Predicting Individual Tree Mortality of Larix gmelinii var. Principis-rupprechtii in Temperate Forests Using Machine Learning Methods
Abstract
1. Introduction
2. Materials and Methods
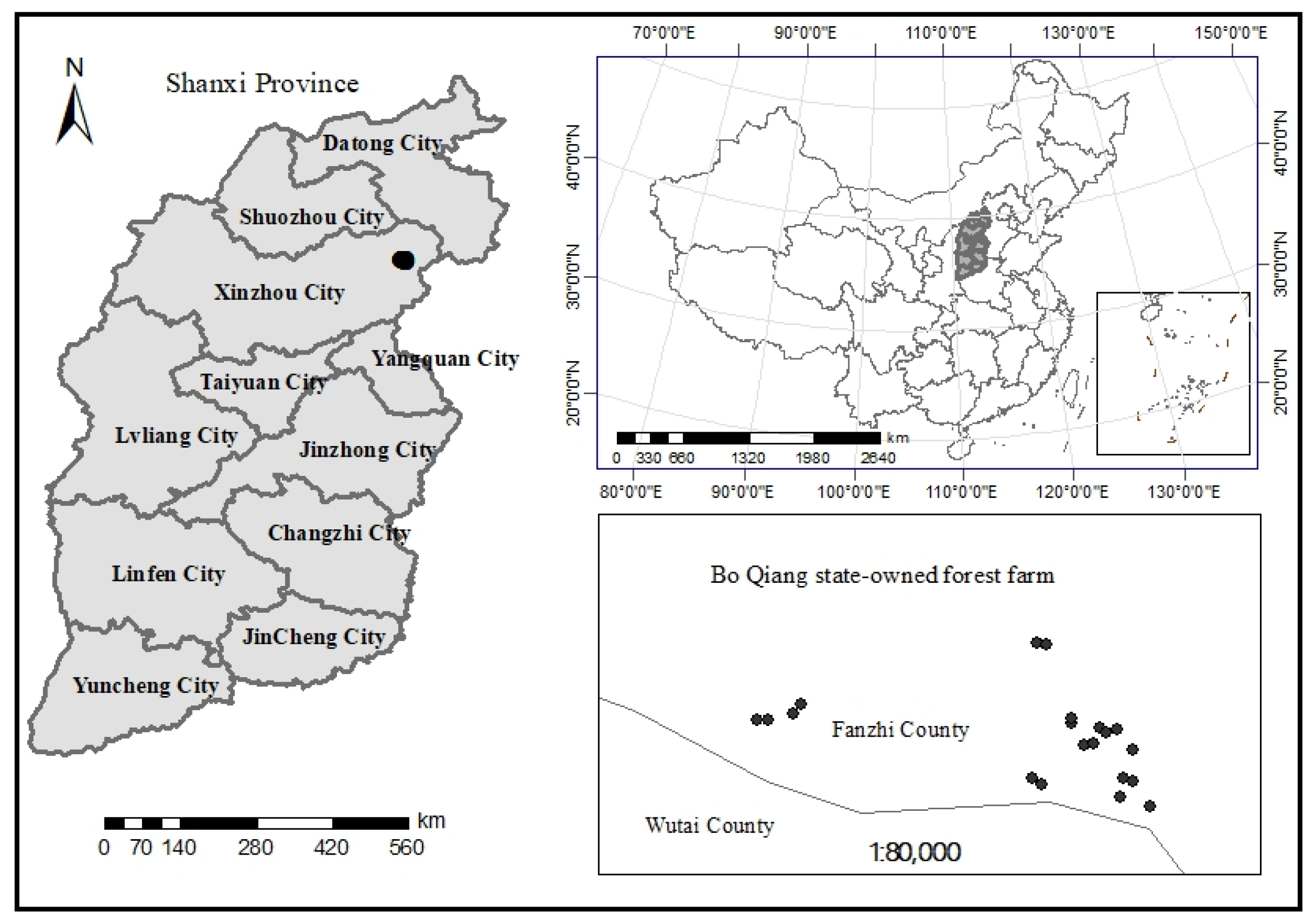
2.1. Study Area
2.2. Data Collection
2.3. Mortality Data Pre-Processing
2.4. Model Selection
2.4.1. Random Forest
2.4.2. Logistic Regression
2.4.3. Support Vector Machines
2.4.4. Generalized Additive Models
2.4.5. K-Nearest Neighbors
2.4.6. Naive Bayes
2.4.7. Gradient Boosting Machine
2.4.8. Artificial Neural Networks
2.5. Model Validation
2.6. Feature Importance
2.7. Model Evaluation
- (1)
- Accuracy: represents the proportion of correctly predicted samples to the total number of samples. It gauges the overall correctness of the model’s classifications. It can be calculated as follows:
- (2)
- Sensitivity: Referred to as the recall or true-positive rate, quantifies the proportion of accurately predicted positive samples relative to the total actual positive samples. It provides insight into the model’s capacity to correctly identify instances belonging to the positive class. It can be calculated as follows:
- (3)
- Specificity: Specificity denotes the proportion of correctly predicted negative samples to the total actual negative samples. It underscores the model’s capacity to differentiate negative class samples. It can be calculated as follows:
- (4)
- Cohen’s Kappa: Cohen’s Kappa is a statistic that quantifies the agreement between predicted and actual results, while considering the difference between classification outcomes and random chance.
- (5)
- Precision: Precision denotes the ratio of correctly predicted positive samples to the total samples predicted as positive. It assesses the accuracy of the model’s positive class predictions. It can be calculated as follows:
- (6)
- F1 Score: The F1 score is the harmonic mean of precision and recall, offering a balanced assessment of the model’s accuracy and coverage. It can be calculated as
- (7)
- Area under the ROC curve (AUC-ROC): The ROC curve is a graphical representation of the true-positive rate plotted against the false-positive rate. It illustrates the balance between sensitivity and specificity. AUC-ROC serves as a metric indicating the effectiveness of a parameter in discriminating between two diagnostic groups (diseased/normal). A higher AUC value corresponds to a superior ability of the model to distinguish between trees that perished and those that endured.
3. Results
3.1. Model Fitting Accuracy
3.2. Model Prediction Accuracy Evaluation on Test Dataset
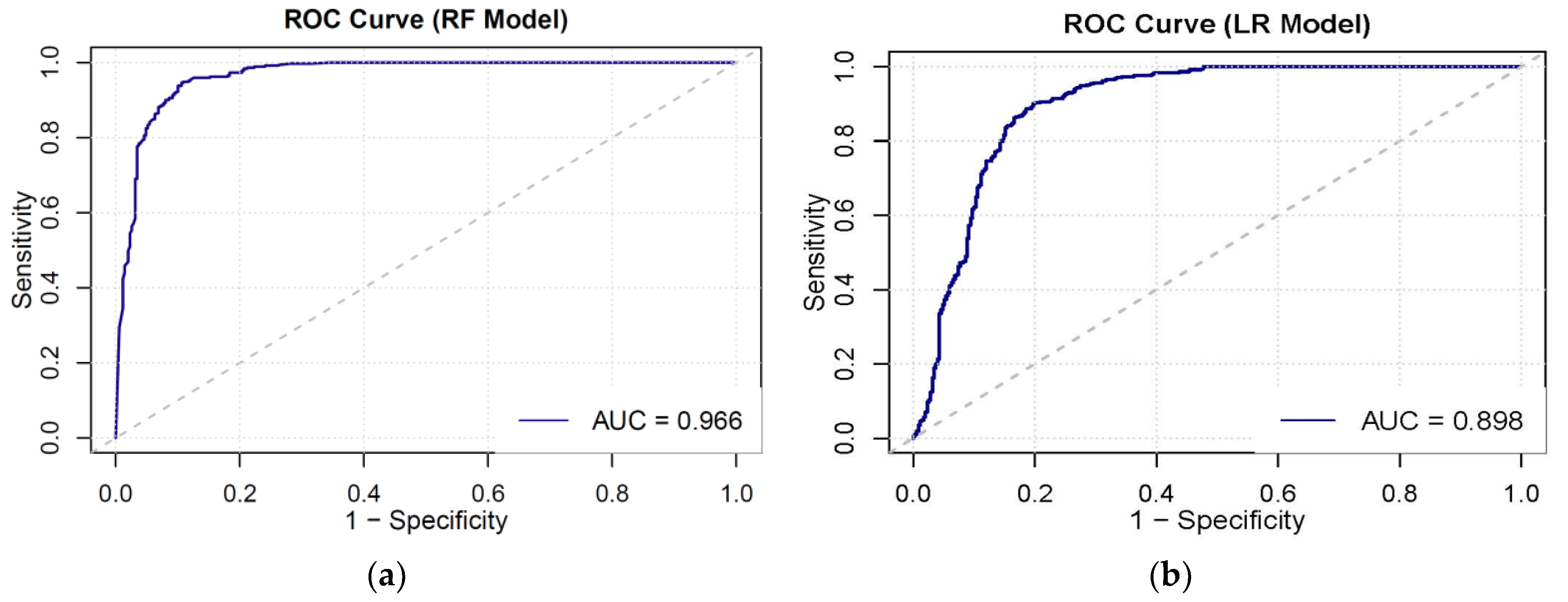
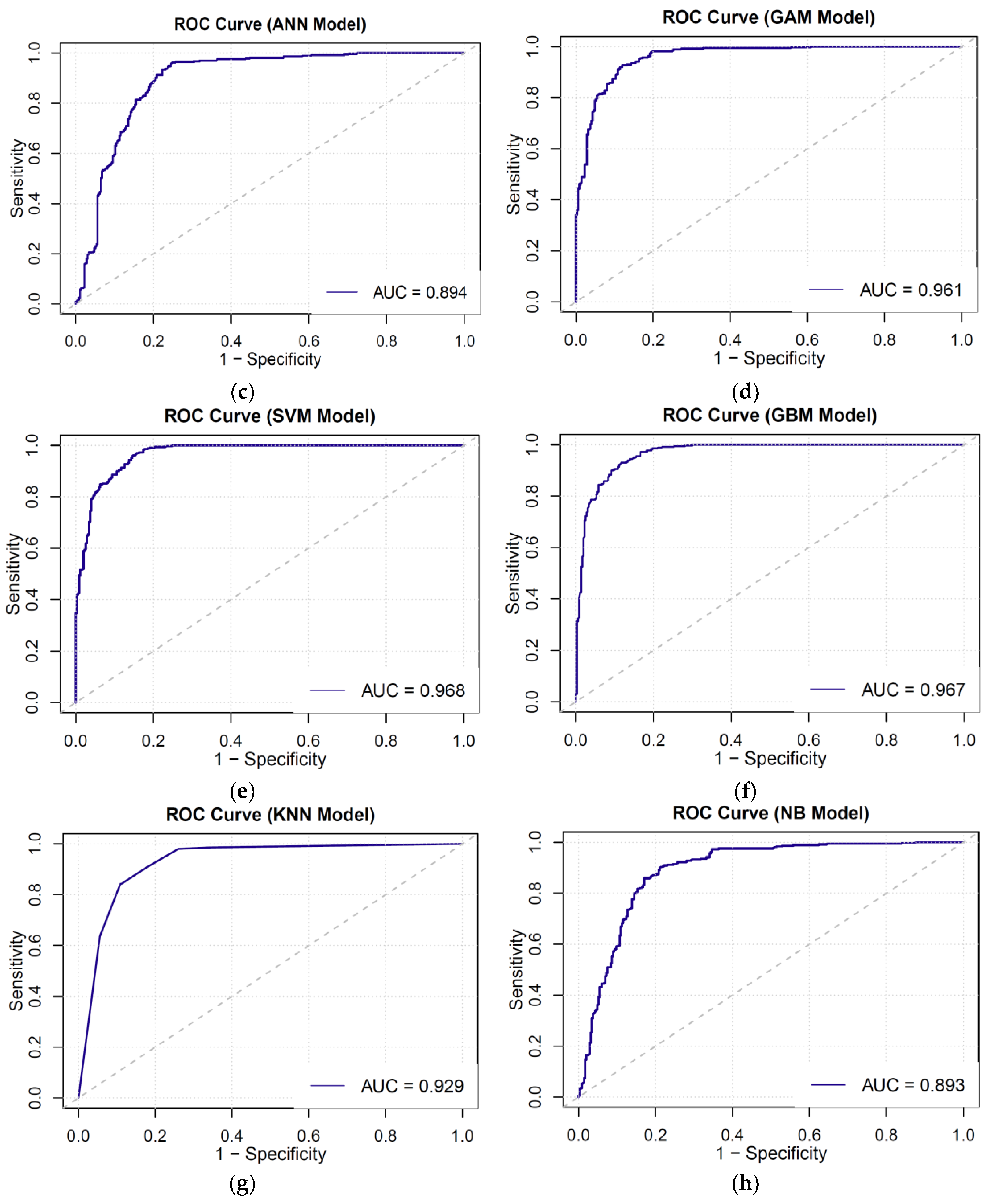
3.3. AUC-ROC Curve
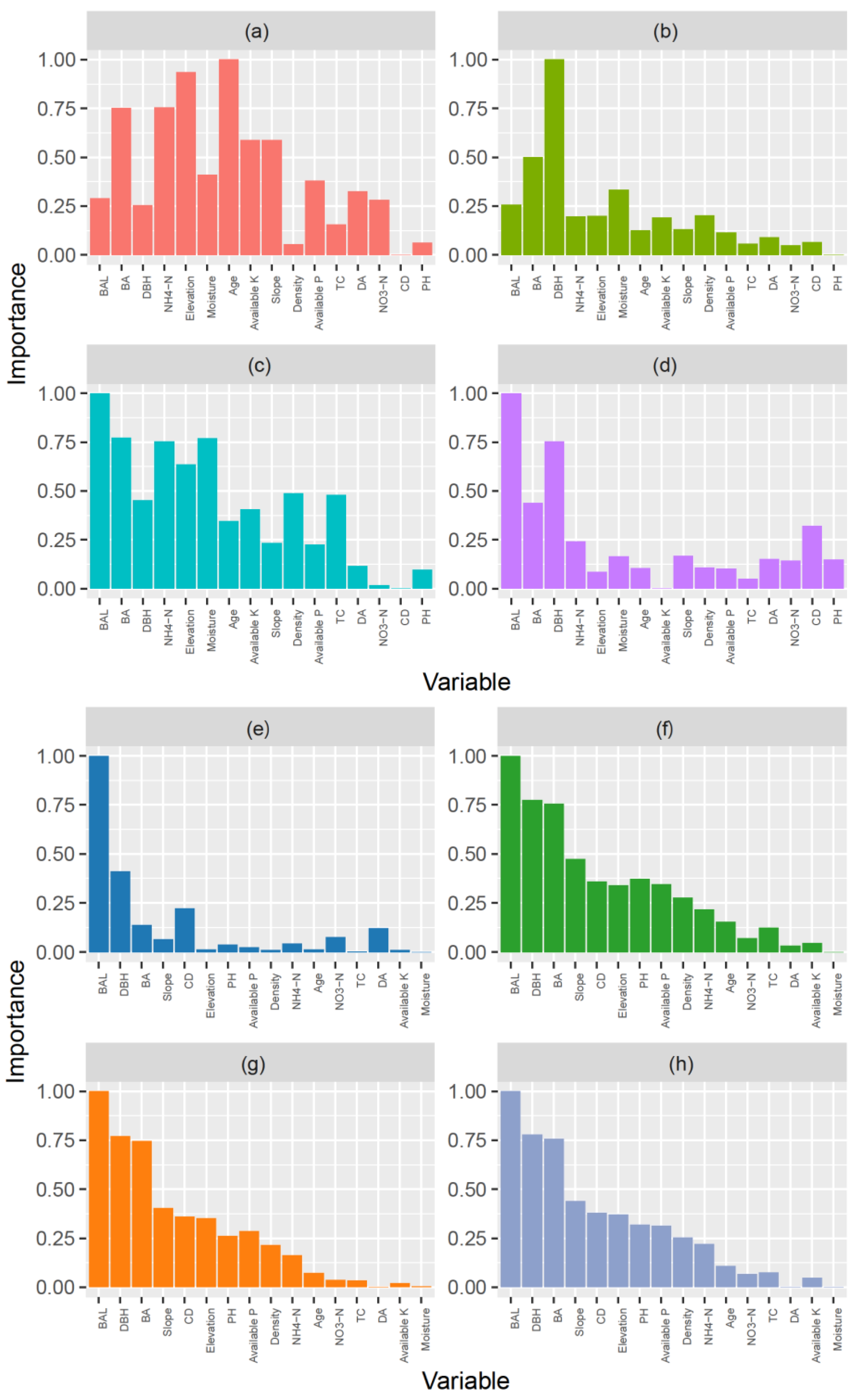
3.4. Variables Importance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Global Ecological Zoning for the Global Forest Resources Assessment Key Findings; FAO: Rome, Italy, 2020. [Google Scholar]
- Millar, C.I.; Stephenson, N.L. Temperate forest health in an era of emerging megadisturbance. Science 2015, 349, 823–826. [Google Scholar] [CrossRef]
- Nowak, D.J.; Crane, D.E.; Stevens, J.C.; Hoehn, R.E.; Walton, J.T.; Bond, J. A ground-based method of assessing urban forest structure and ecosystem services. Arboric. Urban. For. 2008, 34, 347–358. [Google Scholar] [CrossRef]
- Breshears, D.D.; Cobb, N.S.; Rich, P.M.; Price, K.P.; Allen, C.D.; Balice, R.G.; Romme, W.H.; Kastens, J.H.; Floyd, M.L.; Belnap, J.; et al. Regional vegetation die-off in response to global-change-type drought. Proc. Natl. Acad. Sci. USA 2005, 102, 15144–15148. [Google Scholar] [CrossRef]
- Hawkes, C. Woody plant mortality algorithms: Description, problems and progress. Ecol. Modell. 2000, 126, 225–248. [Google Scholar] [CrossRef]
- Searle, E.B.; Chen, H.Y.; Paquette, A. Higher tree diversity is linked to higher tree mortality. Proc. Natl. Acad. Sci. USA 2022, 119, e2013171119. [Google Scholar] [CrossRef]
- Rita, A.; Borghetti, M. Linkage of forest productivity to tree diversity under two different bioclimatic regimes in Italy. Sci. Total Environ. 2019, 687, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Q.; Lei, Y.-C.; Liu, X.-Z. Modeling stand mortality using Poisson mixture models with mixed-effects. iForest 2014, 8, 333–338. [Google Scholar] [CrossRef]
- Ruiz-Benito, P.; Ratcliffe, S.; Zavala, M.A.; Martínez-Vilalta, J.; Vilà-Cabrera, A.; Lloret, F.; Madrigal-González, J.; Wirth, C.; Greenwood, S.; Kändler, G.; et al. Climate- and successional-related changes in functional composition of European forests are strongly driven by tree mortality. Glob. Chang. Biol. 2017, 23, 4162–4176. [Google Scholar] [CrossRef] [PubMed]
- Muscarella, R.; Lohbeck, M.; Martínez-Ramos, M.; Poorter, L.; Rodríguez-Velázquez, J.E.; van Breugel, M.; Bongers, F. Demographic drivers of functional composition dynamics. Ecology 2017, 98, 2743–2750. [Google Scholar] [CrossRef] [PubMed]
- Larson, A.J.; Lutz, J.A.; Donato, D.C.; Freund, J.A.; Swanson, M.E.; HilleRisLambers, J.; Sprugel, D.G.; Franklin, J.F. Spatial aspects of tree mortality strongly differ between young and old-growth forests. Ecology 2015, 96, 2855–2861. [Google Scholar] [CrossRef]
- Thom, D.; Seidl, R. Natural disturbance impacts on ecosystem services and biodiversity in temperate and boreal forests. Biol. Rev. 2016, 91, 760–781. [Google Scholar] [CrossRef]
- Bugmann, H.; Seidl, R.; Hartig, F.; Bohn, F.; Brůna, J.; Cailleret, M.; François, L.; Heinke, J.; Henrot, A.; Hickler, T.; et al. Tree mortality submodels drive simulated long-term forest dynamics: Assessing 15 models from the stand to global scale. Ecosphere 2019, 10, e02616. [Google Scholar] [CrossRef] [PubMed]
- Korner, C. A matter of tree longevity. Science 2017, 355, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Sandén, H.; Rewald, B.; Godbold, D.L.; Katzensteiner, K. Increase in heterotrophic soil respiration by temperature drives decline in soil organic carbon stocks after forest windthrow in a mountainous ecosystem. Funct. Ecol. 2017, 31, 1163–1172. [Google Scholar] [CrossRef]
- Weng, Z.; Van Zwieten, L.; Singh, B.P.; Tavakkoli, E.; Joseph, S.; Macdonald, L.M.; Rose, T.J.; Rose, M.T.; Kimber, S.W.L.; Morris, S.; et al. Biochar built soil carbon over a decade by stabilizing rhizodeposits. Nat. Clim. Chang. 2017, 7, 371–376. [Google Scholar] [CrossRef]
- Thorn, S.; Seibold, S.; Leverkus, A.B.; Michler, T.; Müller, J.; Noss, R.F.; Stork, N.; Vogel, S.; Lindenmayer, D.B. The living dead: Acknowledging life after tree death to stop forest degradation. Front. Ecol. Environ. 2020, 18, 505–512. [Google Scholar] [CrossRef]
- Liu, Q.; Peng, C.; Schneider, R.; Cyr, D.; McDowell, N.G.; Kneeshaw, D. Drought-induced increase in tree mortality and corresponding decrease in the carbon sink capacity of Canada’s boreal forests from 1970 to 2020. Glob. Chang. Biol. 2023, 29, 2274–2285. [Google Scholar] [CrossRef]
- Lewis, S.L.; Phillips, O.L.; Sheil, D.; Vinceti, B.; Baker, T.R.; Brown, S.; Graham, A.W.; Higuchi, N.; Hilbert, D.W.; Laurance, W.F.; et al. Tropical forest tree mortality, recruitment and turnover rates: Calculation, interpretation and comparison when census intervals vary. J. Ecol. 2004, 92, 929–944. [Google Scholar] [CrossRef]
- Hember, R.A.; Kurz, W.A.; Coops, N.C. Relationships between individual-tree mortality and water-balance variables indicate positive trends in water stress-induced tree mortality across North America. Glob. Chang. Biol. 2017, 23, 1691–1710. [Google Scholar] [CrossRef]
- Fortin, M.; Bédard, S.; DeBlois, J.; Meunier, S. Predicting individual tree mortality in northern hardwood stands under uneven-aged management in southern Québec, Canada. Ann. For. Sci. 2008, 65, 205. [Google Scholar] [CrossRef]
- Temesgen, H.; Mitchell, S.J. An individual-tree mortality model for complex stands of southeastern British Columbia. West. J. Appl. For. 2005, 20, 101–109. [Google Scholar] [CrossRef]
- Dobbertin, M.; Biging, G.S. Using the non-parametric classifier CART to model forest tree mortality. For. Sci. 1998, 44, 507–516. [Google Scholar]
- Clark, P.; Wyckoff, J. Predicting tree mortality from diameter growth: A comparison of maximum likelihood and Bayesian approaches. Can. J. For. Res. 2000, 30, 156–167. [Google Scholar] [CrossRef]
- Affleck, D.L.R. Poisson mixture models for regression analysis of stand-level mortality. Can. J. For. Res. 2006, 36, 2994–3006. [Google Scholar] [CrossRef]
- Vieilledent, G.; Courbaud, B.; Kunstler, G.; Dhôte, J.-F.; Clark, J.S. BBiases in the estimation of size-dependent mortality models: Advantages of a semiparametric approach. Can. J. For. Res. 2009, 39, 1430–1443. [Google Scholar] [CrossRef]
- Adame, P.; Del Río, M.; Cañellas, I. Modeling individual-tree mortality in Pyrenean oak (Quercus pyrenaica Willd. ) stands. Ann. For. Sci. 2010, 67, 810. [Google Scholar] [CrossRef]
- Neumann, M.; Mues, V.; Moreno, A.; Hasenauer, H.; Seidl, R. Climate variability drives recent tree mortality in Europe. Glob. Chang. Biol. 2017, 23, 4788–4797. [Google Scholar] [CrossRef]
- Vanclay J, K. Modelling Forest Growth and Yield: Applications Tomixed and Tropical Forests; CAB International: Wallingford, UK, 1994. [Google Scholar]
- Han, P. Climate-Sensitive Growth and Mortality Model of Changbai Larch (Larix olgensis) Forests. Ph.D. Thesis, Beijing Forestry University, Beijing, China, 2020. (In Chinese). [Google Scholar]
- Ferry, B.; Morneau, F.; Bontemps, J.D.; Blanc, L.; Freycon, V. Higher treefall rates on slopes and waterlogged soils result in lower stand biomass and productivity in a tropical rain forest. J. Ecol. 2010, 98, 106–116. [Google Scholar] [CrossRef]
- Kuster, T.; Arend, M.; Bleuler, P.; Günthardt-Goerg, M.S.; Schulin, R. Water regime and growth of young oak stands subjected to air-warming and drought on two different forest soils in a model ecosystem experiment. Plant Biol. 2013, 15, 138–147. [Google Scholar] [CrossRef]
- Arend, M.; Gessler, A.; Schaub, M. The influence of the soil on spring and autumn phenology in European beech. Tree Physiol. 2016, 36, 78–85. [Google Scholar] [CrossRef][Green Version]
- Buchman, R.; Pederson, S.; Walters, N. A tree survival model with application to species of the Great Lakes region. Can. J. For. Res. 1983, 13, 601–608. [Google Scholar] [CrossRef]
- Brang, D. Crown defoliation improves tree mortality models. For. Ecol. Manag. 2001, 141, 271–284. [Google Scholar]
- Zhao, D.; Borders, B.; Wilson, M. Individual-tree diameter growth and mortality models for bottomland mixed-species hardwood stands in the lower Mississippi alluvial valley. For. Ecol. Manag. 2004, 199, 307–322. [Google Scholar] [CrossRef]
- Coyea, M.; Margolis, H. The historical reconstruction of growth efficiency and its relationship to tree mortality in balsam fir ecosystems affected by spruce budworm. Can. J. For. Res. 1994, 24, 2208–2221. [Google Scholar] [CrossRef]
- Crecente-Campo, F.; Marshall, P.; Rodríguez-Soalleiro, R. Modeling non-catastrophic individual-tree mortality for Pinus radiata plantations in northwestern Spain. For. Ecol. Manag. 2009, 257, 1542–1550. [Google Scholar] [CrossRef]
- Holzwarth, F.; Kahl, A.; Bauhus, J.; Wirth, C. Many ways to die–partitioning tree mortality dynamics in a near-natural mixed deciduous forest. J. Ecol. 2013, 101, 220–230. [Google Scholar] [CrossRef]
- Peng, C.; Ma, Z.; Lei, X.; Zhu, Q.; Chen, H.; Wang, W.; Liu, S.; Li, W.; Fang, X.; Zhou, X. A drought-induced pervasive increase in tree mortality across Canada’s boreal forests. Nat. Clim. Chang. 2011, 1, 467–471. [Google Scholar] [CrossRef]
- Zhu, K.; Ran, Y.; Ma, M.; Li, W.; Mir, Y.; Ran, J.; Wu, S.; Huang, P. Ameliorating soil structure for the reservoir riparian: The influences of land use and dam-triggered flooding on soil aggregates. Soil. Tillage Res. 2020, 216, 105263. [Google Scholar] [CrossRef]
- Mao, Q.; Lu, X.; Wang, C.; Zhou, K.; Mo, J. Responses of understory plant physiological traits to a decade of nitrogen addition in a tropical reforested ecosystem. For. Ecol. Manag. 2017, 401, 65–74. [Google Scholar] [CrossRef]
- Bu, W.-S.; Chen, F.-S.; Wang, F.-C.; Fang, X.-M.; Mao, R.; Wang, H.-M. The species-specific responses of nutrient resorption and carbohydrate accumulation in leaves and roots to nitrogen addition in a subtropical mixed plantation. Can. J. For. Res. 2019, 49, 826–835. [Google Scholar] [CrossRef]
- Sarkar, D.; Bali, R.; Sharma, T. Machine Learning Basics. In Practical Machine Learning with Python; Springer: Apress, Berkeley, CA, 2018. [Google Scholar] [CrossRef]
- Wang, J.; Taylor, A.R.; D’Orangeville, L. Warming-induced tree growth may help offset increasing disturbance across the Canadian boreal forest. Proc. Natl. Acad. Sci. USA 2023, 120, e2212780120. [Google Scholar] [CrossRef]
- Alenius, V.; Hökkä, H.; Salminen, H.; Jutras, S. Evaluating estimation methods for logistic regression in modelling individual-tree mortality. In Modelling Forest Systems; CABI Publishing: Wallingford, UK, 2003; pp. 225–236. [Google Scholar]
- Thomas, F.; Petzold, R.; Becker, C.; Werban, U. Usage of visual and near-infrared spectroscopy to predict soil properties in forest stands. In Proceedings of the EGU General, Assembly Conference Abstracts, Online, 4–8 May 2020; p. 9107. [Google Scholar]
- Wang, Z.; Zhang, X.; Chhin, S.; Zhang, J.; Duan, A. Disentangling the effects of stand and climatic variables on forest productivity of Chinese fir plantations in subtropical China using a random forest algorithm. Agric. For. Meteorol. 2021, 304, 108412. [Google Scholar] [CrossRef]
- Lou, X.W.; Weng, Y.H.; Fang, L.M.; Gao, H.L.; Grogan, J.; Hung, I.K.; Oswald, B.P. Predicting stand attributes of loblolly pine in West Gulf Coastal Plain using gradient boosting and random forests. Can. J. For. Res. 2021, 51, 807–816. [Google Scholar] [CrossRef]
- Walia, N.K.; Kalra, P.; Mehrotra, D. Prediction of carbon stock available in forest using naive Bayes approach. In Proceedings of the 2016 Second International Conference on Computational Intelligence & Communication Technology (CICT), Ghaziabad, India, 12–13 February 2016; pp. 275–279. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, H.; Sharma, R.P.; Pang, L.; Wang, G. A generalized nonlinear mixed-effects height to crown base model for Mongolian oak in northeast China. For. Ecol. Manag. 2017, 384, 34–43. [Google Scholar] [CrossRef]
- Rozas, V. Tree age estimates in Fagus sylvatica and Quercus robur: Testing previous and improved methods. Plant Ecol. 2003, 167, 193–212. [Google Scholar] [CrossRef]
- Du, J.S.; Wang, H.L.; Tang, S.Z. Update models of forest resource data for subcompartments in natural forest. Sci. Silvae Sin. 2000, 36, 26–32. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: https://www.R-project.org/ (accessed on 1 June 2023).
- Cutler, D.R.; Edwards, T.C., Jr.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- McRoberts, R.E.; Næsset, E.; Gobakken, T. Optimizing the k-Nearest Neighbors technique for estimating forest aboveground biomass using airborne laser scanning data. Remote Sens. Environ. 2015, 163, 13–22. [Google Scholar] [CrossRef]
- King, S.L.; Bennett, K.P.; List, S. Modeling noncatastrophic individual tree mortality using logistic regression, neural networks, and support vector methods. Comput. Electron. Agric. 2000, 27, 401–406. [Google Scholar] [CrossRef]
- Hasenauer, H.; Merkl, D.; Weingartner, M. Estimating tree mortality of Norway spruce stands with neural networks. Adv. Environ. Res. 2001, 5, 405–414. [Google Scholar] [CrossRef]
- Castro, R.V.O.; Soares, C.P.B.; Leite, H.G.; de Souza, A.L.; Nogueira, G.S.; Martins, F.B. Individual growth model for Eucalyptus stands in Brazil using artificial neural network. ISRN For. 2013, 196832. [Google Scholar] [CrossRef]
- Reis, L.P.; de Souza, A.L.; dos Reis, P.C.M.; Mazzei, L.; Soares, C.P.B.; Torres, C.M.M.E.; da Silva, L.F.; Ruschel, A.R.; Rêgo, L.J.S.; Leite, H.G. Estimation of mortality and survival of individual trees after harvesting wood using artificial neural networks in the amazon rain forest. Ecol. Eng. 2018, 112, 140–147. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Cox, D.R. The regression analysis of binary sequences. J. R. Stat. Soc. Ser. B Stat. Methodol. 1958, 20, 215–232. [Google Scholar] [CrossRef]
- Artin, N.; Chung, K.L. Theory of Probability and its Applications. Vol. III—1958. In An English Translation of the Soviet Journal Teoriya Veroyatnosteĭ i ee Primeneniya; Geological Society Publishing House: Bath, UK, 1990. [Google Scholar]
- Hastie, T.J.; Tibshirani, R.J. Generalized Additive Models. Stat. Sci. 1986, 1, 297–318. [Google Scholar] [CrossRef]
- Zhang, Z. Introduction to machine learning: K-nearest neighbors. Ann. Transl. Med. 2016, 4, 218. [Google Scholar] [CrossRef] [PubMed]
- Rish, I. An empirical study of the naive Bayes classifier. In Proceedings of the IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Seattle, WA, USA, 4–6 August 2001; Volume 4, pp. 41–46. [Google Scholar]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 1, 1189–1232. [Google Scholar] [CrossRef]
- Krenker, A.; Bešter, J.; Kos, A. Introduction to the artificial neural networks. In Artificial Neural Networks: Methodological Advances and Biomedical Applications; InTech: London, UK, 2011; pp. 1–18. [Google Scholar]
- Hamidi, S.K.; Zenner, E.K.; Bayat, M.; Fallah, A. Analysis of plot-level volume increment models developed from machine learning methods applied to an uneven-aged mixed forest. Ann. For. Sci. 2021, 78, 4. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, Q.; Sharma, R.P.; Wang, Y.; He, P.; Guo, J.; Lei, Y.; Fu, L. A climate sensitive mixed-effects diameter class mortality model for Prince Rupprecht larch (Larix gmelinii var. principis-rupprechtii) in northern China. For. Ecol. Manag. 2021, 491, 119091. [Google Scholar] [CrossRef]
- Zhou, X.; Fu, L.; Sharma, R.P.; He, P.; Lei, Y.; Guo, J. Generalized or general mixed-effect modelling of tree mortality of Larix gmelinii subsp. principis-rupprechtii in Northern China. J. For. Res. 2021, 32, 2447–2458. [Google Scholar] [CrossRef]
- Xie, L.; Chen, X.; Zhou, X.; Sharma, R.P.; Li, J. Developing tree mortality models using bayesian modeling approach. Forests 2022, 13, 604. [Google Scholar] [CrossRef]
- Yao, X.; Titus, S.J.; Macdonald, S.E. A generalized logistic model of individual tree mortality for aspen, white spruce, and lodgepole pine in Alberta mixedwood forests. Can. J. For. Res. 2001, 31, 283–291. [Google Scholar] [CrossRef]
- Lutz, J.A.; Halpern, C.B. Tree mortality during early forest development: A long-term study of rates causes, and consequences. Ecol. Monogr. 2006, 76, 257–275. [Google Scholar] [CrossRef]
- Bałazy, R.; Kamińska, A.; Ciesielski, M.; Socha, J.; Pierzchalski, M. Modeling the effect of environmental and topographic variables affecting the height increment of Norway spruce stands in mountainous conditions with the use of LiDAR data. Remote Sens. 2019, 11, 2407. [Google Scholar] [CrossRef]
- Stage, A.R.; Christian, S. Interactions of Elevation, Aspect, and Slope in Models of Forest Species Composition and Productivity. For. Sci. 2007, 53, 486–492. [Google Scholar]
- Li, C.; Zhao, L.; Li, L. Modeling stand-Level mortality of Mongolian Oak (Quercus mongolica) based on mixed effect model and zero-inflated model methods. For. Sci. 2019, 55, 27–36. (In Chinese) [Google Scholar]
- Morin, R.S.; Randolph, K.C.; Steinman, J. Mortality rates associated with crown health for eastern forest tree species. Environ. Monit. Assess. 2015, 187, 87. [Google Scholar] [CrossRef]
- Zhu, Y.; Qi, B.; Hao, Y.S.; Liu, H.; Sun, G.; Chen, R.; Song, S. Appropriate NH4+/NO3− ratio triggers plant growth and nutrient uptake of flowering Chinese cabbage by optimizing the pH value of nutrient solution. Front. Plant Sci. 2021, 12, 656144. [Google Scholar] [CrossRef]
- Richardson, A.E.; Lynch, J.P.; Ryan, P.R.; Delhaize, E.; Smith, F.A.; Smith, S.E.; Harvey, P.R.; Ryan, M.H.; Veneklaas, E.J.; Lambers, H.; et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil. 2011, 349, 121–156. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Porder, S.; Houlton, B.Z.; Chadwick, O.A. Terrestrial phosphorus limitation: Mechanisms, implications, and nitrogen–phosphorus interactions. Ecol. Appl. 2010, 20, 5–15. [Google Scholar] [CrossRef] [PubMed]
| Variable | Min | Max | Mean | Std |
|---|---|---|---|---|
| DBH (cm) | 0.90 | 44.50 | 13.1910 | 8.7541 |
| BA (cm2) | 0.64 | 1554.50 | 196.6600 | 226.2700 |
| BAL (cm2) | 0.00 | 19,543.61 | 9698.3700 | 4396.1000 |
| Thickness (cm) | 1.80 | 12.00 | 5.8732 | 2.8754 |
| CD | 0.54 | 0.92 | 0.7678 | 0.1200 |
| Elevation (m) | 2079 | 2438 | 2239 | 97.4420 |
| DA | 26.00 | 71.00 | 53.0115 | 14.3212 |
| Slope (degree) | 8.00 | 38.50 | 22.9313 | 6.8836 |
| Moisture (%) | 12.38 | 37.46 | 26.0389 | 6.5818 |
| Density (g/m3) | 0.72 | 1.71 | 1.0107 | 0.2129 |
| PH | 6.40 | 6.77 | 6.6103 | 0.1012 |
| TC (g/kg) | 1.29 | 4.25 | 2.5476 | 0.8757 |
| NO3-N (mg/kg) | 6.81 | 18.98 | 10.5527 | 3.7009 |
| NH4-N (mg/kg) | 9.92 | 64.67 | 21.6761 | 10.9171 |
| Available K (mg/kg) | 48.35 | 90.62 | 65.9163 | 9.4617 |
| Available P (mg/kg) | 3.31 | 10.88 | 5.5186 | 1.9669 |
| Age | 16 | 70 | 39.7297 | 16.4208 |
| Model | Accuracy | Sensitivity | Specificity | Kappa | Precision | F1 Score |
|---|---|---|---|---|---|---|
| RF | 0.9793 | 0.9623 | 0.9975 | 0.9585 | 0.9976 | 0.9796 |
| LR | 0.8488 | 0.8039 | 0.8969 | 0.6984 | 0.8930 | 0.8461 |
| ANN | 0.8660 | 0.8179 | 0.9176 | 0.7326 | 0.9142 | 0.8634 |
| GAM | 0.8946 | 0.8510 | 0.9355 | 0.7884 | 0.9252 | 0.8866 |
| SVM | 0.9028 | 0.8681 | 0.9399 | 0.8059 | 0.9392 | 0.9023 |
| GBM | 0.9455 | 0.9339 | 0.9576 | 0.8909 | 0.9583 | 0.9459 |
| K-NN | 0.9052 | 0.8601 | 0.9526 | 0.8107 | 0.9502 | 0.9029 |
| NB | 0.8399 | 0.7979 | 0.8852 | 0.6805 | 0.8826 | 0.8381 |
| Model | Accuracy | Sensitivity | Specificity | Kappa | Precision | F1 Score |
|---|---|---|---|---|---|---|
| RF | 0.9291 | 0.9277 | 0.9303 | 0.8580 | 0.9251 | 0.9259 |
| LR | 0.8472 | 0.8143 | 0.8784 | 0.6937 | 0.8636 | 0.8386 |
| ANN | 0.8599 | 0.7908 | 0.9247 | 0.7184 | 0.9079 | 0.8459 |
| GAM | 0.8946 | 0.8510 | 0.9355 | 0.7884 | 0.9252 | 0.8867 |
| SVM | 0.8972 | 0.8657 | 0.9270 | 0.7940 | 0.9182 | 0.8916 |
| GBM | 0.9042 | 0.8861 | 0.9222 | 0.8083 | 0.9193 | 0.9033 |
| K-NN | 0.8653 | 0.8207 | 0.9091 | 0.7303 | 0.8988 | 0.8588 |
| NB | 0.8391 | 0.8150 | 0.8613 | 0.6773 | 0.8443 | 0.8294 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Duan, G.; Sharma, R.P.; Peng, W.; Zhou, L.; Fan, Y.; Zhang, M. Predicting Individual Tree Mortality of Larix gmelinii var. Principis-rupprechtii in Temperate Forests Using Machine Learning Methods. Forests 2024, 15, 374. https://doi.org/10.3390/f15020374
Yang Z, Duan G, Sharma RP, Peng W, Zhou L, Fan Y, Zhang M. Predicting Individual Tree Mortality of Larix gmelinii var. Principis-rupprechtii in Temperate Forests Using Machine Learning Methods. Forests. 2024; 15(2):374. https://doi.org/10.3390/f15020374
Chicago/Turabian StyleYang, Zhaohui, Guangshuang Duan, Ram P. Sharma, Wei Peng, Lai Zhou, Yaru Fan, and Mengtao Zhang. 2024. "Predicting Individual Tree Mortality of Larix gmelinii var. Principis-rupprechtii in Temperate Forests Using Machine Learning Methods" Forests 15, no. 2: 374. https://doi.org/10.3390/f15020374
APA StyleYang, Z., Duan, G., Sharma, R. P., Peng, W., Zhou, L., Fan, Y., & Zhang, M. (2024). Predicting Individual Tree Mortality of Larix gmelinii var. Principis-rupprechtii in Temperate Forests Using Machine Learning Methods. Forests, 15(2), 374. https://doi.org/10.3390/f15020374








