Development of a Real-Time PCR Assay for the Early Detection of the Eucalyptus Pathogen Quambalaria eucalypti
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Plant Materials
2.2. Genomic DNA Extraction and Quantification
2.3. Primer Design and cPCR Amplification
2.4. SYBR Green Real-Time PCR Optimization
2.5. Real-Time PCR Assay’s Specificity and Sensitivity
2.6. Interlaboratory Tests
3. Results
3.1. Primer Design and cPCR Amplification
3.2. SYBR Green Real-Time PCR Optimization
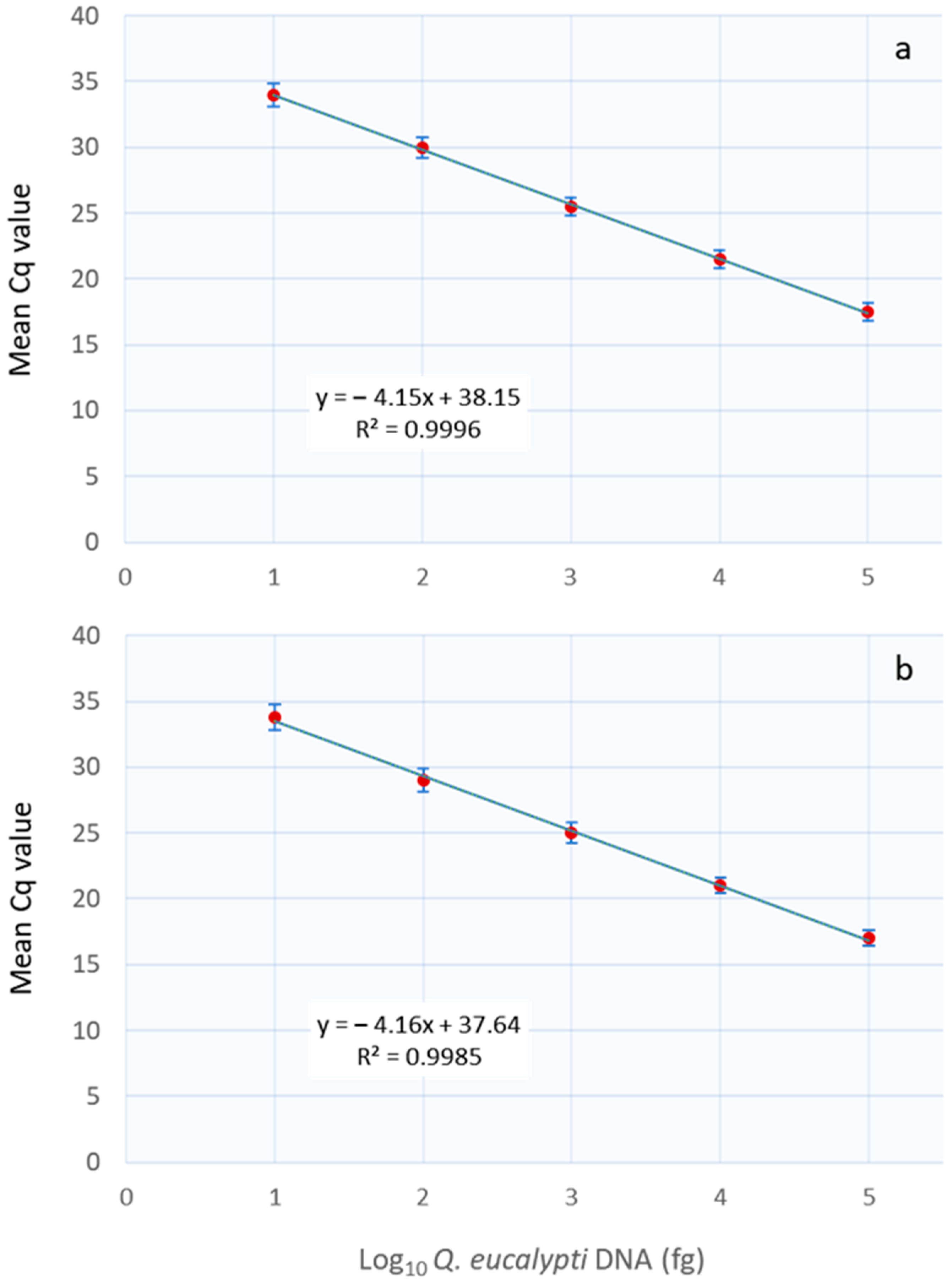
3.3. Real-Time PCR Assay Specificity and Sensitivity
3.4. Interlaboratory Tests
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naithani, H.B. Botany of genus Eucalyptus. In Eucalyptus in India; Bhojvaid, P.P., Kaushik, S., Singh, Y.P., Kumar, D., Thapliyal, M., Barthwal, S., Eds.; ICFRE: Dehra Dun, India, 2014; pp. 1–20. [Google Scholar]
- Trabado, G.I.; Wilstermann, D. Eucalyptus Global Map 2008: Cultivated Forests Worldwide—EUCALYPTOLOGICS: GIT Forestry Consulting Information Resources on Eucalyptus Cultivation Worldwide. Available online: http://git-forestry-blog.blogspot.com/2008/09/eucalyptus-global-map-2008-cultivated.html (accessed on 4 January 2024).
- Wen, Y.; Zhou, X.; Yu, S.; Zhu, H. The predicament and countermeasures of development of global eucalyptus plantations. Guangxi Sci. 2018, 25, 107–116. [Google Scholar]
- Salehi, B.; Sharifi-Rad, J.; Quispe, C.; Llaique, H.; Villalobos, M.; Smeriglio, A.; Trombetta, D.; Ezzat, S.M.; Salem, M.A.; Zayed, A.; et al. Insights into Eucalyptus genus chemical constituents, biological activities and health-promoting effects. Trends Food Sci. Technol. 2019, 91, 609–624. [Google Scholar] [CrossRef]
- Duong, H.T.; Williams, B.; White, D.; Burgess, T.I.; Hardy, G.E.S.J. qPCR assays for sensitive and rapid detection of Quambalaria species from plant tissues. Plant Dis. 2022, 106, 107–113. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Crous, P.W.; Swart, W.J. Sporothrix eucalypti (sp. nov.), a shoot and leaf pathogen of Eucalyptus in South Africa. Mycopathologia 1993, 123, 159–164. [Google Scholar] [CrossRef]
- Santos, G.S.; Mafia, R.G.; Zarpelon, T.G.; Soares, T.P.F.; Ferreira, M.A. First report of Quambalaria eucalypti in young eucalyptus plants in field conditions in Brazil. Forest Pathol. 2020, 50, e12633. [Google Scholar] [CrossRef]
- Tarigan, M.; Wingfield, M.J.; Marpaung, Y.M.A.N.; Durán, A.; Pham, N.Q. Quambalaria eucalypti found on Eucalyptus in Indonesia. Forest Pathol. 2023, 53, e12829. [Google Scholar] [CrossRef]
- Alfenas, A.C.; Zauza, E.A.V.; Mafia, R.G.; Assis, T.F. Clonagem e Doenças do Eucalipto, 2nd ed.; Universidade Federal de Viçosa: Viçosa, Brazil, 2009; p. 500. Available online: https://www.researchgate.net/publication/280577416_Clonagem_e_doencas_do_eucalipto (accessed on 4 January 2024).
- Bettucci, L.; Alonso, R.; Tiscornia, S. Endophytic mycobiota of healthy twigs and the assemblage of species associated with twig lesions of Eucalyptus globulus and E. grandis in Uruguay. Mycol. Res. 1999, 103, 468–472. [Google Scholar] [CrossRef]
- Alfenas, A.C.; Zauza, E.A.; Rosa, O.P.; Assis, T.F. Sporothrix eucalypti, a new pathogen of eucalyptus in Brazil. Fitopatol. Bras. 2001, 26, 221. [Google Scholar] [CrossRef][Green Version]
- Berbee, M.L.; Taylor, J.W. 18S Ribosomal RNA gene sequence characters place the human pathogen Sporothrix schenckii in the genus Ophiostoma. Exp. Mycol. 1992, 16, 87–91. [Google Scholar] [CrossRef]
- Simpson, J.A. Quambalaria, a new genus of eucalypt pathogens. Australas. Mycol. 2000, 19, 57–62. [Google Scholar]
- Pegg, G.S.; O’Dwyer, C.; Carnegie, A.J.; Burgess, T.I.; Wingfield, M.J.; Drenth, A. Quambalaria species associate with plantation and native eucalypts in Australia. Plant Pathol. 2008, 57, 702–714. [Google Scholar] [CrossRef]
- Bragança, H.; Diogo, E.L.F.; Neves, L.; Valente, C.; Araujo, C.; Bonifácio, L.; Phillips, A.J.L. Quambalaria eucalypti a pathogen of Eucalyptus globulus newly reported in Portugal and in Europe. For. Pathol. 2016, 46, 67–75. [Google Scholar] [CrossRef]
- Chen, S.F.; Liu, Q.; Li, G.; Wingfield, M.J. Quambalaria species associated with eucalypt diseases in southern China. Front. Agric. Sci. Eng. 2017, 4, 433–447. [Google Scholar] [CrossRef]
- Schena, L.; Li Destri Nicosia, M.G.; Sanzani, S.M.; Faedda, R.; Ippolito, A.; Cacciola, S.O. Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. J. Plant Pathol. 2013, 95, 7–24. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for Phylogenetics. In PCR Protocols: A Sequencing Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bact. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR To Amplify Con-served Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, B.D.; Liu, M.; Nguyen, H.D.T.; Lane, F.A.; Morgan, S.W.; De Vos, L.; Wilken, P.M.; Duong, T.A.; Aylward, J.; Coetzee, M.P.A.; et al. Nine draft genome sequences of Claviceps purpurea s.lat., including C. arundinis, C. humidiphila, and C. cf. spartinae, pseudomolecules for the pitch canker pathogen Fusarium circinatum, draft genome of Davidsoniella eucalypti, Grosmannia galeiformis, Quambalaria eucalypti, and Teratosphaeria destructans. IMA Fungus 2018, 9, 401–418. [Google Scholar] [CrossRef]
- de Oliveira, L.A.; Breton, M.C.; Bastolla, F.M.; Camargo, S.d.S.; Margis, R.; Frazzon, J.; Pasquali, G. Reference genes for the normalization of gene expression in Eucalyptus species. Plant Cell Physiol. 2012, 53, 405–422. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene amplification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef] [PubMed]
- McCartney, H.A.; Poster, S.J.; Fraaije, B.A.; Ward, E. Molecular diagnostics for fungal plant palhogens. Pest Manage. Sci. 2003, 59, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Zijlstra, C.; Lund, I.; Justesen, A.F.; Nicolaisen, M.; Jensen, P.K.; Bianciotto, V.; Posta, K.; Balestrini, R.; Przetakiewicz, A.; Czembor, E.; et al. Combining novel monitoring tools and precision application technologies for integrated high-tech crop protection in the future (a discussion document). Pest Manage. Sci. 2011, 67, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Schena, L.; Nigro, F.; Ippolito, A.; Gallitelli, D. Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. Eur. J. Plant Pathol. 2004, 110, 893–908. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Li Destri Nicosia, M.G.; Faedda, R.; Cacciola, S.O.; Schena, L. Use of quantitative PCR detection methods to study biocontrol agents and phytopathogenic fungi and oomycetes in environmental samples. J. Phytopathol. 2014, 162, 1–13. [Google Scholar] [CrossRef]
- Hariharnn, G.; Prasannath, K. Recent advances in molecular diagnostics of fungal plant pathogens: A mini review. Front. Cell. lnfect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- Baskarathevan, J.; Taylor, R.K.; Ho, W.; McDougal, R.L.; Shivas, R.G.; Alexander, B.J.R. Real-time PCR assays for the detection of Puccinia psidii. Plant Dis. 2016, 100, 617–624. [Google Scholar] [CrossRef]
- Arya, M.; Shergill, I.S.; Williamson, M.; Gommersall, L.; Arya, N.; Patel, H.R. Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 2005, 5, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Navarro, E.; Serrano-Heras, G.; Castaño, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta 2015, 1439, 231–250. [Google Scholar] [CrossRef] [PubMed]

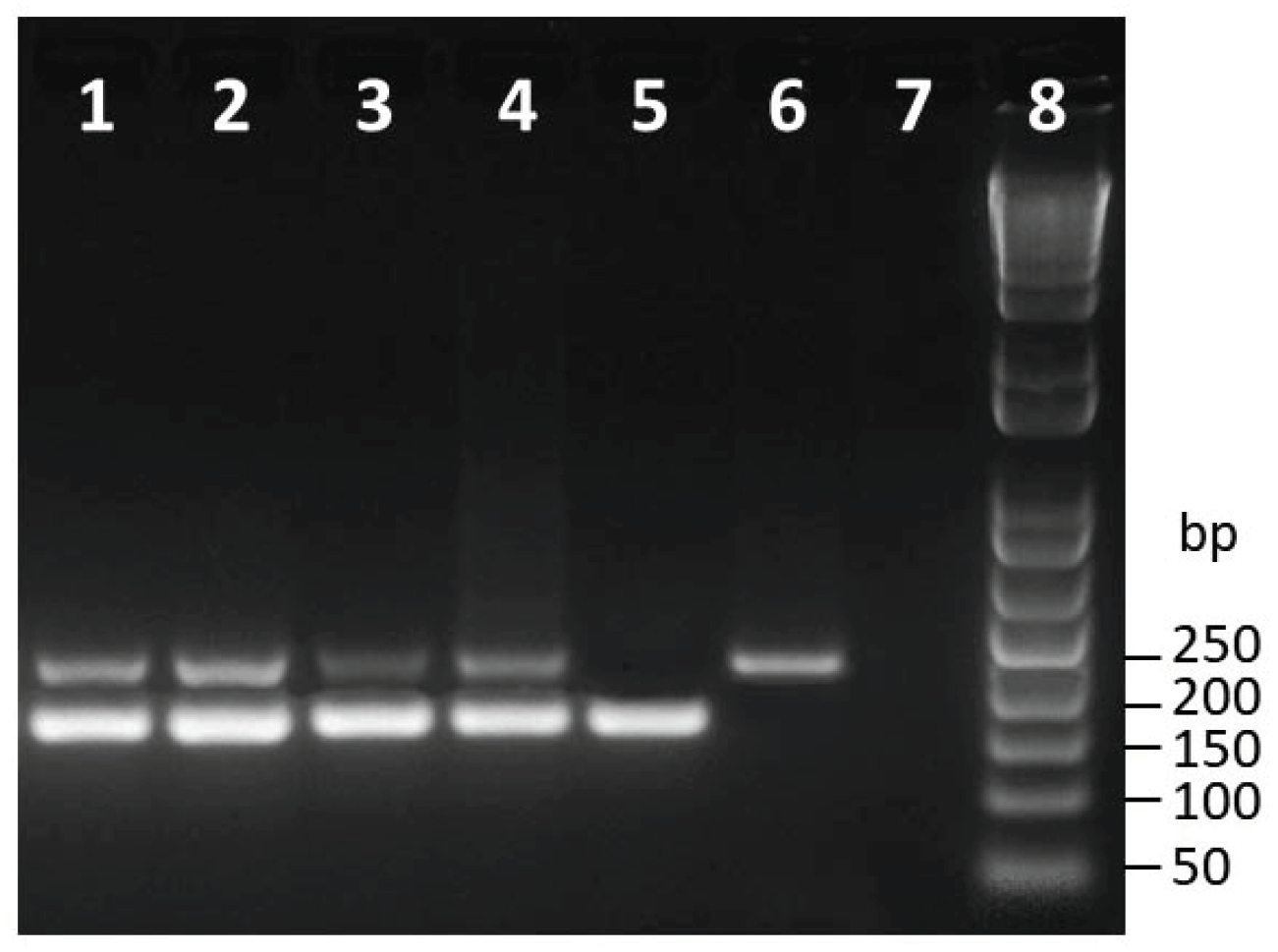
| Species | Isolate a | Host | Origin | Cq b Value ± SD c | Tm (°C) d |
|---|---|---|---|---|---|
| Quambalaria eucalypti | CBS 118844 e | Eucalyptus grandis | South Africa | 21.54 ± 0.71 | 87.2 |
| CBS 119680 f | 21.30 ± 0.52 | 87.0 | |||
| ITEF/1937 | 20.48 ± 0.35 | 86.1 | |||
| ITEF/1999 | 20.43 ± 0.14 | 86.8 | |||
| EGES-1 | Eucalyptus globulus | Brazil | 21.14 ± 0.35 | 86.1 | |
| EGES-2 | 20.87 ± 0.62 | 87.1 | |||
| EGES-5 | 20.91 ± 0.53 | 87.0 | |||
| EGES-10 | 22.07 ± 0.26 | 86.0 | |||
| EGES-11 | 22.09 ± 0.88 | 86.2 | |||
| EGES-13 | 21.30 ± 0.13 | 87.0 | |||
| EGES-17 | 21.58 ± 0.44 | 86.9 | |||
| EGES-19 | 22.43 ± 0.46 | 86.1 | |||
| EGES-20 | 22.46 ± 0.35 | 86.2 | |||
| EGES-22 | 22.45 ± 0.21 | 87.2 | |||
| RGS-1 | 21.41 ± 0.57 | 86.3 | |||
| RGS-2 | 21.35 ± 0.22 | 87.0 | |||
| RGS-3 | 21.47 ± 0.31 | 87.2 | |||
| RGS-4 | 21.29 ± 0.81 | 86.2 | |||
| RGS-5 | 20.93 ± 0.73 | 87.0 | |||
| RGS-6 | 21.02 ± 0,54 | 86.6 | |||
| SUL/01 | Uruguay | 21.27 ± 0.52 | 86.7 | ||
| SUL/03 | 22.05 ± 0.54 | 86.8 | |||
| UY198 | 21.24 ± 0.31 | 86.3 | |||
| UY199 | 21.63 ± 0.61 | 87.0 | |||
| MATY 4665 | 19.98 ± 1.07 | 86.5 | |||
| MATY 4751 | 20.15 ± 0.86 | 86.5 | |||
| MATY 4752 | 21.73 ± 0.24 | 86.7 | |||
| MATY 5001 | 21.65 ± 0.35 | 86.6 | |||
| 3421-S | 19.82 ± 1.13 | 87.0 | |||
| 3422-T | 21.36 ± 0.52 | 87.1 | |||
| Quambalaria cyanescens | CBS 876.73 | Eucalyptus pauciflora | Australia | n/a | n/a |
| IMI 178848 | n/a | n/a | |||
| EU-PA | n/a | n/a | |||
| Quambalaria pitereka | CERC/03/08 | Eucalyptus sp. | China | n/a | n/a |
| CERC/04/08 | n/a | n/a | |||
| CERC/05/05 | n/a | n/a | |||
| Quambalaria simpsonii | CBS 124772 f | Eucalyptus tintinnans | Australia | n/a | n/a |
| EU-TI3 | n/a | n/a | |||
| Quambalaria tasmaniae | CBS 145602 a | Eucalyptus sp. | Australia | n/a | n/a |
| 7608 | n/a | n/a | |||
| Alternaria alternata | CYC-60 | Eucalyptus grandis | Uruguay | n/a | n/a |
| Calonectria spathulata | CS11340 | Eucalyptus grandis | Brazil | n/a | n/a |
| Colletotrichum boninense | MWJ-43 | Eucalyptus grandis | South Africa | n/a | n/a |
| Cryphonectria havanensis | GSEG_95 | Eucalyptus grandis | Brazil | n/a | n/a |
| Cylindrocladium candelabrum | 11356 | Eucalyptus sp. | Brazil | n/a | n/a |
| Cylindrocladium pteridis | IMI 354530 | Eucalyptus grandis | Brazil | n/a | n/a |
| Neopestalotiopsis eucalyptorum | 912/88 | Eucalyptus globulus | Portugal | n/a | n/a |
| Phomopsis arnoldiae | CR 345-96 | Eucalyptus grandis | Uruguay | n/a | n/a |
| Pseudocercospora eucalyptorum | BBR 5689 | Eucalyptus globulus | Spain | n/a | n/a |
| Isolate | Disease Severity | Tm (°C) | Cq a Value ± SD b | DNA Conc. (fg) ± SD b |
|---|---|---|---|---|
| CBS 118844 | 0 | 87.2 | 33.10 ± 0.02 | 16.48 ± 2.40 |
| 1 | 86.9 | 27.70 ± 0.05 | 329.66 ± 5.24 | |
| 2 | 87.3 | 26.75 ± 0.03 | 558.45 ± 11.07 | |
| 3 | 87.0 | 24.89 ± 0.02 | 1567.40 ± 14.85 | |
| EGES-5 | 0 | 87.3 | 32.87 ± 0.18 | 18.72 ± 0.65 |
| 1 | 86.8 | 27.43 ± 0.27 | 382.94 ± 5.18 | |
| 2 | 87.1 | 26.73 ± 0.02 | 564.69 ± 11.67 | |
| 3 | 86.9 | 24.95 ± 0.03 | 1516.08 ± 12.36 | |
| UY198 | 0 | 87.0 | 32.84 ± 0.26 | 19.03 ± 0.15 |
| 1 | 87.0 | 27.53 ± 0.02 | 362.27 ± 4.73 | |
| 2 | 86.9 | 26.71 ± 0.04 | 570.99 ± 10.45 | |
| 3 | 87.1 | 24.81 ± 0.06 | 1638.54 ± 21.74 |
| DNA Conc. (pg) | Cq a Value ± SD b (Intra-Assay) | Cq a Value ± SD b (Inter-Assay) | Coefficient of Variance (%) | |||
|---|---|---|---|---|---|---|
| Operator 1 | Operator 2 | Operator 3 | Intra-Assay | Inter-Assay | ||
| 10 | 21.49 ± 0.15 | 21.40 ± 0.54 | 20.98 ± 0.66 | 20.78 ± 0.15 | 0.3 | 0.9 |
| 1 | 25.50 ± 0.11 | 25.18 ± 0.36 | 25.34 ± 0.12 | 26.03 ± 0.24 | 0.4 | 1.5 |
| 0.1 | 30.01 ± 0.12 | 29.87 ± 0.21 | 29.91 ± 0.15 | 30.05 ± 0.45 | 0.6 | 1.3 |
| 0.01 | 34.01 ± 0.13 | 33.85 ± 0.18 | 34.13 ± 0.11 | 33.97 ± 0.21 | 0.8 | 2.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faedda, R.; Silva, G.B. Development of a Real-Time PCR Assay for the Early Detection of the Eucalyptus Pathogen Quambalaria eucalypti. Forests 2024, 15, 375. https://doi.org/10.3390/f15020375
Faedda R, Silva GB. Development of a Real-Time PCR Assay for the Early Detection of the Eucalyptus Pathogen Quambalaria eucalypti. Forests. 2024; 15(2):375. https://doi.org/10.3390/f15020375
Chicago/Turabian StyleFaedda, Roberto, and Gabriela B. Silva. 2024. "Development of a Real-Time PCR Assay for the Early Detection of the Eucalyptus Pathogen Quambalaria eucalypti" Forests 15, no. 2: 375. https://doi.org/10.3390/f15020375
APA StyleFaedda, R., & Silva, G. B. (2024). Development of a Real-Time PCR Assay for the Early Detection of the Eucalyptus Pathogen Quambalaria eucalypti. Forests, 15(2), 375. https://doi.org/10.3390/f15020375






