Transcriptional Profiling Reveals Key Regulatory Roles of the WUSCHEL-Related Homeobox Gene Family in Yellowhorn (Xanthoceras sorbifolia Bunge)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of WOX Genes in Yellowhorn
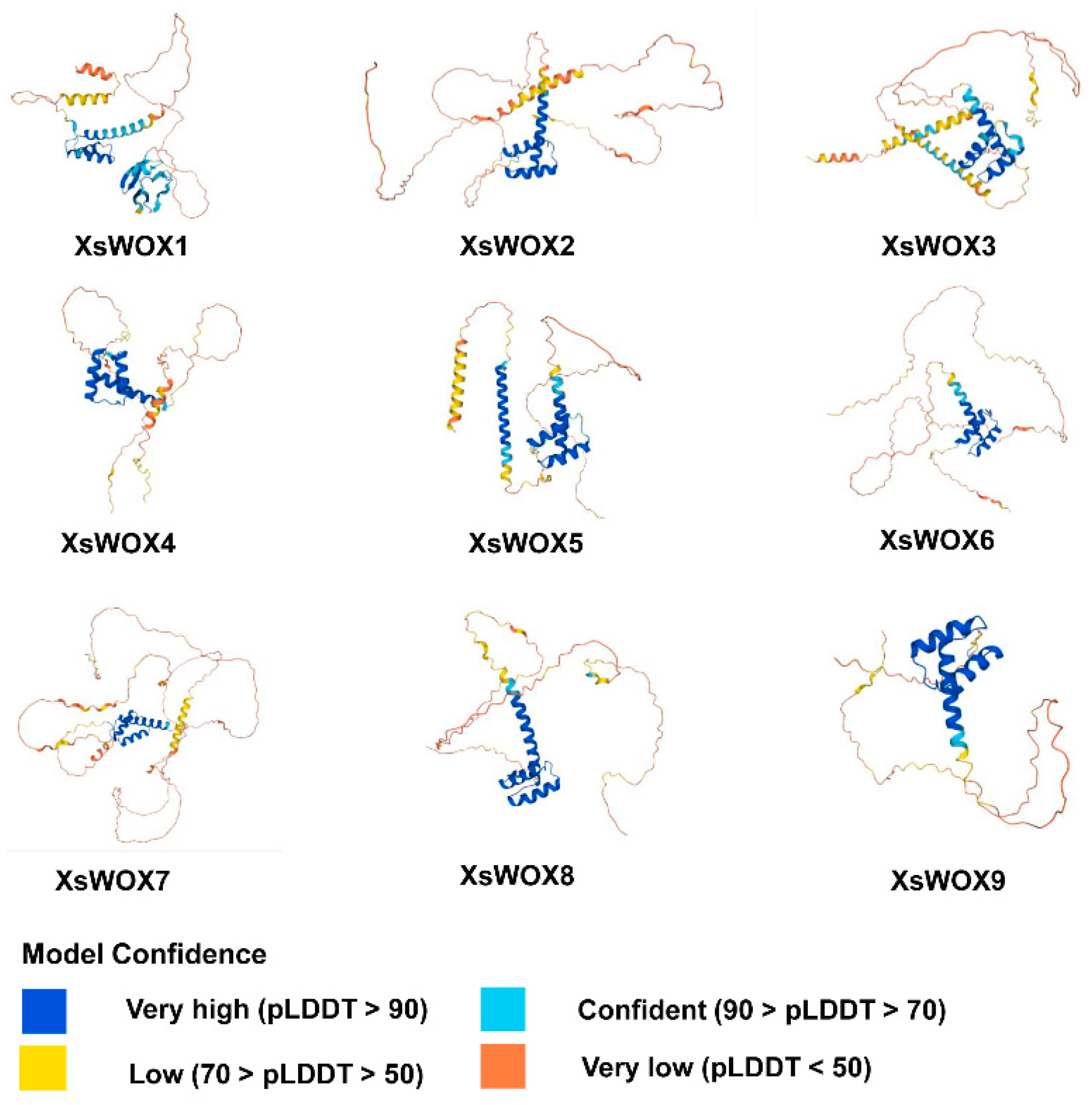
2.2. Structural Analysis of WOX Proteins
2.3. Phylogenetic Analyses of the WOX Proteins
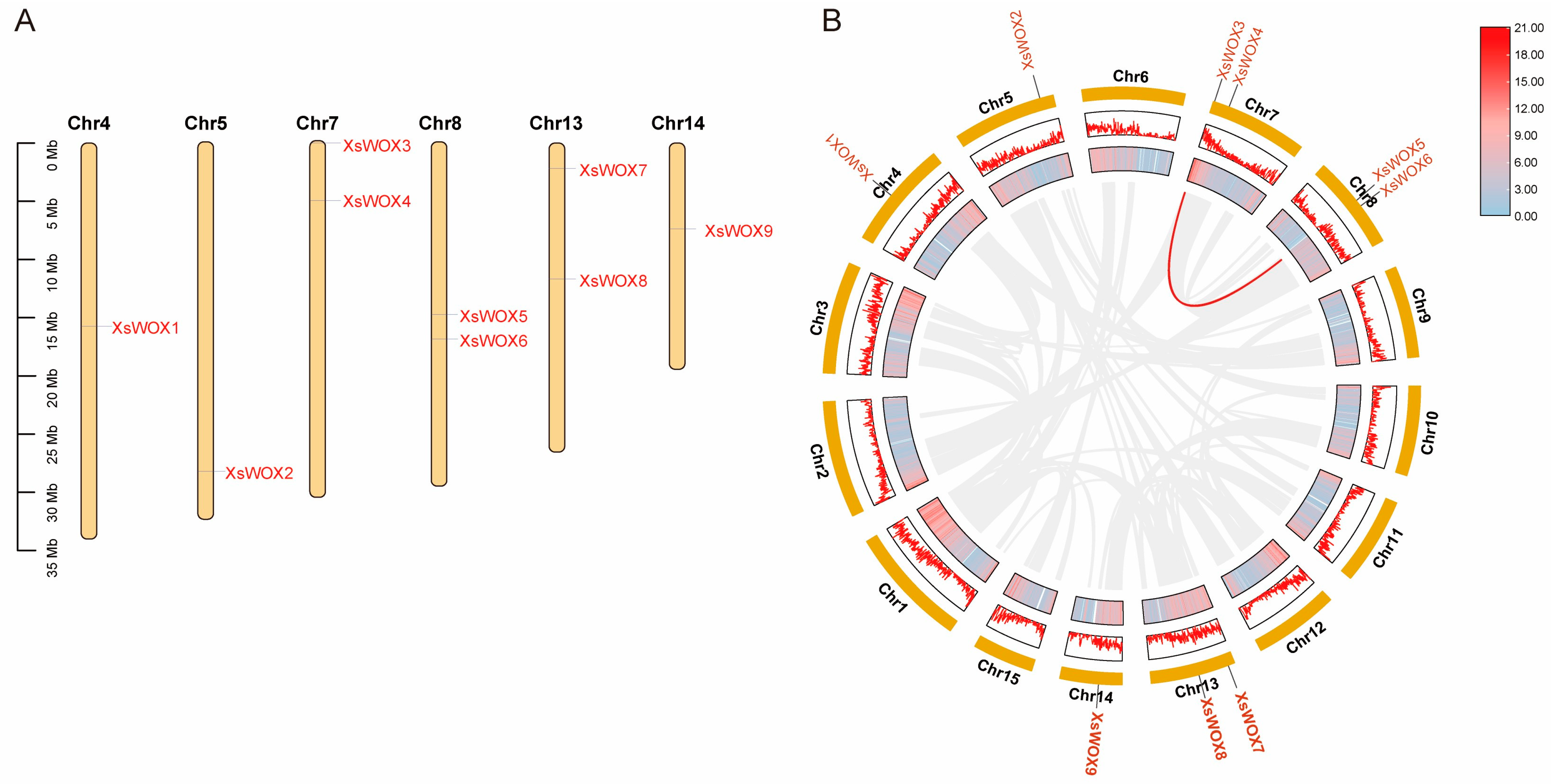
2.4. Chromosome Localization and Synteny Analysis
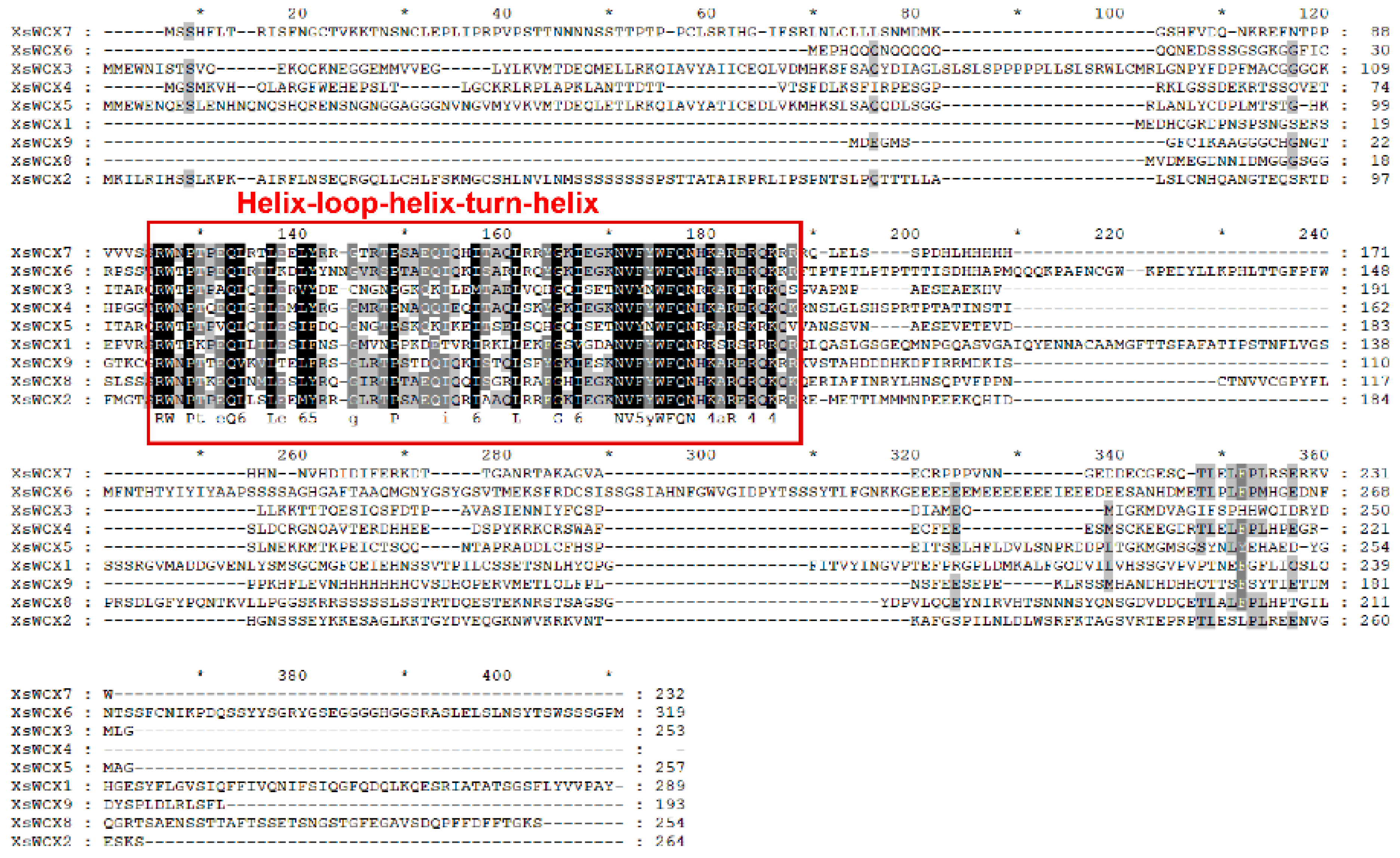
2.5. Multiple Sequence Alignment, Gene Structure, and Motif Feature Analysis
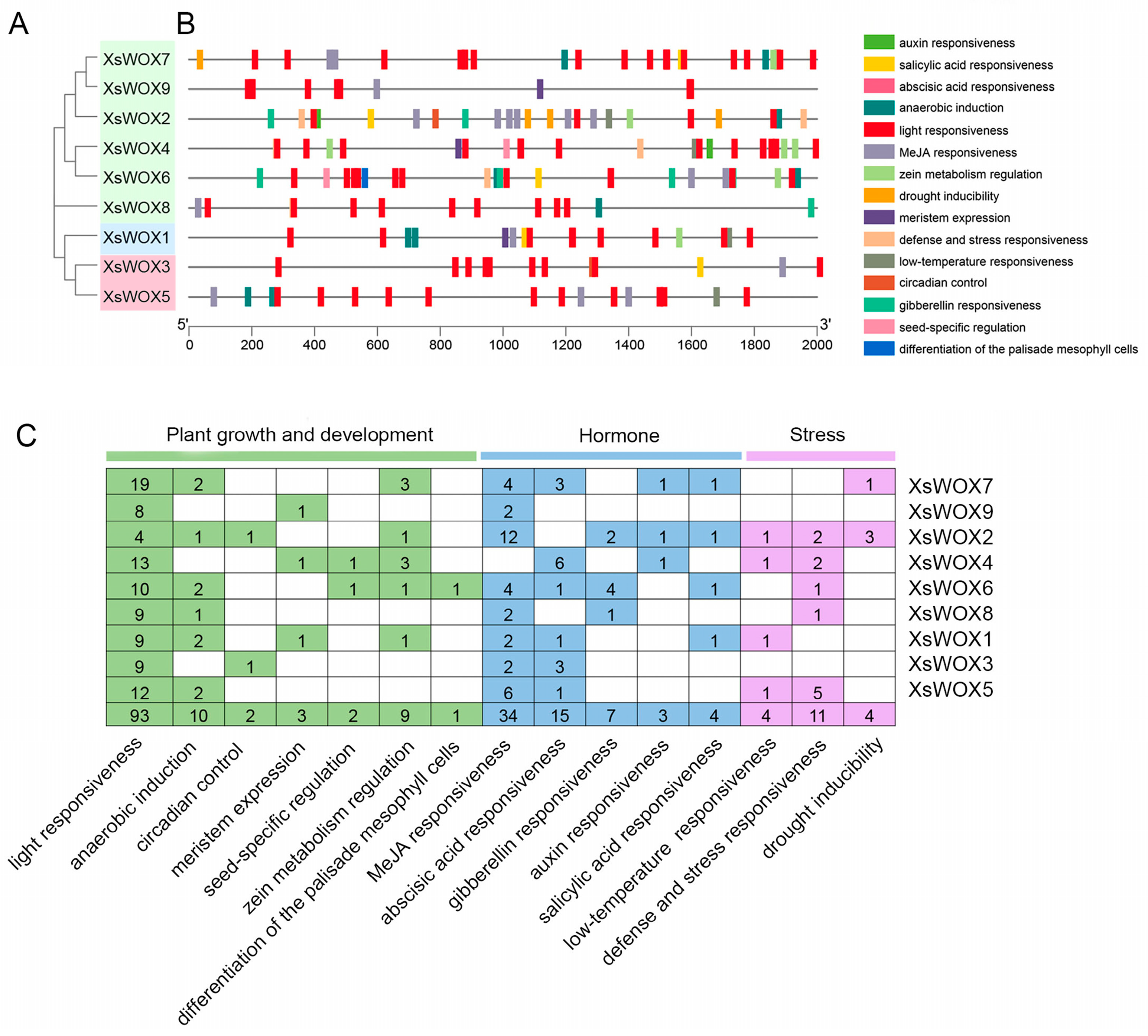
2.6. Cis-Elements Analysis
2.7. Plant Materials and Culture Conditions
2.8. Expression Profile Analysis of XsWOX Genes
2.9. Quantitative RT-PCR Analysis
3. Results
3.1. Characterization of XsWOX Genes in Yellowhorn
3.2. Phylogenetic Analysis of XsWOX Genes in Yellowhorn
3.3. Chromosomal Localization and Syntenic Analysis of XsWOX Genes in Yellowhorn
3.4. Domain, Motif Identification, and Gene Structure Analysis
3.5. Analysis of Cis-Acting Elements in the XsWOX Promoters
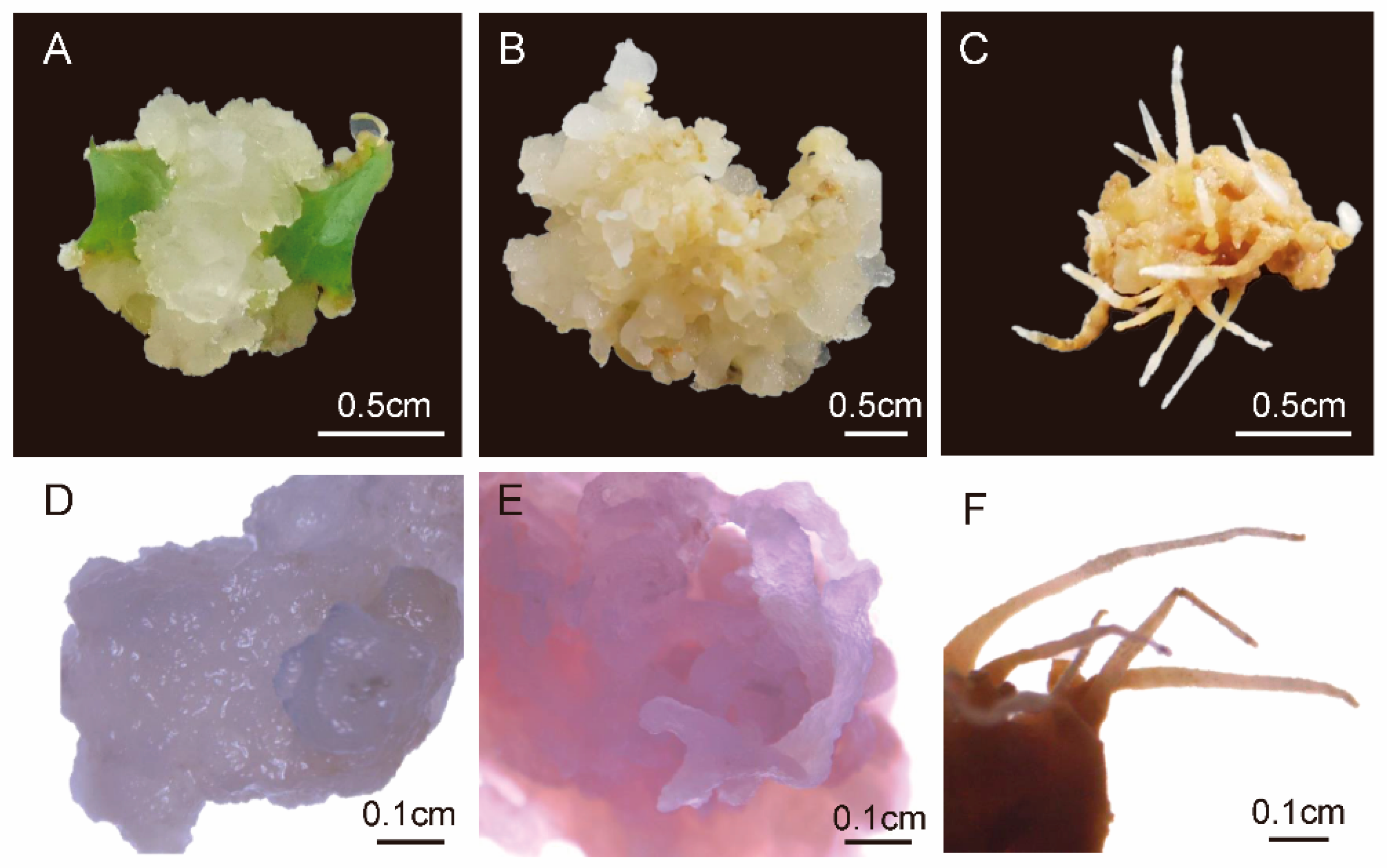
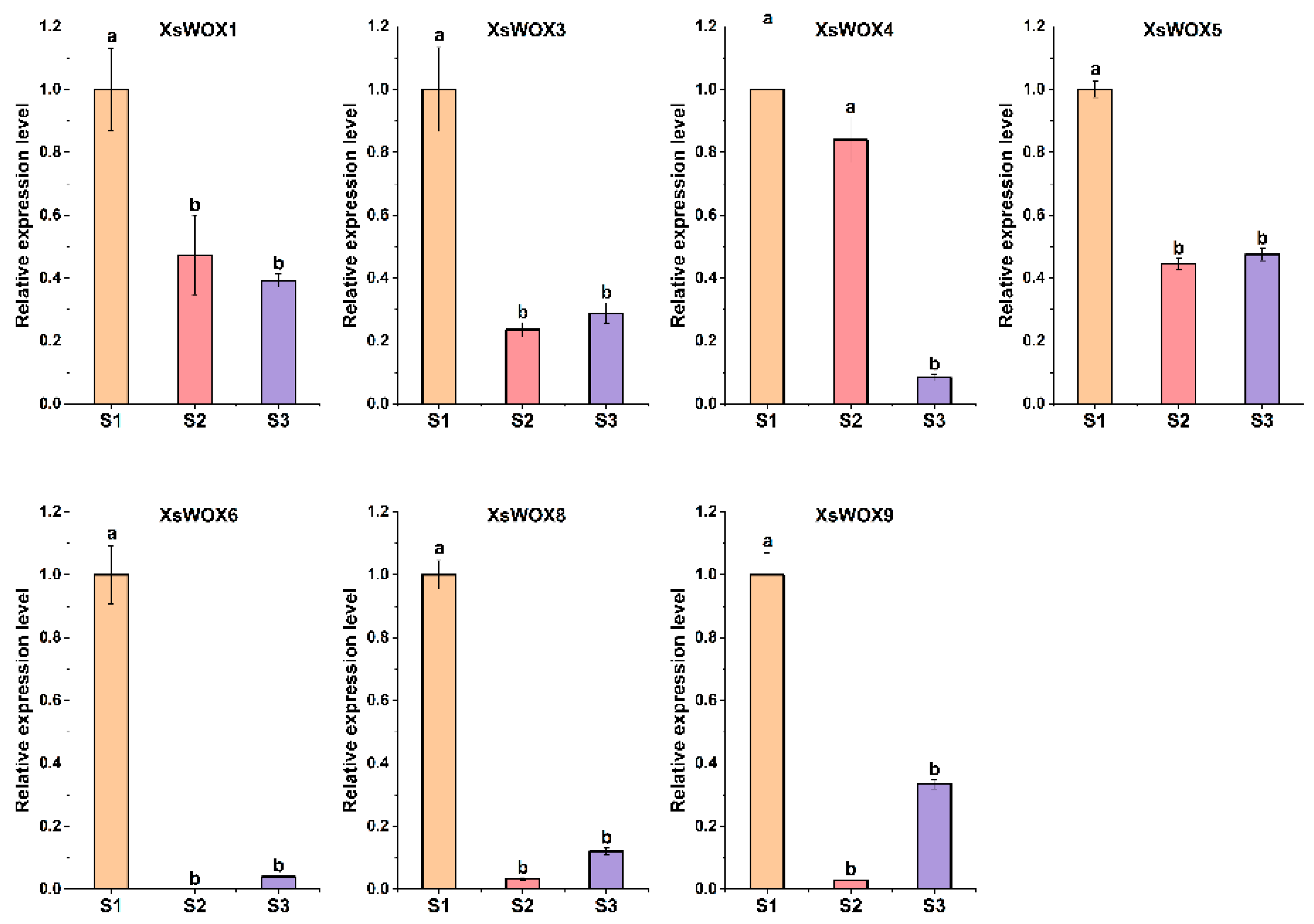
3.6. Phenotypes of Callus at Different Developmental Stages of Yellowhorn and Transcriptome Analysis
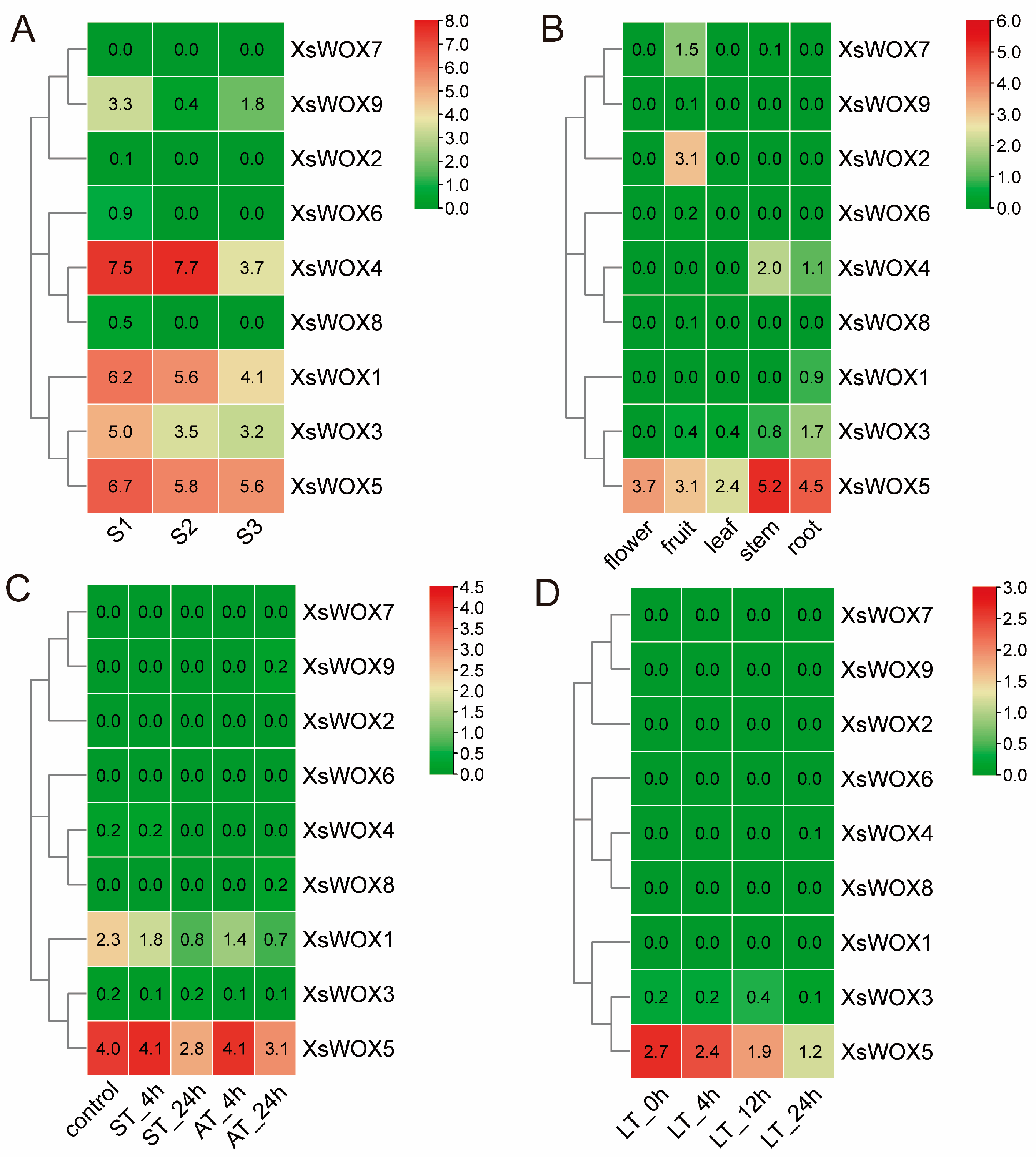
3.7. Transcriptome Analysis of Yellowhorn in Different Organs and under Different Stress Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, C.-Y.; Ha, W.; Lin, Y.; Jiang, K.; Yang, J.-L.; Shi, Y.-P. Polyphenols isolated from Xanthoceras sorbifolia husks and their anti-tumor and radical-scavenging activities. Molecules 2016, 21, 1694. [Google Scholar] [CrossRef]
- Jin, H.; Zou, J.; Li, L.; Bai, X.; Zhu, T.; Li, J.; Xu, B.; Wang, Z. Physiological responses of yellow-horn seedlings to high temperatures under drought condition. Plant Biotechnol. Rep. 2020, 14, 111–120. [Google Scholar] [CrossRef]
- Yao, Z.-Y.; Qi, J.-H.; Yin, L.-M. Biodiesel production from Xanthoceras sorbifolia in China: Opportunities and challenges. Renew. Sustain. Energy Rev. 2013, 24, 57–65. [Google Scholar] [CrossRef]
- Wang, H.; Yao, X.; Sui, G.; Yin, L.; Wang, L. Properties of Xanthoceras sorbifolia husk fibers with chemical treatment for applications in polymer composites. J. Mater. Sci. Technol. 2015, 31, 164–170. [Google Scholar] [CrossRef]
- Chen, X.; Lei, Z.; Cao, J.; Zhang, W.; Wu, R.; Cao, F.; Guo, Q.; Wang, J. Traditional uses, phytochemistry, pharmacology and current uses of underutilized Xanthoceras sorbifolium bunge: A review. J. Ethnopharmacol. 2022, 283, 114747. [Google Scholar] [CrossRef]
- Huang, Y.; Yin, Z.; Guo, J.; Wang, F.; Zhang, J. Oil extraction and evaluation from yellow horn using a microwave-assisted aqueous saline process. Molecules 2019, 24, 2598. [Google Scholar] [CrossRef] [PubMed]
- Egertsdotter, U.; Ahmad, I.; Clapham, D. Automation and scale up of somatic embryogenesis for commercial plant production, with emphasis on conifers. Front. Plant Sci. 2019, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Yarra, R.; Jin, L.; Zhao, Z.; Cao, H. Progress in tissue culture and genetic transformation of oil palm: An overview. Int. J. Mol. Sci. 2019, 20, 5353. [Google Scholar] [CrossRef] [PubMed]
- Von Arnold, S.; Sabala, I.; Bozhkov, P.; Dyachok, J.; Filonova, L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult. 2002, 69, 233–249. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X. Regulation of somatic embryogenesis in higher plants. Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Jiménez, V.M.; Bangerth, F. Endogenous hormone concentrations and embryogenic callus development in wheat. Plant Cell Tissue Organ Cult. 2001, 67, 37–46. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Zuo, J.; Niu, Q.W.; Frugis, G.; Chua, N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Jha, P.; Van Staden, J. LEAFY COTYLEDONs (LECs): Master regulators in plant embryo development. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 475–487. [Google Scholar] [CrossRef]
- Wei, X.; Geng, M.; Li, J.; Duan, H.; Li, F.; Ge, X. GhWOX11 and GhWOX12 promote cell fate specification during embryogenesis. Ind. Crop. Prod. 2022, 184, 115031. [Google Scholar] [CrossRef]
- Zhao, Q.-h.; Fisher, R.; Auer, C. Developmental phases and STM expression during Arabidopsis shoot organogenesis. Plant Growth Regul. 2002, 37, 223–231. [Google Scholar] [CrossRef]
- Sugimoto, K.; Meyerowitz, E.M. Regeneration in Arabidopsis tissue culture. Plant Organog. Methods Protoc. 2013, 959, 265–275. [Google Scholar]
- Yang, Z.; Gong, Q.; Qin, W.; Yang, Z.; Cheng, Y.; Lu, L.; Ge, X.; Zhang, C.; Wu, Z.; Li, F. Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017, 17, 113. [Google Scholar] [CrossRef]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Li, F.-W.; Kramer, E.M. Large-scale phylogenomic analysis suggests three ancient superclades of the WUSCHEL-RELATED HOMEOBOX transcription factor family in plants. PLoS ONE 2019, 14, e0223521. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Chang, Y.; Song, X.; Zhang, Q.; Liu, H.; Bai, Y.; Lei, X.; Pei, D. Genome-wide identification of WOX gene family and expression analysis during rejuvenational rhizogenesis in walnut (Juglans regia L.). Forests 2019, 11, 16. [Google Scholar] [CrossRef]
- Shafique Khan, F.; Zeng, R.-F.; Gan, Z.-M.; Zhang, J.-Z.; Hu, C.-G. Genome-wide identification and expression profiling of the WOX gene family in Citrus sinensis and functional analysis of a CsWUS member. Int. J. Mol. Sci. 2021, 22, 4919. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The role of WOX genes in flower development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef]
- Hu, X.; Xu, L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis. Plant Physiol. 2016, 172, 2363–2373. [Google Scholar] [CrossRef]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef]
- Tian, H.; Jia, Y.; Niu, T.; Yu, Q.; Ding, Z. The key players of the primary root growth and development also function in lateral roots in Arabidopsis. Plant Cell Rep. 2014, 33, 745–753. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsis homeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef]
- Xu, M.; Xie, W.; Huang, M. Two WUSCHEL-related HOMEOBOX genes, PeWOX11a and PeWOX11b, are involved in adventitious root formation of poplar. Physiol. Plant. 2015, 155, 446–456. [Google Scholar] [CrossRef]
- Zhang, A.; Li, Y.; Yarra, R.; Li, R.; Cao, H.; Jin, L. Genome-Wide Identification of WUSCHEL-Related Homeobox Gene Family and their Expression Analysis During Somatic Embryogenesis in Oil Palm (Elaeis guineensis). Trop. Plant Biol. 2022, 15, 55–64. [Google Scholar] [CrossRef]
- Tang, Y.; Li, H.; Guan, Y.; Li, S.; Xun, C.; Dong, Y.; Huo, R.; Guo, Y.; Bao, X.; Pei, E. Genome-wide identification of the physic nut WUSCHEL-related homeobox gene family and functional analysis of the abiotic stress responsive gene JcWOX5. Front. Genet. 2020, 11, 670. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chen, X.; Lou, X.; Zhang, Y.; Han, X.; Yang, Q.; Tong, Z.; Zhang, J. Identification of WUSCHEL-related homeobox (WOX) gene family members and determination of their expression profiles during somatic embryogenesis in Phoebe bournei. For. Res. 2023, 3, 1581. [Google Scholar] [CrossRef]
- Liu, H.; Yan, X.-M.; Wang, X.-r.; Zhang, D.-X.; Zhou, Q.; Shi, T.-L.; Jia, K.-H.; Tian, X.-C.; Zhou, S.-S.; Zhang, R.-G. Centromere-specific retrotransposons and very-long-chain fatty acid biosynthesis in the genome of yellowhorn (Xanthoceras sorbifolium, sapindaceae), an oil-producing tree with significant drought resistance. Front. Plant Sci. 2021, 12, 2546. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef]
- Feng, C.; Zou, S.; Gao, P.; Wang, Z. In silico identification, characterization expression profile of WUSCHEL-Related Homeobox (WOX) gene family in two species of kiwifruit. PeerJ 2021, 9, e12348. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- KB, N. GeneDoc: Analysis and visualization of genetic variation. EMBnet News 1997, 4, 1–4. [Google Scholar]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Wang, W. The molecular detection of Corynespora Cassiicola on cucumber by PCR assay using DNAman software and NCBI. In Proceedings of the Computer and Computing Technologies in Agriculture IX: 9th IFIP WG 5.14 International Conference, CCTA 2015, Beijing, China, 27–30 September 2015; Revised Selected Papers, Part II 9. pp. 248–258. [Google Scholar]
- Liu, Z.; Saiyinduleng; Chang, Q.; Cheng, C.; Zheng, Z.; Yu, S. Identification of yellowhorn (Xanthoceras sorbifolium) WRKY transcription factor family and analysis of abiotic stress response model. J. For. Res. 2021, 32, 987–1004. [Google Scholar] [CrossRef]
- Park, H.M. Comparing Group Means: T-Tests and One-Way ANOVA Using Stata, SAS, R, and SPSS; The University Information Techology Services (UITS) Center for Statistical and Mathematical Computing, Indiana University: Bloomington, IN, USA, 2009. [Google Scholar]
- Tvorogova, V.; Krasnoperova, E.; Potsenkovskaia, E.; Kudriashov, A.; Dodueva, I.; Lutova, L. What does the WOX say? Review of regulators, targets, partners. Mol. Biol. 2021, 55, 362–391. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.-a. Genome-wide analysis of the WOX gene family and function exploration of GmWOX18 in soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yao, L.; Sun, L.; Zhu, Z. ETHYLENE INSENSITIVE 3 suppresses plant de novo root regeneration from leaf explants and mediates age-regulated regeneration decline. Development 2020, 147, dev179457. [Google Scholar] [CrossRef]
- Nic-Can, G.I.; López-Torres, A.; Barredo-Pool, F.; Wrobel, K.; Loyola-Vargas, V.M.; Rojas-Herrera, R.; De-la-Peña, C. New insights into somatic embryogenesis: LEAFY COTYLEDON1, BABY BOOM1 and WUSCHEL-RELATED HOMEOBOX4 are epigenetically regulated in Coffea canephora. PLoS ONE 2013, 8, e72160. [Google Scholar] [CrossRef]
- Gambino, G.; Minuto, M.; Boccacci, P.; Perrone, I.; Vallania, R.; Gribaudo, I. Characterization of expression dynamics of WOX homeodomain transcription factors during somatic embryogenesis in Vitis vinifera. J. Exp. Bot. 2011, 62, 1089–1101. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar] [CrossRef]
- Liu, Z.; Dai, X.; Li, J.; Liu, N.; Liu, X.; Li, S.; Xiang, F. The type-B cytokinin response regulator ARR1 inhibits shoot regeneration in an ARR12-dependent manner in Arabidopsis. Plant Cell 2020, 32, 2271–2291. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Yarra, R.; Zhou, L.; Zhao, Z.; Cao, H. miRNAs as key regulators via targeting the phytohormone signaling pathways during somatic embryogenesis of plants. 3 Biotech 2020, 10, 495. [Google Scholar] [CrossRef]
- Zhou, S.; Jiang, W.; Long, F.; Cheng, S.; Yang, W.; Zhao, Y.; Zhou, D.-X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 2017, 29, 1088–1104. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Y.; Chen, X.; Zheng, Y.; Sun, Y.; Yang, J.; Ye, N. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 2019, 33, 1129–1142. [Google Scholar] [CrossRef]
- Li, Y.; Jin, C.; Liu, Y.; Wang, L.; Li, F.; Wang, B.; Liu, G.; Jiang, J.; Li, H. Global Analysis of the WOX Transcription Factor Gene Family in Populus× xiaohei TS Hwang et Liang Reveals Their Stress−Responsive Patterns. Forests 2022, 13, 122. [Google Scholar] [CrossRef]


| Genome ID | Gene Name | Protein Length (aa) | MW (Da) | pI | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|
| XS04G0106200.1 | XsWOX1 | 289 | 31,923.78 | 5.77 | −0.32 | Nuclear |
| XS05G0232400.1 | XsWOX2 | 264 | 30,132.33 | 10.11 | −0.814 | Nuclear |
| XS07G0006100.1 | XsWOX3 | 253 | 28,800.05 | 5.79 | −0.426 | Nuclear |
| XS07G0075600.1 | XsWOX4 | 221 | 25,333.47 | 9.11 | −0.992 | Nuclear |
| XS08G0109100.1 | XsWOX5 | 257 | 28,795.94 | 5.41 | −0.814 | Nuclear |
| XS08G0130200.1 | XsWOX6 | 319 | 35,920.22 | 5.33 | −0.983 | Nuclear |
| XS13G0025800.1 | XsWOX7 | 232 | 26,842.18 | 9.5 | −0.931 | Nuclear |
| XS13G0128800.1 | XsWOX8 | 254 | 28,201.88 | 7.79 | −0.827 | Nuclear |
| XS14G0083400.1 | XsWOX9 | 193 | 22,498.07 | 7.00 | −0.975 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Xie, X.; Le, L.; Cao, F. Transcriptional Profiling Reveals Key Regulatory Roles of the WUSCHEL-Related Homeobox Gene Family in Yellowhorn (Xanthoceras sorbifolia Bunge). Forests 2024, 15, 376. https://doi.org/10.3390/f15020376
Zhang W, Xie X, Le L, Cao F. Transcriptional Profiling Reveals Key Regulatory Roles of the WUSCHEL-Related Homeobox Gene Family in Yellowhorn (Xanthoceras sorbifolia Bunge). Forests. 2024; 15(2):376. https://doi.org/10.3390/f15020376
Chicago/Turabian StyleZhang, Wentao, Xinyao Xie, Linlin Le, and Fuliang Cao. 2024. "Transcriptional Profiling Reveals Key Regulatory Roles of the WUSCHEL-Related Homeobox Gene Family in Yellowhorn (Xanthoceras sorbifolia Bunge)" Forests 15, no. 2: 376. https://doi.org/10.3390/f15020376
APA StyleZhang, W., Xie, X., Le, L., & Cao, F. (2024). Transcriptional Profiling Reveals Key Regulatory Roles of the WUSCHEL-Related Homeobox Gene Family in Yellowhorn (Xanthoceras sorbifolia Bunge). Forests, 15(2), 376. https://doi.org/10.3390/f15020376






