Abstract
The rubber elongation factor (REF) is the most abundant protein in the latex of Hevea brasiliensis, which is closely related to natural rubber biosynthesis. In order to gain a deeper understanding of the transcriptional regulation mechanism of HbREF1, a 1758 bp genomic DNA fragment of the HbREF1 promoter was isolated. Promoter sequence analysis revealed several transcription factor binding sites in the HbREF1 promoter, such as bZIP, bHLH, EIL, AP2/ERF, MYB, and Trihelix. To assess the promoter activity, a series of HbREF1 promoter deletion derivatives were created and fused with firefly luciferase (LUC). The LUC image demonstrated that all of the HbREF1 promoters exhibited transcriptional activity. Furthermore, the assay revealed the presence of multiple regulatory elements within the promoter region that negatively regulate the transcriptional activity. Subsequent analysis of the transcriptional activity following treatment with phytohormones identified an ABA-responsive element located between −583 bp and −200 bp, an ET-responsive element between −718 bp and −583 bp, a JA-responsive element between −1758 bp and −1300 bp, and a SA-responsive element between −1300 bp and −718 bp. These results were largely consistent with the predictions of cis-acting elements. This study has established significant groundwork for future investigations into the regulatory mechanism of HbREF1.
1. Introduction
Natural rubber (NR) is a vital industrial raw material. It possesses excellent properties such as good toughness, elasticity, insulation, and high-temperature resistance. It cannot be completely replaced by synthetic rubber in important engineering fields, especially in high-end manufacturing, such as the production of aviation tires. Among more than 2500 rubber-producing plant species, the rubber tree (Hevea brasiliensis Mull. Arg.) is the primary commercial source of NR [1,2].
The main component of NR is cis-1,4-polyisoprene, which is biosynthesized in the laticifer cells of the rubber tree. Nature latex is the cytoplasm of the laticifer cells, and it contains 30–50% NR. The biosynthesis of NR is a key factor in determining its production, and its biosynthetic pathway has been relatively clear [3]. The precursor to NR biosynthesis is isopentenyl pyrophosphate (IPP), which is generated through the mevalonate (MVA) pathway and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway. It then goes through three phases: initiation of rubber precursors, extension of the hydrocarbon chain, and termination of the hydrocarbon chain. Following these three phases, the cis-1,4-polyisoprene is finally formed [4]. The extension of the hydrocarbon chain is catalyzed by the rubber transferase complex, which is bound to rubber particles. The rubber elongation factor (REF) is an important component of the rubber transferase complex and is involved in regulating the extension of the rubber hydrocarbon chain and the formation of high molecular weight.
The REF is the most abundant protein in rubber latex and is essential for the biosynthesis of NR [5,6,7]. In vitro studies have shown that the REF can promote the incorporation of IPP into whole latex or washed rubber particles, and this activity can be inhibited by the corresponding antibodies [5,8,9]. The content of REF protein in latex was positively correlated with rubber content [5]. In Taraxacum brevicorniculatum, the rubber content has significantly decreased in the RNAi-silenced lines of TbREF, indicating that this protein plays a key role in NR biosynthesis [10]. The abundance of HbREF1 in the latex of high-yield rubber trees was three to five times higher than that of low-yield rubber trees, indicating that there is a positive correlation between the HbREF1 expression pattern and the latex yield [11,12,13]. Compared with other genes in the REF gene family, HbREF1 has the highest expression level in various tissues of the rubber tree and is highly expressed in latex [14]. It is one of the three pivotal genes in the rubber biosynthetic pathway [15]. Although many studies have been identified, the REF plays a positive role in rubber biosynthesis. No catalytic domain was found in the REF protein, indicating that the REF may function as a structural protein, stabilizing the rubber biosynthetic machinery on rubber particles [3].
Plant hormones are a group of disparate small molecules that act as chemical messengers to coordinate the activities of plant cells [16]. The use of plant hormones and other stimulants to increase NR yield by chemical stimulation has always been an important component in rubber production and theoretical research. Ethylene (ET) has been widely used to increase NR production. ET stimulation typically increases latex yield 1.5–2 fold [17], but overstimulation with ET may induce tapping panel dryness [18,19,20,21]. Tungngoen et al. [22] found that bark treatments with ET, auxin, abscisic acid (ABA), and salicylic acid (SA) in mature, untapped rubber trees could increase latex production. The role mechanisms of ET, ABA, and SA are still not well understood. Methyl jasmonate (MeJA) has been reported to increase natural rubber production by inducing laticifer differentiation, regulating natural rubber synthesis, and extending the duration of latex flow [23,24,25,26,27,28]. The jasmonate-signaling pathway has been identified as COI1-JAZ3-MYC2 in laticifer cells. Its activation is associated with enhanced rubber biosynthesis via the upregulation of the expression of a farnesyl pyrophosphate synthase gene and a small rubber particle protein gene [29].
Previous research mainly focuses on the functional characterization of the REF protein and the identification of interacting proteins [30,31,32,33], while the transcriptional regulatory mechanism of HbREF1 in rubber trees has not been reported. Here, transcription factors binding sites (TFBS) related to phytohormones in the HbREF1 promoter were predicted and their function identified, and the regulation of transcriptional activity was studied after treatment with ABA, ET, MeJA, and SA, which laid a foundation for further study of the regulatory mechanism of HbREF1.
2. Materials and Methods
2.1. Plant Materials
The rubber tree leaves used for DNA extraction were harvested from the Hevea brasiliensis clone Reyan 7-33-97, which was grown at the experimental plantation of the Chinese Academy of Tropical Agricultural Sciences (Hainan, China). Tobacco plants (Nicotiana benthamiana) for agroinfiltration were grown in growth chambers with a 16 h/8 h light/dark cycle at 26 °C for six weeks. Escherichia coli strain DH5a was used to clone and propagate all recombinant plasmid vectors. Agrobacterium tumefaciens strain GV3101 (psoup19) was used for tobacco leaf infiltration.
2.2. Cloning and Sequence Analysis of the HbREF1 Promoter
Leaves of the Hevea brasiliensis clone Reyan 7-33-97 were collected and ground to a fine powder in liquid nitrogen, and genomic DNA was extracted by using a plant genomic DNA extraction kit (Foregene, Chengdu, China). A pair of special primers, REF-F: 5′-AATTTTAGCTCTCTAATCTCGATGC-3′ and REF-R: 5’-AGCCATAATCGAAGATTTCCTTTTGC-3′, was designed based on the HbREF1 promoter sequence in the NCBI database (Accession NO. AB861873). PCR was performed in 20 μL reaction volume, which was composed of 1 μL (25 ng) template DNA, 10 μL 2 × Tap-HS PCR Mix, 0.2 μL of each 10 μM primer, and 8.6 μL ddH2O. The PCR amplification profile consisted of a first cycle at 94 °C for 3 min, followed by 35 cycles at 94 °C for 20 s/58 °C 40 s/72 °C 40 s, and a last cycle at 72 °C for 5 min. The PCR product was purified and cloned into a pMD19-T easy vector (TaKaRa, Dalian, China). The cloned vector was sequenced and named pMD19-REF1P.
TSSPlant (http://www.softberry.com/berry.phtml?topic=tssplant&group=programs&subgroup=promoter) (accessed on 12 January 2024) was used for predicting the transcription start site of HbREF1 [34]. The JASPAR core plantae motifs (https://jaspar.elixir.no/) (accessed on 12 January 2024) [35,36] and PlantRegMap (http://plantregmap.gao-lab.org/binding_site_prediction.php) (accessed on 26 January 2024) [37] were used to identify the TFBS in the HbREF1 promoter.
2.3. Construction of the HbREF1 Promoter Deletion Constructs
The 1758 bp HbREF1 promoter region and a series of 5′ deletions fragments (1300, 718, 583, and 200 bp) were generated using PCR amplification from pMD19-REF1P. The forward primers REFP-1758F (5′-GCCTCGAGATTTTAGCTCTCTAATCTCGATGCT-3′), REFP-1300F (5′-GCCTCGAGATATTTTTGCAGGGTTTTTTTTTT-3′), REFP-718 (5′-GCCTCGAGGTAATTTTGGAAGCCATTTAAATA-3′), REFP-583F (5′-GCCTCGAGTTTTAAAAAGGAACAGATTGTTAA-3′), and REFP-200F (5′-GCCTCGAGACACGCTCCTTTTCTTAACAG-3′) were designed to correspond to the −1758, −1300, −718, −583, and −200 sequences of the HbREF1 promoter, and the reverse primer REFP-R (5′-GCGGATCCTAATCGAAGATTTCCTTTTGCA-3′) was located in the 3′ end of the HbREF1 promoter. A XhoI restriction site (underlined sequences) was introduced into the forward primers and a BamHI restriction site (underlined sequences) was introduced into the reverse primer. Each of the five promoter fragments was digested by XhoI and BamHI and ligated into the XhoI/BamHI digested binary vector pGreenII0800-LUC.
2.4. Agrobacterium-Mediated Transient Expression Assays
The promoter constructs were transferred into Agrobacterium tumefaciens strain GV3101(psoup19). The individual colony of transformed A. tumefaciens GV3101 harboring a promoter construct was inoculated into 3 mL LB liquid medium supplemented with rifampicin (50 μg/mL) and kanamycin (50 μg/mL) and grown for 20 h at 28 °C with an agitation of 200 rpm. Post-cultures were diluted 1:25 into 25 mL of fresh LB liquid medium containing the same antibiotics and incubated at 28 °C for 6 h with an agitation of 200 rpm. Agrobacterium cells were harvested after centrifugation at 4000× g for 10 min and resuspended in infiltration solution (10 mM MES, pH 5.7, 10 mM MgCl2, and 100 μM acetosyringone) to a final OD600 of 0.8 for agroinfiltration. The solution was infiltrated into the abaxial surfaces of fully expanded young tobacco leaves by the use of a needleless syringe [38]. The tobacco plants were maintained in a moist chamber under a 16/8 h day/night cycle at 25 °C.
2.5. Plant Hormone Treatments
Tobacco leaves were evenly sprayed with 100 µM ABA, 100 µM ethylene, 100 µM MeJA, and 100 µM SA, respectively, 48 h after agroinfiltration. The control plants were sprayed with water. Treated tobacco plants were then incubated in a vinyl bag. The infiltrated leaves were collected and the protein was extracted for LUC activities assay 24 h after treatment.
2.6. Imaging of Luciferase Bioluminescence
The same position of the infiltrated tobacco leaves was sprayed with a D-luciferin potassium salt working solution after 72 h, and the luciferase signals in the infiltrated region were detected by a Lumazone Sophia 2048B (TELEDYNE, Trenton, NJ, USA) imaging apparatus 10 min later.
2.7. Dual-luciferase Activity Assays
The infiltrated leaves were collected for total protein extraction to measure LUC activities up until 72 h after infiltration. The supernatant of total proteins was treated with the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) following the manufacturer’s instructions, and the fluorescent values of LUC and REN were detected using a GloMax®-Multi+ Detection System (Promega, Madison, WI, USA). The LUC value was normalized to that of REN. Three biological repeats were measured for each combination.
3. Results
3.1. Cloning of HbREF1 Promoter
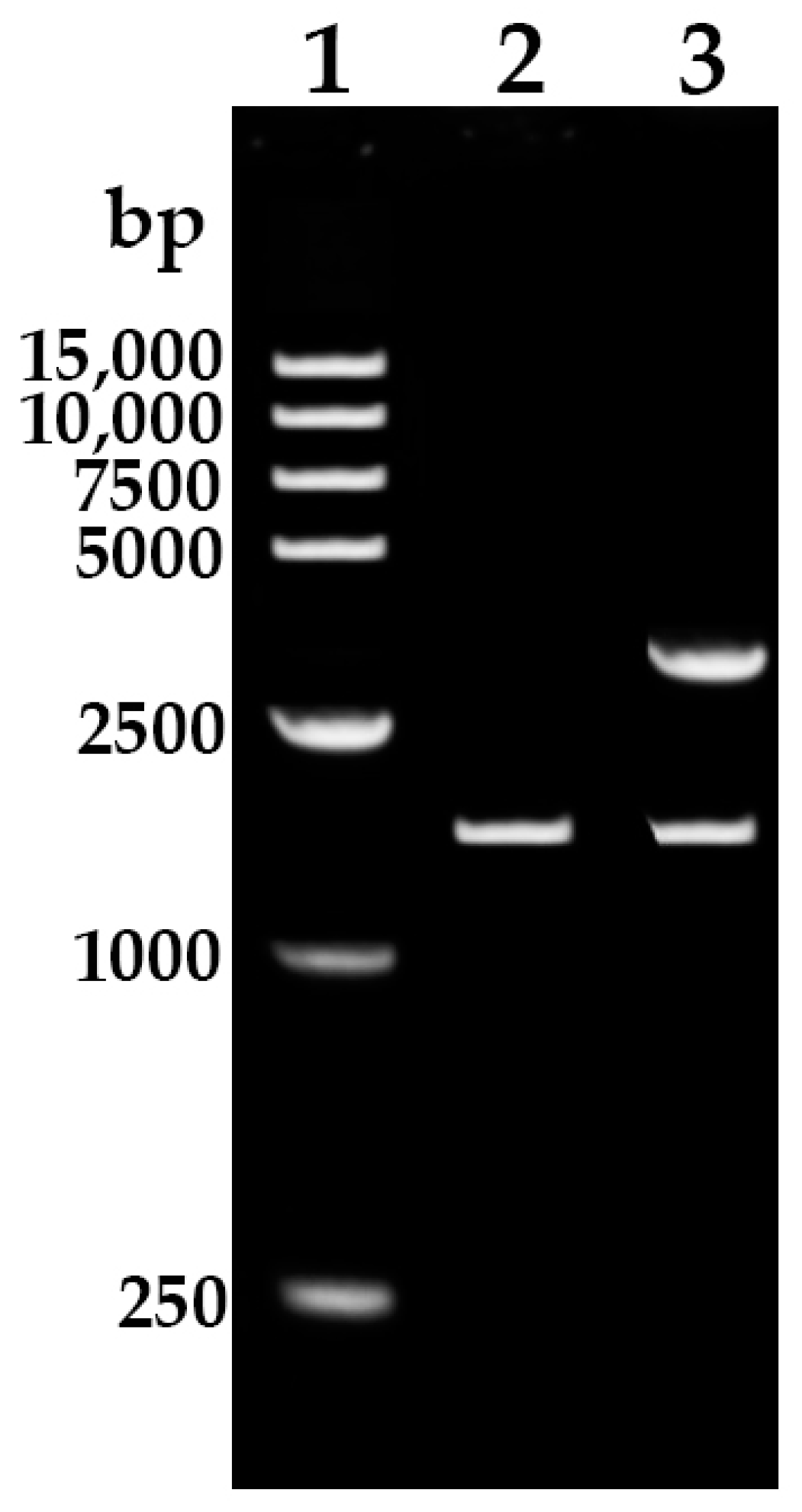
Using specific primers REF-F and REF-R, a fragment of about 1700 bp was amplified from the rubber tree’s genomic DNA by PCR (Figure 1). Sequencing results showed that the sequence length was 1758 bp, which is consistent with the genomic sequence of the HbREF1 (Gene NO. LOC110644928) promoter in NCBI, indicating that the HbREF1 promoter was successfully cloned.
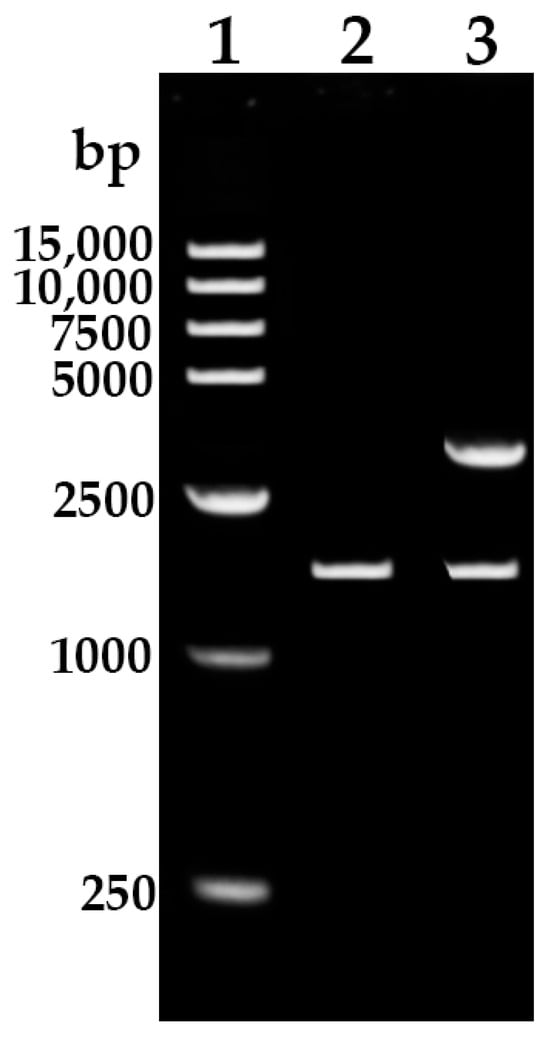
Figure 1.
PCR amplification of HbREF1 promoter and construction of pMD19-REF1P. Lane 1, 15,000 DNA ladder; lane 2, the PCR product of HbREF1 promoter amplified from rubber tree genomic; lane 3, pMD19-REF1P double digested with XhoI and BamHI to release the HbREF1 promoter insert fragment.
3.2. HbREF1 Promoter Sequence Analysis
The prediction result from the TSSPlant tool shows that the transcription start site of HbREF1 was located 118 bp upstream of the translation start site (the translation start site was designated as +1) and the promoter core sequence TATA-box was located at −150 bp. The HbREF1 promoter sequence was scanned to identify the TFBS by using the JASPAR core plantae motifs (Threshold:0.9) to concurrently perform a binding site prediction on the PlantRegMap website (threshold p ≤ 1 × 10−5). Combined, the predicted binding sites from both websites equal the result of the TFBSs. This result shows that six types of transcription factors related to plant abiotic stress response or phytohormone signal have binding sites in the HbREF1 promoter (Table 1). Among the different TF families, MYB has the most with four binding sites, bZIP has three binding sites, bHLH and ERF/DREB each have two binding sites, and EIL and Trihelix each have one binding site.

Table 1.
Identification of transcription factor binding sites (TFBS) in the HbREF1 promoter.
3.3. Construction and Identification of the 5′ Deletion Fragments of HbREF1 Promoter
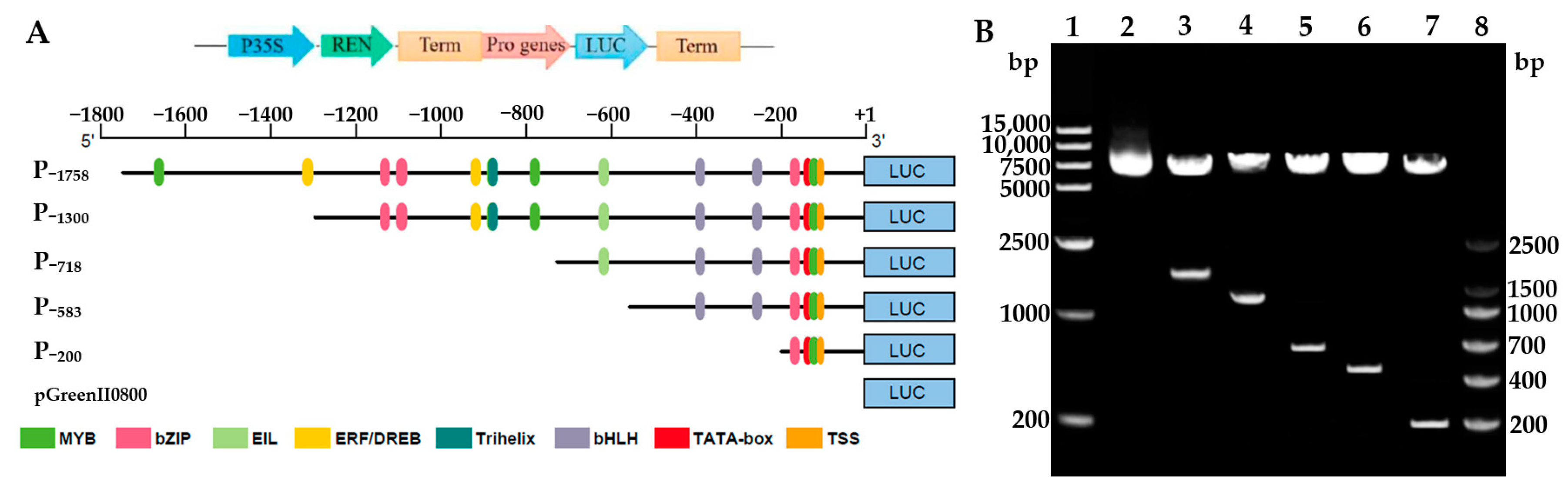
To clarify the core regulatory region and hormone response element distribution of the HbREF1 promoter, a series of 5’ promoter deletions were performed and five derivatives of the HbREF1 promoter fragments were fused to firefly luciferase (LUC) according to the cis-acting elements analysis (Figure 2A). The constructs were digested by restriction enzymes and generated exogenous fragments of expected size, respectively (Figure 2B). This indicates that the HbREF1 promoter deletion fragment expression vectors have been successfully constructed. The recombinant vectors were named P−1758, P−1300, P−718, P−583, and P−200, respectively.
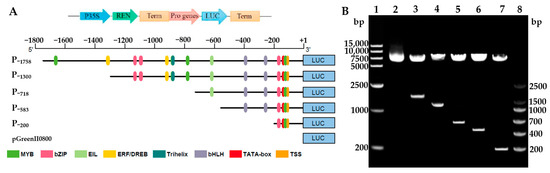
Figure 2.
Construction of the HbREF1 promoter deletion constructs. (A) Schematic diagram of the HbREF1 promoter deletion structures used for assay of transcriptional activity. The translation start site is indicated as +1. The numbers on each structure indicate the distance from the HbREF1 translation start site. Plasmids pGreenII0800-LUC were used as negative controls. (B) Identification of HbREF1 promoter deletion constructs by double digestion. Lanes 1 and 8, 15,000 DNA ladder and DNA marker V; lanes 2–7, double digested with XhoI and BamHI in order of pGreenII0800-LUC, P−1758, P−1300, P−718, P−583, and P−200.
3.4. Expression Analysis of Luciferase Driven by HbREF1 Promoter
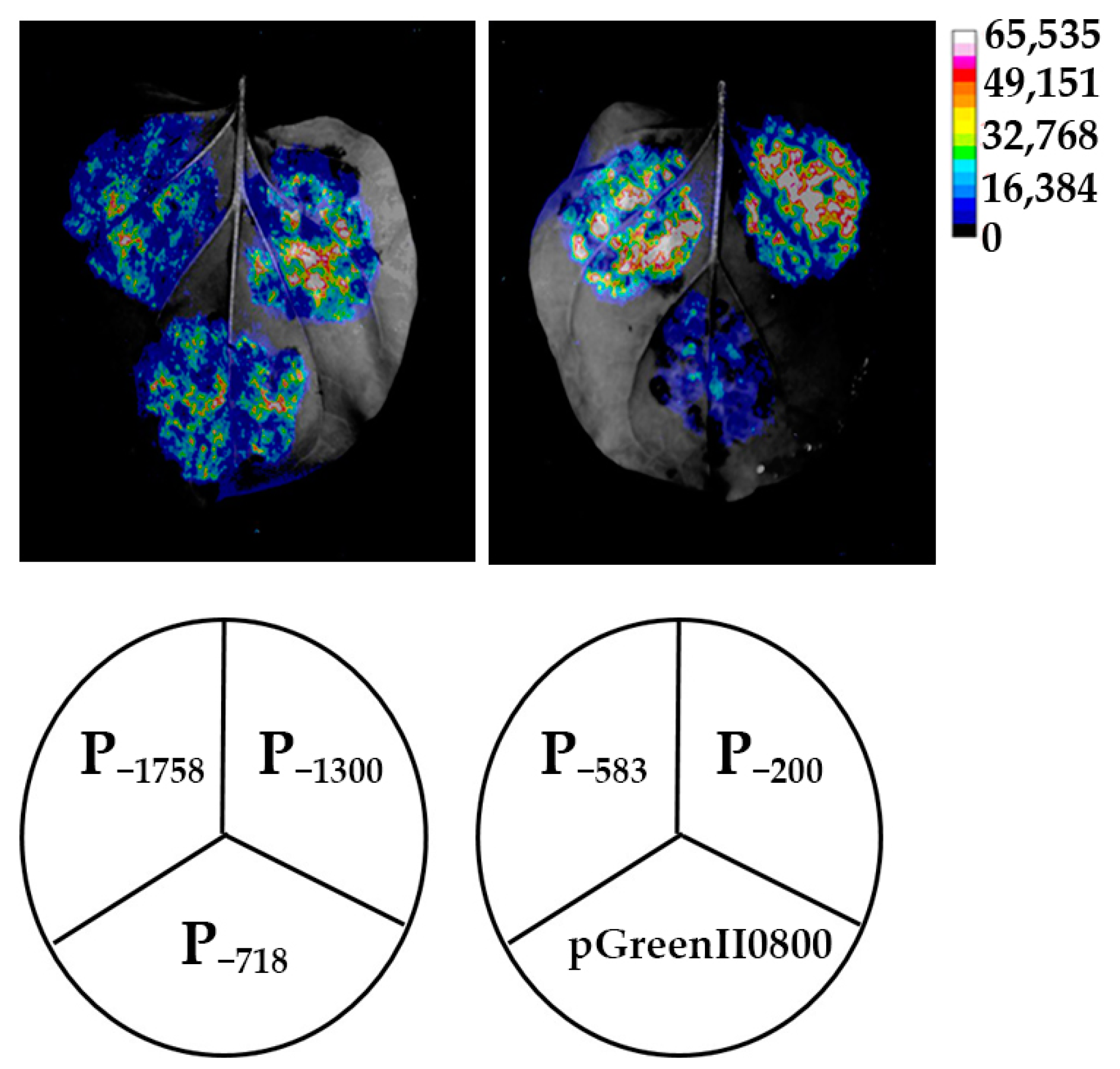
To identify whether the HbREF1 promoter fragments have transcriptional activity, each vector was transiently expressed in tobacco leaves through Agrobacterium-mediated transformation, with the pGreenII0800-LUC vector as a negative control. The LUC in vivo bioluminescence imaging showed that strong bioluminescence signals were detected in all five promoters, P−1758, P−1300, P−718, P−583, and P−200, while almost no light signal was detected in the negative control (Figure 3). This result suggests that all of the HbREF1 promoter fragments have transcriptional activity.
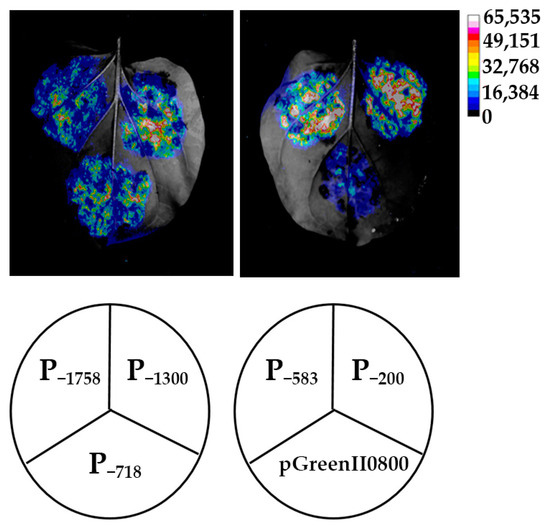
Figure 3.
Firefly bioluminescence imaging of HbREF1 promoter transient expression in tobacco leaves.
3.5. Basal Transcriptional Activity Analysis of HbREF1 Promoter
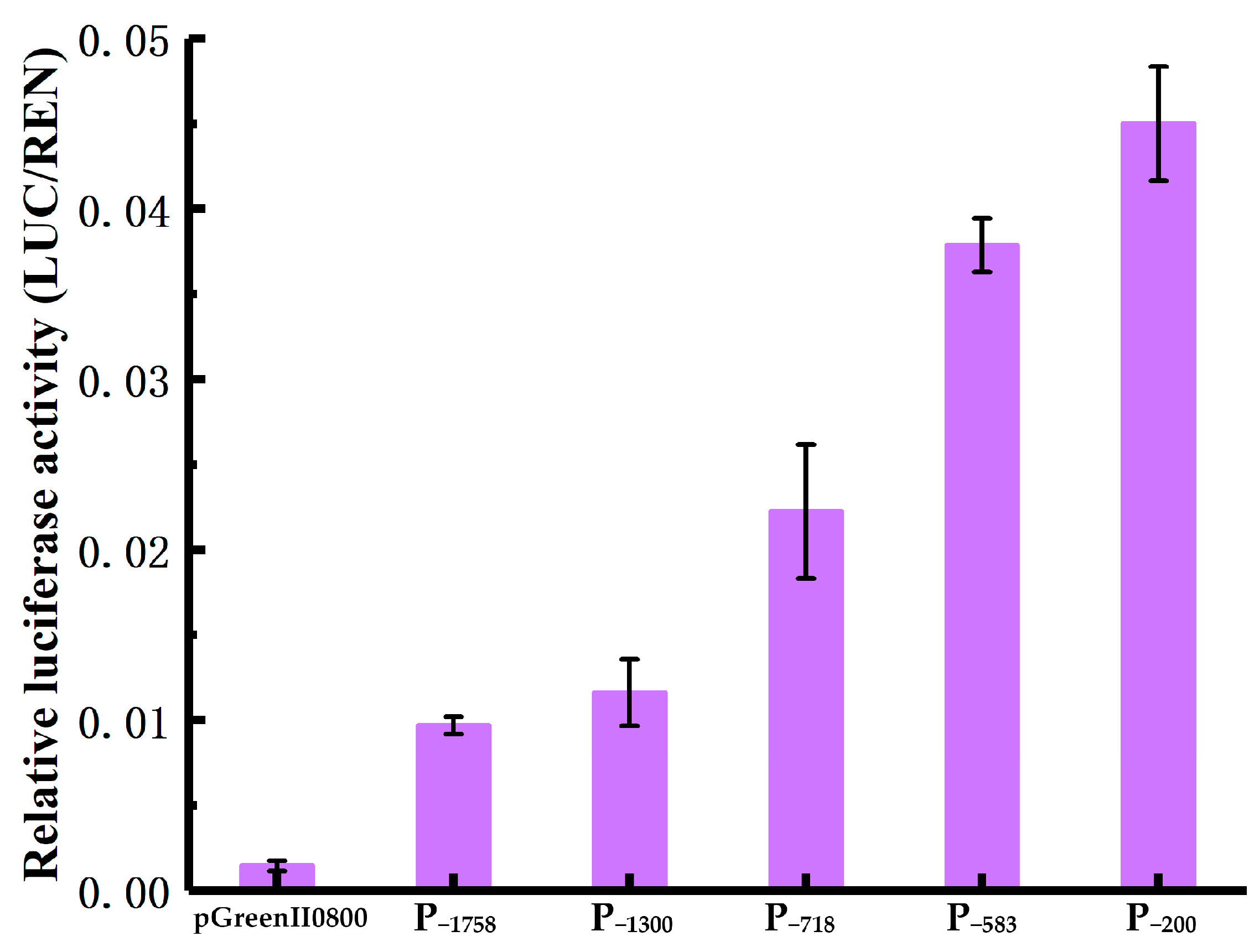
Using a dual-luciferase reporter system, the basal transcriptional activity of the HbREF1 promoter fragments was measured. The results are shown in Figure 4. Compared with the transcriptional activity of the promoter, the negative control had almost no luciferase activity. Among the five promoter fragments, P−1758 had the lowest luciferase activity and P−200 had the highest luciferase activity. As the 5′ terminal deletion fragments of the HbREF1 promoter increased, the transcriptional activity of the promoter was enhanced. The transcriptional activity of P−718 showed a greater increase than P−1300, and P−583 showed a greater increase than P−718. These results indicate that the HbREF1 promoter region of the 5′ terminal deletion fragments contained multiple elements that suppressed the transcriptional activity of the promoter, and there were strong elements that suppressed the transcriptional activity in the −1300 bp~−718 bp and −718 bp~−583 bp regions.
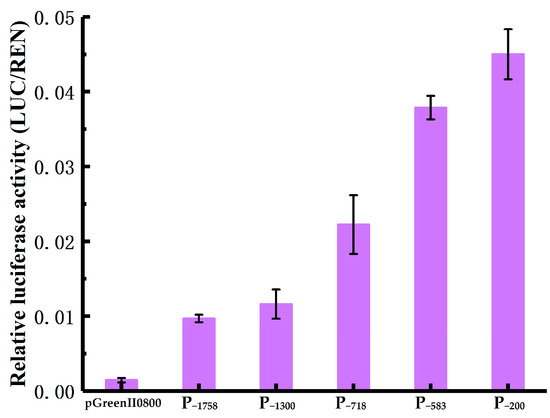
Figure 4.
Luciferase activity assays in tobacco leaves transiently transformed with HbREF1 promoter deletion constructs. Data are means ± standard deviations from three independent experiments.
3.6. Responsiveness Analysis of HbREF1 Promoter to Phytohormone Treatment
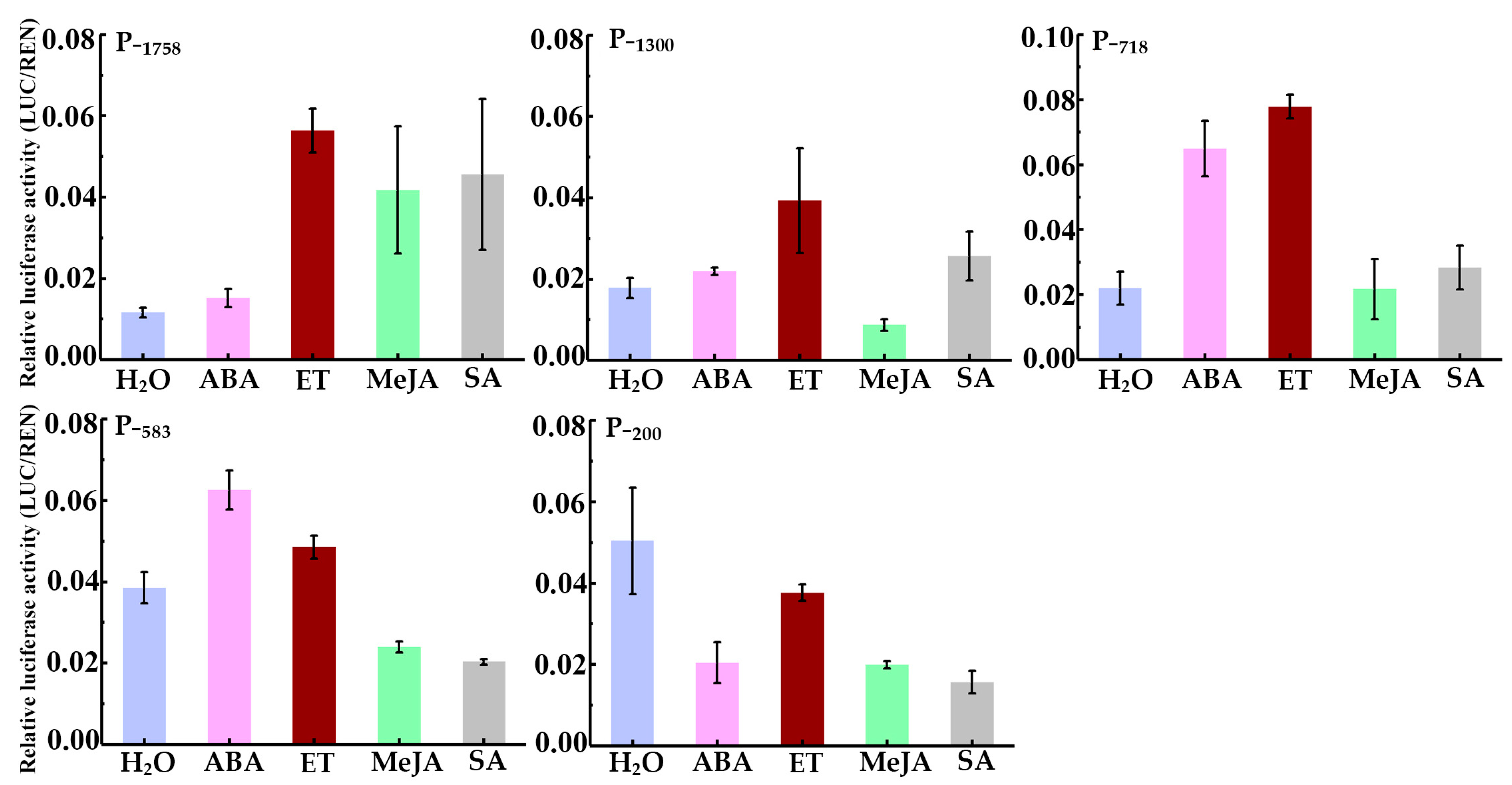
To study the transcriptional regulation by phytohormone, the transcriptional activity of the HbREF1 promoter fragments was measured after ABA, ET, MeJA, and SA treatment and after being sprayed with ddH2O as a control (Figure 5). ABA increased the transcriptional activity of P−1300, P−718, and P−583, ET increased the transcriptional activity of P−1758, P−1300, and P−718, MeJA increased the transcriptional activity of P−1300, and SA increased the transcriptional activity of P−1758 and P−1300. The transcriptional activity of P−200 showed a decrease after ABA, MeJA, and SA treatment. These results suggest that the −583 bp to −200 bp region of the HbREF1 promoter contained ABA-responsive elements, the −718 bp to −583 bp region contained ET-responsive elements, the −1758 bp to −1300 bp region contained JA-responsive elements, and the −1300 bp to −718 bp region contained SA-responsive elements.
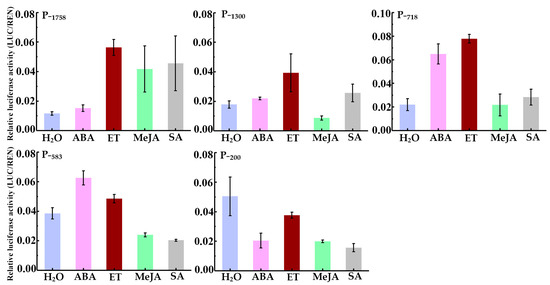
Figure 5.
HbREF1 promoter activity after treatment with ABA, ET, MeJA, and SA. Data are means ± standard deviations from three independent experiments.
4. Discussion
Promoter sequences are located upstream of the 5′ end of the structural gene, which can be recognized and bound by RNA polymerase and initiate gene transcription. Eukaryotic RNA polymerase requires interaction with transcription factors in order to initiate the process of transcription. These transcription factors possess the ability to locate the specific DNA elements for binding and to attract other proteins to the transcription site [39]. Eukaryotic promoters mainly include two types of regulatory elements: relatively conservative basic elements and cis-elements with great differences between genes [40]. The composition of promoter elements, especially cis-acting elements, is crucial for the temporal and spatial expression of genes and the regulation of transcription levels. In this study, the cloned HbREF1 promoter sequence of 1758 bp has an AT content as high as 75%. The analysis of the promoter sequence indicates that the core element of the TATA box required for transcription is located upstream of the translation start site at −150 bp. There are binding sites for nine types of transcription factors associated with plant abiotic stress and hormone responses in the HbREF1 promoter region. Transcription factor families such as MYB, bHLH, EIL, ERF/DREB, Trihelix, and bZIP play crucial roles in plant growth and development, secondary metabolism regulation, response to hormones, and regulation of abiotic stress [41,42,43,44,45,46]. MYC2, a member of the bHLH family, is a major regulatory factor in the JA signaling pathway. ABA is one of the important hormones involved in plant tolerance of nonbiological stress, and bZIP transcription factors play a significant role in ABA signal transduction. EIL and ERF/DREB are important transcription factors in the ethylene signaling pathway. TGA transcription factors (D subfamily of the bZIP transcription factor) have been reported to participate in SA response. Previous studies have shown that the expression of HbREF1 in latex is induced by ABA, MeJA, and tapping [29,47]. It is speculated that these predicted transcription factors regulate the transcription of HbREF1 in response to hormones and tapping damage.
Deng et al. [48] isolated the sequence of 378 bp upstream of the HbREF1 translation initiation site using the chromosome walking method. Priya et al. [12] fused the promoter sequence of 378 bp upstream of the translation initiation site with GUS and transformed it into tobacco; the results showed that the promoter sequence of 378 bp has the activity to initiate GUS transcription. However, these sequences provide too little information to study the detailed regulatory mechanism of HbREF1, and it was necessary to isolate longer promoter sequences. Transient expression analysis provides a rapid and effective method to identify whether the promoter has the active function of initiating downstream gene expression [49]. The transcriptional activity of the promoter was qualitatively and quantitatively analyzed by detecting the luciferase reporter gene. In this study, five HbREF1 promoter vectors fused to the luciferase reporter gene were constructed, with lengths of 1758 bp, 1300 bp, 718 bp, 583 bp, and 200 bp, respectively. The LUC in vivo bioluminescence imaging result showed that all five HbREF1 promoter vectors exhibited transcriptional activity. The basal transcriptional activity analysis result showed that as the deletion length of the HbREF1 promoter increases, the transcriptional activity becomes stronger. It is speculated that there are multiple elements that inhibited the promoter transcriptional activity in the −1758 bp to −200 bp region, especially in the −1300 bp to −583 bp region, where the inhibitory effect is stronger.
Tungngoen et al. [22] found that treating mature untapped rubber tree bark with ET, auxin, ABA, and SA can induce a transient increase in latex yield. Deng et al. [29] found that there is a JA signal transduction module COI1-JAZ3-MYC2 in the laticifer of rubber trees, which promotes the biosynthesis of NR by upregulating the expression of rubber biosynthesis genes. Previous studies have shown that treatment with ET, JA, and ABA can increase the expression of HbREF1 [11,29,47], while the effect of SA treatment on HbREF1 expression has not been reported. Sequence analysis reveals that the promoter sequence of HbREF1 contains multiple TFBSs for transcription factors that may respond to hormones such as ABA, ET, JA, and SA. In Hevea brasiliensis, the MYC transcription factors, as a core component of the JA signal, bind to the G-box motif of the HbPSK5 promoter and activate the HbPSK5 promoter. Overexpression of HbMYC26 and HbPSK5 in Taraxacum kok-saghyz could promote the formation of latex vessels and increase rubber content [50]. In Taraxacum brevicorniculatum, TbbZIP.1 binds to the ABRE elements of the TbSRPP promoter and regulates the expression of the TbSRPP in an ABA-dependent manner, increasing the latex content [51]. The results of this study suggest that the transcriptional activity of the HbREF1 promoter can be induced by ABA, ET, JA, and SA. An analysis of transcriptional activity after treatment by ABA shows that the ABA response element is located upstream of the −200 bp region (Figure 5). The two TFBSs of bZIP were also predicted to exist upstream of 200 bp (Table 1). It was speculated that HbREF1 regulated by ABA is related to these two motifs. In response to ET treatment, the promoter fragments P−1758, P−1300, and P−718 were strongly activated to induce the LUC gene expression, but the promoter fragments P−583 and P−200 were not effective for the promoter activity (Figure 5). It is shown that the ET response element is located upstream of −583 bp. As shown in Table 1, the TFBSs of EIL and ERF/ERDB are located upstream of −585 bp, and they appear to serve as regulatory factors in the HbREF1 promoter that responds to ET. In response to JA treatment, only P−1758 was strongly activated to induce LUC gene expression, but its deletion derivatives (P−1300, P−718, P−583, and P−200) had no significant effect on promoter activity (Figure 5). It is shown that the JA response element is located in the upstream region of −1300 bp. However, the binding site of bHLH is located in the −1144 bp~−202 bp region. Previous studies have indicated that MYB and ERF/DREB are important regulatory factors in the JA signaling pathway [41,44]. One MYB binding site is located at −1677 bp, and one ERF/DREB binding site is located at −1328 bp (Table 1). The induction of HbREF1 by JA may be subject to collaborative regulation by multiple transcription factors. In SA treatment, it is shown that there is an SA response element located upstream of −718 bp (Figure 5), and one bZIP binding site (TGA) situated at −1107 bp (Table 1) that seems to function as a regulatory element within the HbREF1 promoter, responding to SA.
5. Conclusions
REF is the most abundant protein found on rubber particles, and it is an important component of the NR biosynthesis system [8]. It plays an important role in the stability of rubber particles and natural rubber biosynthesis [52]. In this study, the HbREF1 promoter was cloned, and the regulatory roles of each region of the HbREF1 promoter and the response positions of core elements to corresponding hormones were preliminarily determined by analyzing the basal transcriptional activity and the change after phytohormone treatment. This lays a solid foundation for the in-depth study of the molecular mechanism of HbREF1 expression regulation, further revealing the mechanism of NR biosynthesis and providing a theoretical basis for the high-yield genetic improvement of rubber trees through biotechnology methods in the future.
Author Contributions
Conceptualization, D.G., S.-Q.P. and Y.-Q.T.; investigation, D.G., L.-T.C., J.-H.Z., Y.W. and H.-L.L.; writing—original draft, D.G., L.-T.C. and F.A.; writing—review and editing, D.G., Y.-Q.T., F.A. and S.-Q.P. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Key R&D Program of China (2022YFD2301201), the Hainan Provincial Natural Science Foundation of China (321RC1097), the National Natural Science Foundation of China (31970620), and the Central Public-interest Scientific Institution Basal Research Fund for the Chinese Academy of Tropical Agricultural Sciences (No. 1630052022009).
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Gireesh, T.; Meenakumari, T.; Mydin, K.K. Fast track evaluation and selection of Hevea brasiliensis clones from a clonal nursery. Ind. Crop Prod. 2017, 103, 195–201. [Google Scholar] [CrossRef]
- Silva, M.J.; Cunha Claro, P.I.; Da Silva, J.C.; Scaloppi Junior, E.J.; Goncalves, P.D.S.; Martins, M.A.; Capparelli Mattoso, L.H. Evaluation of the physicochemical properties of natural rubber from Hevea brasiliensis clones. Ind. Crop Prod. 2021, 171, 113925. [Google Scholar] [CrossRef]
- Men, X.; Wang, F.; Chen, G.Q.; Zhang, H.B.; Xian, M. Biosynthesis of natural rubber: Current state and perspectives. Int. J. Mol. Sci. 2019, 20, 50. [Google Scholar] [CrossRef]
- Wei, X.; Peng, P.; Peng, F.; Dong, J. Natural polymer Eucommia ulmoides rubber: A novel material. J. Agric. Food Chem. 2021, 69, 3797–3821. [Google Scholar] [CrossRef]
- Dennis, M.S.; Light, D.R. Rubber elongation factor from Hevea brasiliensis. Identification, characterization, and role in rubber biosynthesis. J. Biol. Chem. 1989, 264, 18608–18617. [Google Scholar] [CrossRef]
- Bahri, A.R.S.; Hamzah, S. Immunocytochemical localisation of rubber membrane protein in Hevea latex. J. Nat. Rubber Res. 1996, 11, 88–95. [Google Scholar]
- Yeang, H.Y.; Cheong, K.F.; Sunderasan, E.; Hamzah, S.; Chew, N.P.; Hamid, S.; Hamilton, R.G.; Cardosa, M.J. The 14.6 kd rubber elongation factor (Hev b 1) and 24 kd (Hev b 3) rubber particle proteins are recognized by IgE from patients with spina bifida and latex allergy. J. Allergy Clin. Immunol. 1996, 98, 628–639. [Google Scholar] [CrossRef]
- Oh, S.K.; Kang, H.; Shin, D.H.; Yang, J.; Chow, K.S.; Yeang, H.Y.; Wagner, B.; Breiteneder, H.; Han, K.H. Isolation, characterization, and functional analysis of a novel cDNA clone encoding a small rubber particle protein from Hevea brasiliensis. J. Biol. Chem. 1999, 274, 17132–17138. [Google Scholar] [CrossRef]
- Kim, I.J.; Ryu, S.B.; Kwak, Y.S.; Kang, H. A novel cDNA from Parthenium argentatum Gray enhances the rubber biosynthetic activity in vitro. J. Exp. Bot. 2004, 55, 377–385. [Google Scholar] [CrossRef]
- Laibach, N.; Hillebrand, A.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. Identification of a Taraxacum brevicorniculatum rubber elongation factor protein that is localized on rubber particles and promotes rubber biosynthesis. Plant J. 2015, 82, 609–620. [Google Scholar] [CrossRef]
- Priya, P.; Venkatachalam, P.; Thulaseedharan, A. Differential expression pattern of rubber elongation factor (REF) mRNA transcripts from high and low yielding clones of rubber tree (Hevea brasiliensis Muell. Arg.). Plant Cell Rep. 2007, 26, 1833–1838. [Google Scholar] [CrossRef]
- Priya, P.; Venkatachalam, P.; Thulaseedharan, A. Molecular cloning and characterization of the rubber elongation factor gene and its promoter sequence from rubber tree (Hevea brasiliensis): A gene involved in rubber biosynthesis. Plant Sci. 2006, 171, 470–480. [Google Scholar] [CrossRef]
- Chow, K.S.; Wan, K.L.; Isa, M.N.; Bahari, A.; Tan, S.H.; Harikrishna, K.; Yeang, H.Y. Insights into rubber biosynthesis from transcriptome analysis of Hevea brasiliensis latex. J. Exp. Bot. 2007, 58, 2429–2440. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Tan, D.; Sun, X.; Zhang, J. An integrative transcriptomic and genomic analysis reveals novel insights into the hub genes and regulatory networks associated with rubber synthesis in H. brasiliensis. Ind. Crop Prod. 2020, 153, 112562. [Google Scholar] [CrossRef]
- Fonseca, S.; Rosado, A.; Vaughan-Hirsch, J.; Bishopp, A.; Chini, A. Molecular locks and keys: The role of small molecules in phytohormone research. Front. Plant Sci. 2014, 5, 709. [Google Scholar] [CrossRef]
- Coupe, M.; Chrestin, H. Physio-chemical and biochemical mechanisms of hormonal (ethylene) stimulation. In Physiology of Rubber Tree Latex; d’Auzac, J., Jacob, J.L., Chrestin, H., Eds.; CRC Press: Boca Raton, FL, USA, 1989; pp. 295–319. [Google Scholar]
- Faridah, Y.; Siti Arija, M.A.; Ghandimathi, H. Changes in some physiological latex parameters in relation to over exploitation and onset of induced tapping panel dryness. J. Nat. Rubber Res. 1996, 10, 182–186. [Google Scholar]
- Putranto, R.A.; Herlinawati, E.; Rio, M.; Leclercq, J.; Piyatrakul, P.; Gohet, E.; Sanier, C.; Oktavia, F.; Pirrello, J.; Kuswanhadi; et al. Involvement of ethylene in the latex metabolism and tapping panel dryness of Hevea brasiliensis. Int. J. Mol. Sci. 2015, 16, 17885–17908. [Google Scholar] [CrossRef]
- Wang, X.; Wang, D.; Sun, Y.; Yang, Q.; Chang, L.; Wang, L.; Meng, X.; Huang, Q.; Jin, X.; Tong, Z. Comprehensive proteomics analysis of laticifer latex reveals new insights into ethylene stimulation of natural rubber production. Sci. Rep. 2015, 5, 13778. [Google Scholar] [CrossRef]
- Montoro, P.; Wu, S.; Favreau, B.; Herlinawati, E.; Labrune, C.; Martin-Magniette, M.L.; Pointet, S.; Rio, M.; Leclercq, J.; Ismawanto, S.; et al. Transcriptome analysis in Hevea brasiliensis latex revealed changes in hormone signalling pathways during ethephon stimulation and consequent tapping panel dryness. Sci. Rep. 2018, 8, 8483. [Google Scholar] [CrossRef]
- Tungngoen, K.; Viboonjun, U.; Kongsawadworakul, P.; Katsuhara, M.; Julien, J.L.; Sakr, S.; Chrestin, H.; Narangajavana, J. Hormonal treatment of the bark of rubber trees (Hevea brasiliensis) increases latex yield through latex dilution in relation with the differential expression of two aquaporin genes. J. Plant Physiol. 2011, 168, 253–262. [Google Scholar] [CrossRef]
- Laosombut, T.; Arreewichit, P.; Nirapathpongporn, K.; Traiperm, P.; Kongsawadworakul, P.; Viboonjun, U.; Narangajavana, J. Differential expression of methyl jasmonate-responsive genes correlates with laticifer vessel proliferation in phloem tissue of rubber tree (Hevea brasiliensis). J. Plant Growth Regul. 2016, 35, 1049–1063. [Google Scholar] [CrossRef]
- Cao, X.; Yan, J.; Lei, J.; Li, J.; Zhu, J.; Zhang, H. De novo transcriptome sequencing of MeJA-induced Taraxacum koksaghyz Rodin to identify genes related to rubber formation. Sci. Rep. 2017, 7, 15697. [Google Scholar] [CrossRef]
- Liu, J.P.; Hu, J.; Liu, Y.H.; Yang, C.P.; Zhuang, Y.F.; Guo, X.L.; Li, Y.J.; Zhang, L. Transcriptome analysis of Hevea brasiliensis in response to exogenous methyl jasmonate provides novel insights into regulation of jasmonate-elicited rubber biosynthesis. Physiol. Mol. Biol. Plants 2018, 24, 349–358. [Google Scholar] [CrossRef]
- Wu, C.; Lan, L.; Li, Y.; Nie, Z.; Zeng, R. The relationship between latex metabolism gene expression with rubber yield and related traits in Hevea brasiliensis. BMC Genom. 2018, 19, 897. [Google Scholar] [CrossRef]
- Wahler, D.; Gronover, C.S.; Richter, C.; Foucu, F.; Twyman, R.M.; Moerschbacher, B.M.; Fischer, R.; Muth, J.; Prüfer, D. Polyphenoloxidase silencing affects latex coagulation in Taraxacum species. Plant Physiol. 2009, 151, 334–346. [Google Scholar] [CrossRef]
- Dai, L.; Kang, G.; Nie, Z.; Li, Y.; Zeng, R. Comparative proteomic analysis of latex from Hevea brasiliensis treated with ethrel and methyl jasmonate using iTRAQ-coupled two-dimensional LC-MS/MS. J. Proteom. 2016, 132, 167–175. [Google Scholar] [CrossRef]
- Deng, X.; Guo, D.; Yang, S.; Shi, M.; Chao, J.; Li, H.; Peng, S.; Tian, W. Jasmonate signalling in the regulation of rubber biosynthesis in laticifer cells of rubber tree, Hevea brasiliensis. J. Exp. Bot. 2018, 69, 3559–3571. [Google Scholar] [CrossRef]
- Yamashita, S.; Yamaguchi, H.; Waki, T.; Aoki, Y.; Mizuno, M.; Yanbe, F.; Ishii, T.; Funaki, A.; Tozawa, Y.; Miyagi-Inoue, Y.; et al. Identification and reconstitution of the rubber biosynthetic machinery on rubber particles from Hevea brasiliensis. eLife 2016, 5, e19022. [Google Scholar] [CrossRef]
- Brown, D.; Feeney, M.; Ahmadi, M.; Lonoce, C.; Sajari, R.; Di Cola, A.; Frigerio, L. Subcellular localization and interactions among rubber particle proteins from Hevea brasiliensis. J. Exp. Bot. 2017, 68, 5045–5055. [Google Scholar] [CrossRef]
- Dai, L.; Nie, Z.; Kang, G.; Li, Y.; Zeng, R. Identification and subcellular localization analysis of two rubber elongation factor isoforms on Hevea brasiliensis rubber particles. Plant Physiol. Biochem. 2017, 111, 97–106. [Google Scholar] [CrossRef]
- Wang, D.; Xie, Q.; Sun, Y.; Tong, Z.; Chang, L.; Yu, L.; Zhang, X.; Yuan, B.; He, P.; Jin, X.; et al. Proteomic landscape has revealed small rubber particles are crucial rubber biosynthetic machines for ethylene-stimulation in natural rubber production. Int. J. Mol. Sci. 2019, 20, 5082. [Google Scholar] [CrossRef]
- Shahmuradov, I.A.; Umarov, R.K.; Solovyev, V.V. TSSPlant: A new tool for prediction of plant Pol II promoters. Nucl. Acids Res. 2017, 45, e65. [Google Scholar] [CrossRef]
- Sandelin, A.; Alkema, W.; Engström, P.; Wasserman, W.W.; Lenhard, B. (2004). JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucl. Acids Res. 2004, 32, D91–D94. [Google Scholar] [CrossRef]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucl. Acids Res. 2024, 52, D174–D182. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucl. Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Yang, Y.; Li, R.; Qi, M. In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J. 2000, 22, 543–551. [Google Scholar] [CrossRef]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors—Being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Majewska, M.; Wysokińska, H.; Kuźma, Ł.; Szymczyk, P. Eukaryotic and prokaryotic promoter databases as valuable tools in exploring the regulation of gene transcription: A comprehensive overview. Gene 2018, 644, 38–48. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic helix-loop-helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene signaling in plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Han, H.; Wang, C.; Yang, X.; Wang, L.; Ye, J.; Xu, F.; Liao, Y.; Zhang, W. Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta 2023, 258, 13. [Google Scholar] [CrossRef]
- Luo, J.L.; Zhao, N.; Lu, C.M. Plant Trihelix transcription factors family. Hereditas 2012, 34, 1551–1560. [Google Scholar] [CrossRef]
- Guo, D.; Zhou, Y.; Li, H.L.; Zhu, J.H.; Wang, Y.; Chen, X.T.; Peng, S.Q. Identification and characterization of the abscisic acid (ABA) receptor gene family and its expression in response to hormones in the rubber tree. Sci. Rep. 2017, 7, 45157. [Google Scholar] [CrossRef]
- Deng, X.D.; Fei, X.W.; Huang, J.S.; Zheng, X.Q. Isolation and analysis of rubber elongation factor gene and its 5′-end promoter region. Acta Agron. Sin. 2002, 28, 528–532. [Google Scholar]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- Chao, J.; Wu, S.; Shi, M.; Xu, X.; Gao, Q.; Du, H.; Gao, B.; Guo, D.; Yang, S.; Zhang, S.; et al. Genomic insight into domestication of rubber tree. Nat. Commun. 2023, 14, 4651. [Google Scholar] [CrossRef]
- Fricke, J.; Hillebrand, A.; Twyman, R.M.; Prüfer, D.; Schulze Gronover, C. Abscisic acid-dependent regulation of small rubber particle protein gene expression in Taraxacum brevicorniculatum is mediated by TbbZIP1. Plant Cell Physiol. 2013, 54, 448–464. [Google Scholar] [CrossRef]
- Yokota, S.; Gotoh, T. Effects of rubber elongation factor and small rubber particle protein from rubber-producing plants on lipid metabolism in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2019, 128, 585–592. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).