reproductive ability of hybrid offspring. The study of the role of the
WOX gene family in the rooting process of asexual lines of eucalyptus hybrid progeny can help to understand the variability and conservatism in the growth and development of the progeny. In the present study, the biological information of the
WOX gene family members from
Corymbia citriodora,
E. pellita,
E. urophylla ×
E. grandis, and
E. grandis was systematically analyzed, and the protein structures of
E. urophylla ×
E. grandis and
E. grandis members were determined by qRT-PCR. In the different organs of rooted seedlings of
E. urophylla ×
E. pellita and at different stages of adventitious root development, the key
WOX genes related to the growth and development of adventitious roots of eucalyptus were screened, and their biological functions in rooting were hypothesized. The present study provides theoretical guidance for improving the rooting efficiency and reproductive success rate of excellent clones of eucalyptus by using the
WOX gene family members for genetic transformation in the later stage.
3. Results
3.1. Identification and Physicochemical Properties of the WOX Family Proteins in Eucalyptus
The amino acid sequence analysis of the encoded proteins of 31 eucalyptus
WOX genes (8
EpWOX, 10
EgWOX, 5
CcWOX, and 7
EugWOX genes) showed that the
WOX genes of the four eucalyptus species encoded 165–408 amino acids, with a wide variation in the number of amino acids. EpWOX11 showed the longest amino acid sequence, while EugWOX3 exhibited the shortest amino acid sequence. The molecular weight of the proteins ranged from 19,471.2 to 41,402.77 Da. The theoretical isoelectric point (pI) of the 31 eucalyptus WOX proteins ranged from 4.76 to 11, and most of the proteins were alkaline. EgWOX11, EgWOX13.1, EgWOX13.2, CcWOX5, CcWOX2, CcWOX13, and EugWOX11 protein have a pI of <7 and are acidic protein. The isoelectric point of the protein is associated with the composition and structure of amino acids, and the hydroxyl group of the side chain of the amino acids that constitute the acidic protein has a stronger hydroxyl dissociation ability than the amino group. The instability coefficients of the 31 proteins ranged from 48.40 to 87.80, thus indicating that all of them were unstable. No signal peptide region was detected, and the subcellular localization predictions showed that all the proteins were localized in the nucleus (
Table S3). This finding was consistent with the subcellular localization characteristics of transcription factors that participate in the transcriptional regulation of genes in the nucleus and play biological roles.
3.2. Protein Structure Analysis of the WOX Gene Family in Eucalyptus
The predicted secondary structures of the 31 WOX proteins showed that the proteins contained α-helices, extended strands, irregular curls, and β-turns, with irregular curls constituting the highest percentage (36.36%–69.72%), followed by α-helices (13.08%–44.44%). Thus, the major secondary structural elements of the eucalyptus WOX proteins were irregular curls and α-helices (
Table S4). The main secondary structure components of EugWOX1 were extended chains and random curls. α-Helix and β-helix were the ordered structures of the proteins with high stability and random curls were the disordered structures of the protein. The highest percentage of random curls in the 31 eucalyptus proteins was consistent with the results of the instability coefficient analysis of their physicochemical properties.
3.3. Phylogenetic Analysis of the WOX Family Proteins
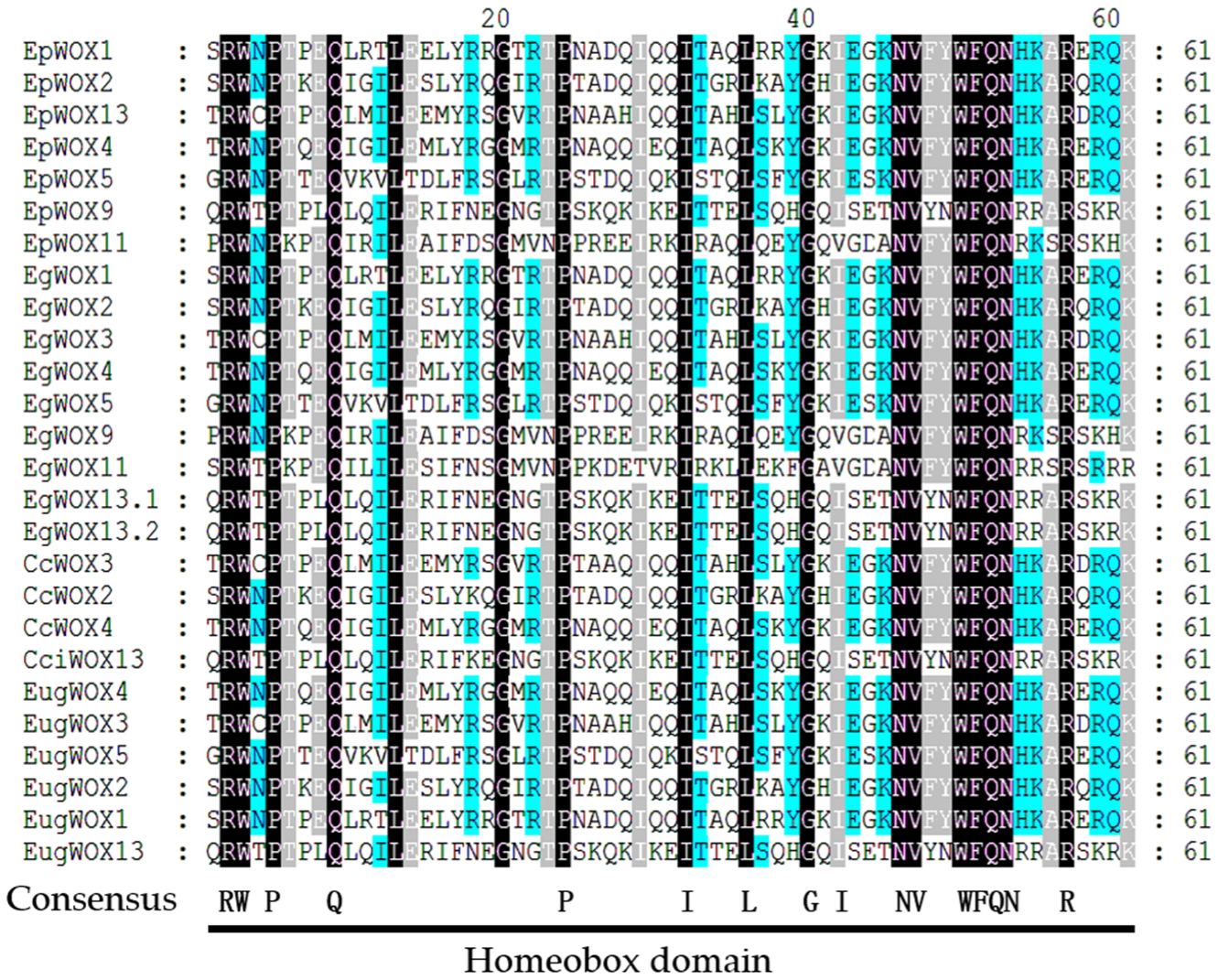
Multiple sequence comparison of the WOX proteins from
E. pellita,
E. grandis,
C. citriodora, and
E. urophylla ×
E. grandis (
Figure 1) indicated that the WOX family proteins from these plants contain the homeobox structural domain. These proteins also contain the 13 conserved sites previously reported [
20]. These conserved sites include the Q and L sites of Helix 1, the G site of Loop, the P and L sites of Helix 2, the G site of Turn, and the N, V, W, F, Q, N, and R sites of Helix 3. Remarkably, these conserved sites are found across different members, thus indicating a high level of conservation among the eucalyptus WOX family proteins. By protein sequence alignment, the similarity between EpWOX11 and EgWOX11 was 99.64%, the similarity between EugWOX11 and EgWOX11 was 100%, and the similarity between CcWOX4 and EgWOX4 was 90%. The similarity of WOX sequences among different eucalyptus species was high.
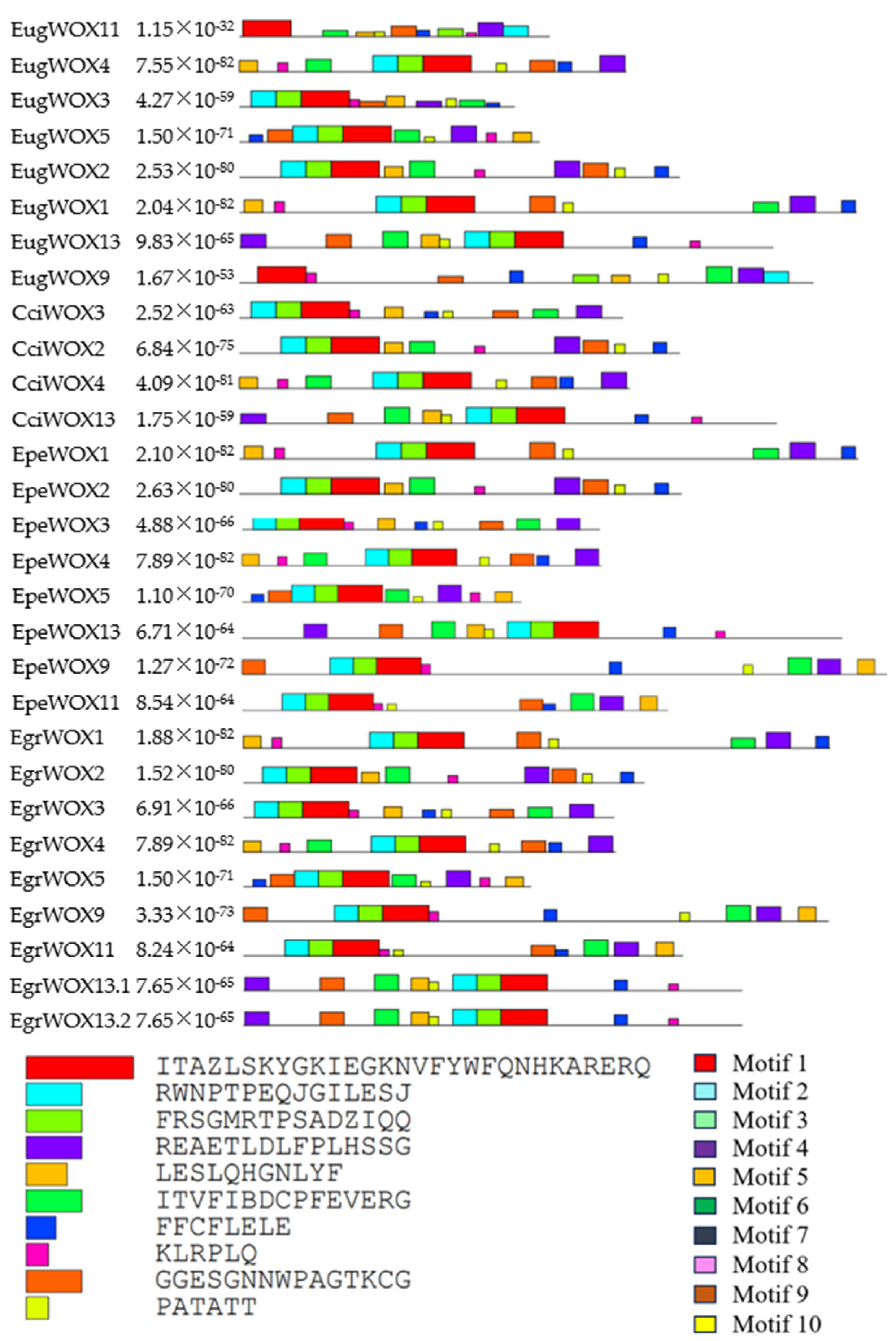
To further investigate the diversity of the eucalyptus
WOX genes, we analyzed WOX protein motifs using the MEME online server. Ten conserved motifs were identified, i.e., motifs 1 to 10 (
Figure 2). An overview of these protein motifs is presented in
Figure S1. All
WOX genes shared the same motifs.
On the basis of domain sequences and referring to the grouping of
WOX family members of
E. grandis [
16], a phylogenetic tree was constructed from the WOX amino acid sequences of 10
E. grandis, 8
E. urophylla ×
E. grandis, 8
E. pellita, and 4
C. citriodora by matching with the WOX amino acid sequences of 12
A. thaliana, 12
P. tomentosa, and 8
P. trichocarpa. Three distinct groups were identified among the 62 WOX protein sequences, which corresponded to the modern, intermediate, and ancestral evolution of plant WOX proteins. Moreover, the evolutionary relationships of
E. pellita,
E. grandis,
C. citriodora, and
E. urophylla ×
E. grandis were similar in the three evolutionary branches (
Figure 3).
These findings indicate a close evolutionary relationship among the WOX proteins of the studied eucalyptus species. In the ancestral evolutionary branch, EgWOX13.1, EgWOX13.2, EugWOX13, EpWOX13, and CcWOX13 exhibited similar evolutionary relationships, with the amino acid sequence of the WOX13 protein being more conserved among E. pellita, E. grandis, C. citriodora, and E. urophylla × E. grandis. In the intermediate evolutionary branch, the amino acid sequences of EugWOX11, EugWOX11, and EpWOX11 were also more conservative. Specifically, the amino acid sequences of the WOX11 protein were more homologous among E. pellita, E. grandis, and E. urophylla × E. grandis. Additionally, the amino acid sequences of EugWOX9, EgWOX9, and EpWOX9 were similar to those of PtoWOX9a and PtoWOX9b, thus indicating greater conservation of the WOX9 protein among the different species. Finally, in the modern evolutionary branch, the amino acid sequences of each WOX9 protein were more conserved among the different species. The amino acid sequences encoded by each WOX gene member were more homologous than those from different species. Furthermore, compared to Arabidopsis, the amino acid sequences of each WOX protein of P. tomentosa and P. trichocarpa showed a higher degree of similarity to those of the WOX proteins of eucalyptus. The amino acid sequences of WOX proteins among woody plants exhibited similarity.
Furthermore, a comparative analysis of the amino acid sequences of WOX proteins among the four eucalyptus species revealed that the amino acid sequences of the individual WOX proteins of E. grandis and E. urophylla × E. grandis exhibited the closest genetic distances. Based on this finding, we predicted that the genomes and proteins of species belonging to the same genus between eucalyptus hybrids and their parent species are more similar than between different genus and the amino acid sequences of WOX proteins were more conserved.
3.4. Expression Patterns of the EupWOX Genes in Different Tissues of E. urophylla × E. pellita
The root development process in plants can be divided into two stages: the induction stage and the formation stage [
14,
17]. Based on the observation of the actual growth (
Figure 3), we found that 10 single asexual
E. urophylla ×
E. pellita group-cultivated seedlings had a healing time ranging from 0 d to 4 d (
Figure 4A,B), adventitious rooting between 5 d and 6 d (
Figure 4C), and adventitious root elongation and lateral root growth between 7 d and 15 d (
Figure 4D–F). In the present study, the rooting process was grouped into three periods: the period of root primordia induction (0–4 d), the adventitious root formation period (5–6 d), and the adventitious root elongation and adventitious root growth period (7–15 d). This sequence of events enables us to comprehensively understand the root development process.
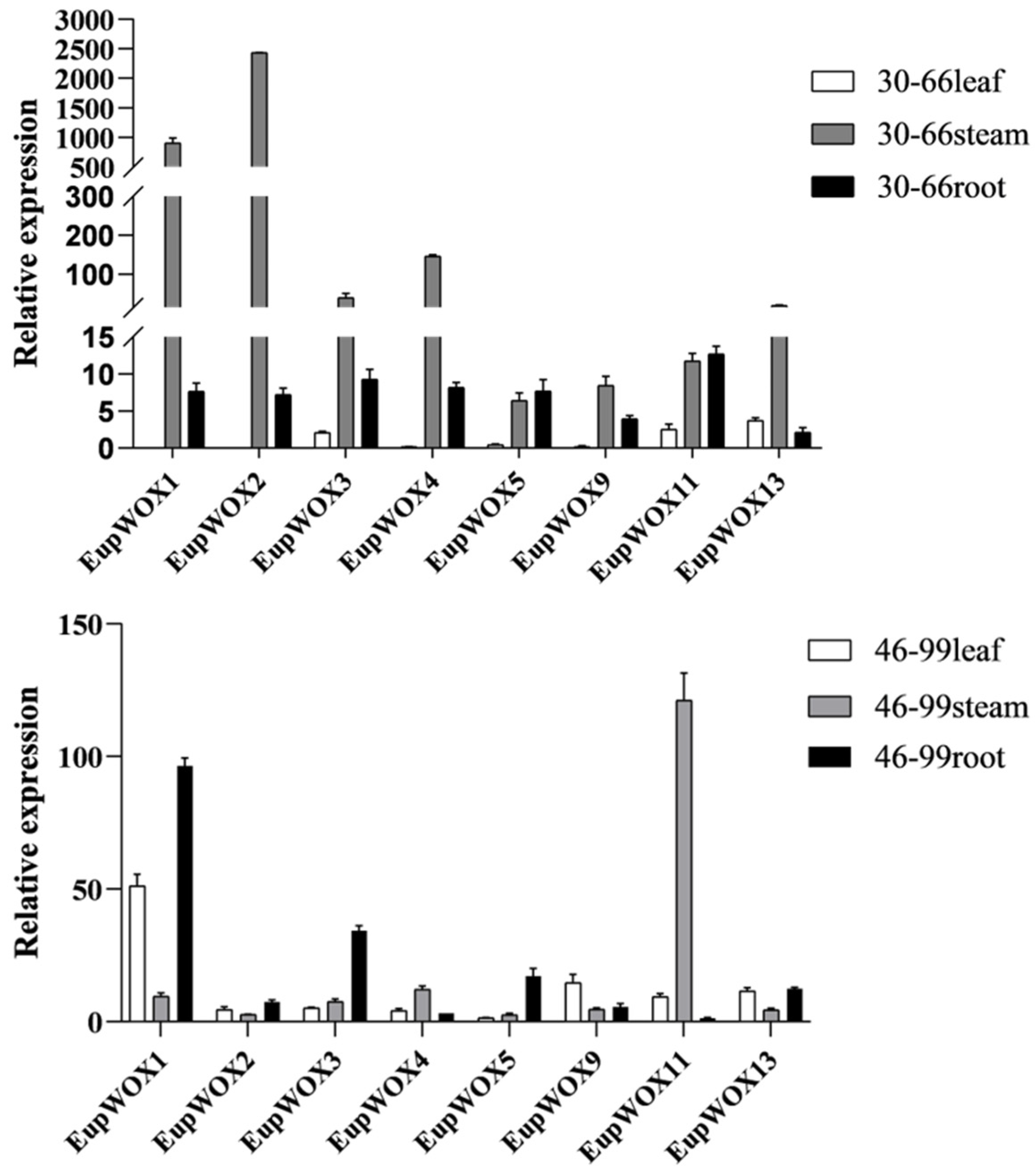
The relative expression trends of the eight
EupWOX genes in the leaves, stem segments, and roots (including rooted healing tissue) of the
E. urophylla ×
E. pellita 18H123 progeny from a single asexual line 30–66 and 46–99 (
Figure 5) showed remarkable differences in the expression patterns of the different members of the
EupWOX genes in different tissues of
E. urophylla ×
E. pellita group-cultivated seedlings. The rooting effect of clones 30–66 was poor and the rooting effect of clones 46–99 was tiptop. The analysis revealed that the
EupWOX genes showed high expression levels in the roots of the 18H123 progeny from a single asexual line 30–66 and 46–99, which exhibited a fair degree of rooting.
EupWOX1,
EupWOX3,
EupWOX5, and
EupWOX13 are highly expressed in the roots of 46–99 and
EupWOX11 are highly expressed in the roots of 30–66.
WOX11 can promote the rooting process [
15], and the rooting rate of line 30–66 is better, which may be related to the high expression of
EupWOX11. The findings also indicated that
EupWOX13,
EupWOX11, and
EupWOX3 showed relatively high expression in leaves, while
EupWOX2 and
EupWOX1 exhibited high expression levels in stems.
EupWOX1,
EupWOX5, and
EupWOX13were expressed in the roots of line 46–99 with excellent rooting condition, thus suggesting a positive role of
EupWOX1 in the rooting development of 46–99. On the other hand,
EupWOX2 and
EupWOX1 showed relatively high expression in leaves, and
EupWOX11 exhibited relative high expression in stems. Overall, based on these results, we hypothesized that
EupWOX1,
EupWOX5,
EupWOX11, and
EupWOX13 played an active role in the rooting development of adventitious roots of
E. urophylla ×
E. pellita group-cultured seedlings.
3.5. Expression Analysis of the EupWOX Family Genes at Different Rooting Stages
Based on the expression characteristics of the eight
EupWOX genes in three different organs, previous studies have indicated that
EupWOX1 and
EupWOX11 are specifically expressed in roots and could serve as the key genes that regulate adventitious rooting in
E. urophylla ×
E. pellita group-cultured seedlings. Furthermore, WOX5 and WOX13 have been shown to play key roles in rooting stage in plants such as
A. thaliana and
P. tremula [
9,
13,
21]. Therefore, in the present study,
WOX1,
WOX5,
WOX11, and
WOX13 were selected as the candidate genes for assessment in the asexual group culture seedlings of the dominant monocot of 10 inbred progenies of the
E. urophylla ×
E. pellita full-sibling line 18H123 at different rooting stages. The relative expression levels of these genes were determined at multiple time points at different rooting stages.
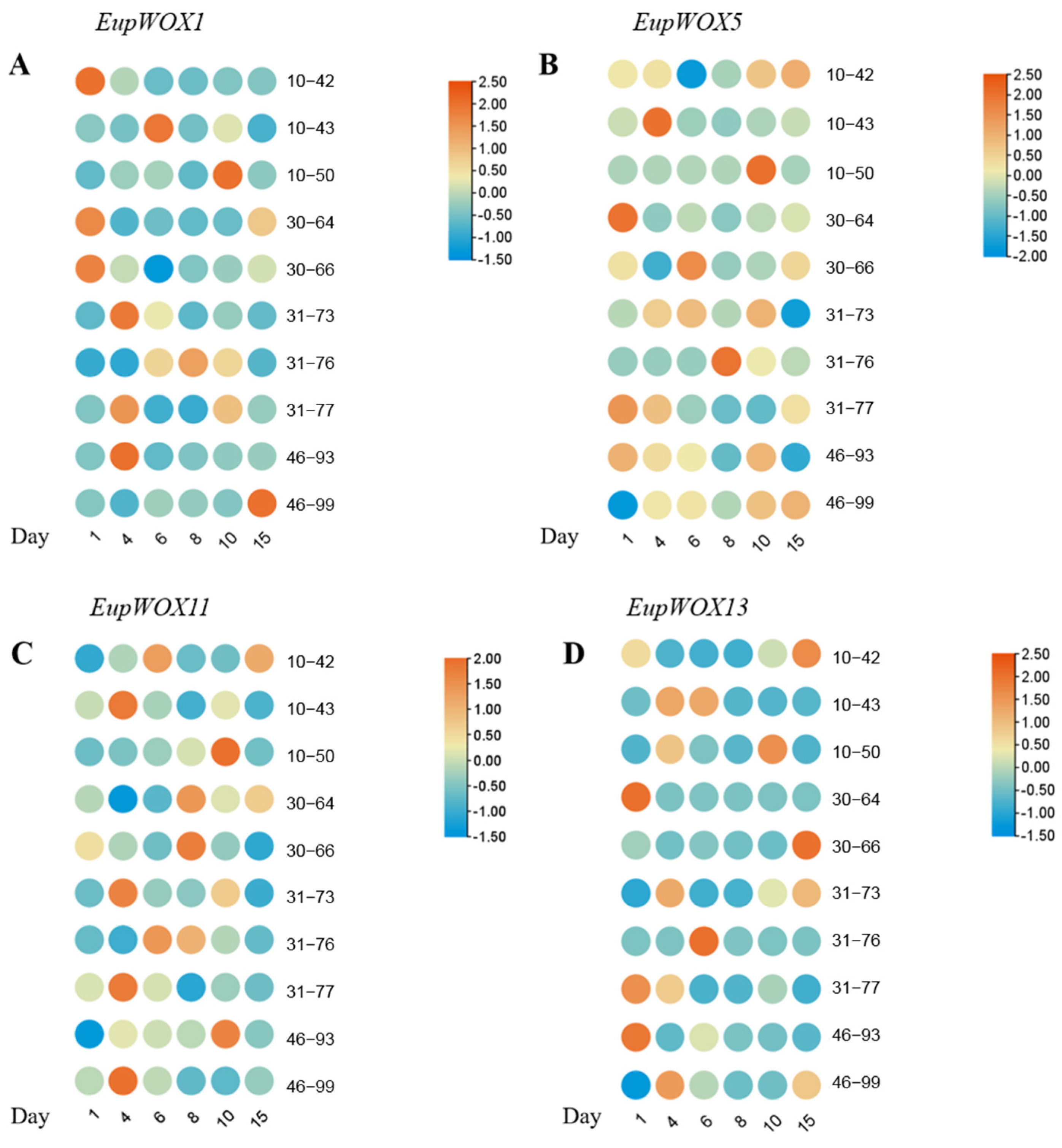
Significant differences were observed in the relative expression levels of the different
EupWOX genes at different time points and different rooting stages (
Figure 6). The heatmap in
Figure 6 was obtained after normalization of the columns, where the relative expression of each gene obtained at different times through the 10 asexual lines was normalized to obtain an array. Combined with the rooting after 24 d (
Table 1), the expression of four
EupWOX (
EupWOX1,
5,
11, and
13) was significantly different among lines, and significant differences were observed in the rooting rate, the average number of roots, and the average root length of each asexual line. Among the 18H123 family lines, 46–93 had the highest rooting rate (100%), 46–99 had the highest average number of roots (4.14), and 10–42 had significantly better average root lengths than the other nine asexual lines, with the average root lengths of 46–99 and 46–93 being 2.38 and 2.32 cm, respectively. Collectively, the best rooting effect was found in line 46–93, the asexual lines with significantly better rooting effect results were 10–42 and 46–99, and those with significantly poorer rooting effect results were 10–43, 31–76, and 31–77.
The formation of root primordia is a critical step in the rooting process of asexual tissue culture seedlings of woody plants. The root primordium is derived from the orderly division of meristem cells in the callus after dedifferentiation, and adventitious rooting starts from the induction of the root primordium [
22]. During the root primordium induction stage (0–4 d), in lines with a good rooting effect (10–42, 46–93, and 46–99),
EupWOX1 exhibited high expression in 10–42 and 46–93,
EupWOX11 in 46–93 and 46–99, and both
EupWOX5 and
EupWOX13 showed high expression levels. In contrast, in lines with a poor rooting effect (10–43 and 31–76), these four genes showed low expression or were barely expressed at the root primordia induction stage in 31–76, while
EupWOX5,
EupWOX11, and
EupWOX13 showed high expression at the root primordia induction stage in 10–43. The significantly lower rooting rate in 10–43 might be related to the low expression level of
EupWOX1 at the root primordia induction stage in conjunction with the remarkably higher rooting rate of
EupWOX1. This is combined with the observation that
EupWOX1 was highly expressed in the root induction phase of the asexual lines with substantially higher rooting rates (10–42, 30–64, 31–73, and 46–93). The difference between 46–93 and 46–99 was only in the rooting rate. Because 46–93 had a higher expression of
EupWOX1 at this rooting stage, while 46–99 had a lower expression, we hypothesized that
EupWOX1 might play a critical role in the root primordia induction and was correlated with the rooting rate of
E. urophylla ×
E. pellita.
During the adventitious root formation period (5–6 d), in well-rooted lines (10–42, 46–93, and 46–99), EupWOX5 was highly expressed in 46–93 and 46–99, EupWOX11 was highly expressed in 10–42 and 46–99, and both EupWOX1 and EupWOX13 showed low expression. In lines with the poor rooting effect (10–43 and 31–76), EupWOX1 and EupWOX13 showed high expression in 10–43, EupWOX11 and EupWOX13 exhibited high expression in 31–76, and EupWOX5 showed low expression. Based on these findings, we hypothesized that EupWOX1 and EupWOX13 were not highly correlated with root formation and that EupWOX5 and EupWOX11 played a crucial role in the root formation process.
During the period of adventitious root elongation and adventitious root growth (7–15 d), in lines with a good rooting effect (10–42, 46–93, and 46–99), EupWOX1 exhibited low expression in 46–93 and 10–42 and high expression at 15 d in 46–99, EupWOX5 showed high expression in both 46–93 and 10–42, EupWOX11 exhibited high expression in both 10–42 and 46–93 but low expression in 46–99, and EupWOX13 exhibited high expression in both 10–42 and 46–99 but low expression in 46–93. In lines with a poor rooting effect (10–43 and 31–76), EupWOX1, EupWOX5, and EupWOX13 showed low expression, and EupWOX11 exhibited low expression in 10–43 but high expression in 31–76 at 8 d. Based on these findings, we hypothesized that EupWOX1, EupWOX5, and EupWOX13 could be the key regulators of adventitious root elongation and adventitious lateral root growth and development in E. urophylla × E. pellita group-cultured seedlings.
In summary, we found that different EupWOX genes played roles in the different stages of adventitious root growth and development of E. urophylla × E. pellita tissue culture seedlings. We hypothesized that EupWOX1, EupWOX5, EupWOX11, and EupWOX13 had spatial and temporal expression variability and that EupWOX1 might be a key regulator of root primordium induction that has a critical relationship with the rooting rate. Furthermore, EupWOX5 and EupWOX11 might be key regulators of adventitious root formation, and EupWOX1 and EupWOX13 might function as key regulators of adventitious root elongation and adventitious lateral root growth. EupWOX11 may also play the role of a key regulatory gene for adventitious root formation. EupWOX1, EupWOX5, and EupWOX13 may serve as key regulatory genes for adventitious root elongation and adventitious lateral root growth. In combination with the asexual lines possessing significantly higher average root numbers (10–42, 30–66, 46–93, and 46–99), EupWOX11 and EupWOX13 exhibited higher expression levels during the induction of root primordia, elongation of adventitious roots, and growth of adventitious lateral roots. Therefore, it was hypothesized that EupWOX11 and EupWOX13 may play a key role in the induction of the root primordial base and indefinite lateral root of tissue culture seedlings and may have an important relationship with the number of roots. Combined with clones with significantly longer average root length (10–42, 10–50, 30–66, 46–93, and 46–99), EupWOX5 was highly expressed during the root elongation period. This finding suggests that EupWOX5 is not only a key regulator of adventitious root formation but also a key regulator of root elongation in adventitious roots and adventitious lateral roots in E. urophylla × E. pellita asexual seedlings. Thus, EupWOX5 may function as a key regulatory gene not only in forming adventitious roots but also in elongating adventitious roots and lateral roots.
4. Discussion
Primary organ regeneration is an indirect plant regeneration process in plant tissue cultures, and adventitious root regeneration from primary roots mainly occurs from healing tissue [
23]. The root development of woody plants is a very complex process. Hence, it is crucial to understand the molecular mechanisms underlying primordial, lateral, and adventitious roots to improve the efficiency of the root regeneration system in histoculture and to conduct an in-depth study of plant regeneration [
4]. Numerous studies have revealed that the WOX gene family plays an important regulatory role in the critical period of plant rooting [
5,
6,
9,
10,
11,
12,
13,
17]. The
WOX gene family has been identified and analyzed in several species. However, to date, most of the studies have focused on herbaceous plants and few studies have been conducted on woody plants.
In the present study, protein sequence analysis of the obtained
WOX gene families of
E. pellita,
E. grandis,
C. citriodora, and
E. urophylla ×
E. grandis revealed that the nine WOX family members of eucalyptus were significantly fewer than those of
A. thaliana (15) [
5],
Dimocarpus longan (13) [
24],
Malus domestica (12) [
25], and
Capsicum annuum (10) [
26] in terms of numbers but similar to that of the WOX families of woody plants. Presumably, during biological evolution, the WOX family members did not undergo large-scale amplification events. This hypothesis is consistent with the findings for
D. longan and
Arabidopsis [
24,
27]. Based on the phylogenetic evolutionary tree analysis, the WOX family of eucalyptus,
P. tomentosa,
P. trichocarpa, and
A. thaliana was divided into the teleomorphic branch, intermediate branch, and WUS branch. The members located in the same branch may have similar functions. The number of members in the WUS branch was the highest, while that in the teleomorphic branch was the lowest. Furthermore, the number of members in the WUS branch gradually increased from that in the teleomorphic branch to that in the intermediate branch. This finding is consistent with the results of the studies that analyzed the genetic evolutionary relationship of WOX proteins in
Malus domestica [
25],
Capsicum annuum [
26], and
Broussonetia kazinoki ×
B. papyrifera [
28]. The amino acid sequences of the WOX proteins were more conserved between eucalyptus species sequences and were less divergent from those of the woody plants
P. tomentosa and
P. trichocarpa. However, the amino acid sequences differed more from those of
A. thaliana. This finding could be attributed to the greater variability of the WOX genes between herbaceous and woody plants. Analysis of the physicochemical properties of the proteins showed that all eucalyptus WOX family members encoded unstable proteins. This finding was consistent with the results of pepper [
26] and
Brassica napus [
29]. The pI of the 31 eucalyptus WOX proteins ranged from 4.76 to 11, and these proteins included both acidic and basic proteins. This result was consistent with that for
Plukenetia volubilis [
30],
Brassica napus [
26], paper mulberry [
28], and pepper [
26]. The differences in the physicochemical properties of the different members of the eucalyptus WOX family might be related to their different biological functions. All eucalyptus WOX proteins are localized in the nucleus. In paper mulberry,
BpWOX7,
BpWOX8, and
BpWOX10 are localized in both the nucleus and cytoplasm, while
BpWOX1,
BpWOX2,
BpWOX3,
BpWOX4,
BpWOX5,
BpWOX9, and
BpWUS are localized in the nucleus. In kale-type oilseed rape [
28],
BnWOX44 and
BnWOX49 are localized in chloroplasts and the remaining
WOX genes are localized in the nucleus [
29]. Thus, the subcellular localization of the
WOX genes is consistent with their biological function of transcriptional regulation of other genes in the nucleus.
In the present study, based on the specific expression of the eucalyptus WOX family members in the different organs of 24 d
E. urophylla ×
E. pellita tissue culture rooted seedlings, we found that the eucalyptus WOX family members were expressed in leaves, stems, and roots. The
EupWOX gene exhibited an apparent tissue specificity. The expression of this gene in the tissue of the different zygotic monoecious asexual lines in the 18H123 family line was somewhat different. The asexual lines of the zygotic offspring of the full sibling crosses exhibited a certain degree of reproductive capacity. The high expression of
EupWOX11 and
EupWOX1 in 30–66 and 46–99 roots, respectively, suggests that these genes could serve as the key regulatory genes for rooting in
E. urophylla ×
E. pellita and may play different roles in the rooting effect. Furthermore, the expression of the WOX gene family member in different organs indicates that different genes are expressed in different parts of the body. The expression levels of the
WOX gene family members in different organs show, to some extent, the expression specificity of the different genes in the different parts of the plant. Combining the rooting results of each asexual line after 24 d and the specific expression results of the four
WOX genes at six key time points from 0 d to 15 d, the development of adventitious roots and adventitious lateral roots of
E. urophylla ×
E. pellita was found to be affected and regulated by
EupWOX1,
EupWOX5,
EupWOX11, and
EupWOX13.
EupWOX1 could serve as a key regulator of the formation of root primordia, while
EupWOX5 and
EupWOX13 may function as key regulators of root formation.
EupWOX1,
EupWOX5, and
EupWOX13 could function as the key regulatory factors of root elongation and adventitious lateral root formation. In
Arabidopsis, root development is co-regulated by
WOX5 [
16],
WOX11 [
27],
WOX12 [
31],
WOX13, and
WOX14 [
9]. This finding is consistent with the present study.
Healing tissue regenerate plants. Members of the WOX family and endogenous cytokines and growth hormones play a key role in regulating the formation and maintenance of healing tissue.
WOX11 is a key gene involved in growth hormone response and cell fate transition, and the expression of
WOX11 transforms regeneration-competent cells into adventitious root founder cells [
4,
32]. The OsADA2–OsGCN5 complex was identified as a WOX11-interacting partner and was shown to activate the expression of downstream genes to maintain the cell division rate [
33].
OsWOX11 is essential for crown root emergence and promotes crown root growth, and its overexpression results in a higher number of lateral roots [
34].
JrWOX11 in poplar plants promotes root crown and root length growth [
13].
AtWOX11 regulates the occurrence of neonatal root organogenesis and the growth of adventitious lateral roots [
32,
35]. Based on these findings, we hypothesized that
EupWOX11 is a key regulator of adventitious root genesis and elongation in
E. urophylla ×
E. pellita and is involved in regulating lateral root growth and development.
WOX11,
WOX5, and
WOX7 are required for root growth and development. However, the expression of
WOX11 is decreased during cell division, while
WOX5 maintains the original role of root stem cells in the indefinite root primordial and plays a regulatory role in root meristem tissue [
36,
37,
38]. During the adventitious root formation (ARF) process, the transcripts of
JrWOX11 in walnut (
Juglans regia L.) and
JrWOX5 were consecutively increased on a significance level;
JrWOX11 was essential for adventitious root primordia formation and
JrWOX5 was crucial for root development [
39].
PtoWOX5a in
P. tomentosa can coordinate with cell division-related genes
CYCD and
CDF4 to regulate adventitious root development [
40].
PtoWUSa,
PtoWOX4a, and
PtoWOX5a are closely associated with the number and length of advents [
11]. The formation of the callus or root primordia in
Arabidopsis requires the activation of the auxin response factor
ARF7/19, which initiates the downstream
WOX11-
LBD16/17/18/19-
WOX5 signaling pathway [
41].
WOX5 is also expressed in the initiation and outward growth of the lateral root primordia, and it exerts a regulatory effect on lateral root growth [
42]. Thus, it can be inferred that
EupWOX5 plays an important regulatory role in the formation of the indefinite root primordia and indefinite lateral roots of eucalyptus and must be synergistic with
EupWOX11.
In
Petunia hybrida, the
WOX1 gene is involved in the development of posterior organs, and mutation in this gene increases the lateral growth of leaves and floral organs [
33].
WOX1 is also associated with the initiation of differentiation of healing tissue and root primordia [
10]. In jasmine,
JsWOX1 is associated with root regeneration, and overexpression of
JsWOX1 promoted the proliferation and number of healing tissues in the rooting stage [
10]. In
Arabidopsis,
WOX1 is probably associated with cell proliferation and plays an important role in meristematic tissue [
43]. Hence, we hypothesized that
EupWOX1 is a key regulatory gene for the induction of adventitious root primordia in
E. urophylla ×
E. pellita.
WOX13 is the most conserved region among the plant
WOX family members, and it is mainly expressed at the initiation and developmental stages of primary and lateral roots [
11].
WOX13 is a regulator of the formation of healing tissue at the wound site after injury to plants.
PpWOX13L-mediated cell elongation is a part of the stem cell initiation process [
44]. In
A. thaliana,
WOX13 is involved in the process of primordial and lateral root genesis and development, and mutation in this gene reduces the number of lateral roots [
45]. We, therefore, hypothesized that
EupWOX13 plays a critical role in regulating the elongation of adventitious roots and the growth of adventitious lateral roots.