Current Achievements and Future Challenges of Genotype-Dependent Somatic Embryogenesis Techniques in Hevea brasiliensis
Abstract
:1. Introduction
2. Somatic Embryogenesis in H. brasiliensis
3. Application of Somatic Embryogenesis Technologies in H. brasiliensis Seedling Production
4. Application of Somatic Embryogenesis Technologies in the Genetic Modification of H. brasiliensis
| Genotype | Explant Tissue | Agrobacterium Strain | Promoter | Plasmid | Transformed Gene | Transformation Efficiency | References |
|---|---|---|---|---|---|---|---|
| GL-1 | Anther-derived callus | Particle gun method | CaMV 35s | pBI221; pMON9793; pDE10; pHP23. | gus; npt II; cat | 80 callus, 2 transgenic plantlets | [50] |
| GL-1 | Anther-derived callus | GV2260 | CaMV 35s | p35SGUSINT | gus; npt II | 35 transgenic callus obtained from 438 callus (8.0% transformation efficiency); 2 regenerated transgenic plantlets obtained from 65 resistant embryoids (3.1% regeneration efficiency) | [56] |
| GL-1 | Anther-derived callus | GV2260 | CaMV 35s | pLGMR: pToK47 | HSA | 423 transgenic callus obtained from 5711 callus (7.4% transformation efficiency); 102 regenerated transgenic plantlets obtained from 1446 resistant embryoids (7.1% regeneration efficiency) | [51] |
| RRII 105 | Anther-derived callus | EHA101 | CaMV 35s | pDU96.2144 | HbSOD | 4% transformation efficiency; 7.1% regeneration efficiency | [48] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | CaMV 35s | pCAMBIA2301 | gus; npt II | More than 1695–5264 transgenic events per gram of callus; 372 transgenic plantlets obtained | [43,57] |
| RRII 105 | Leaf-derived embryogenic callus | LBA4404 | CaMV 35s | - | TB antigen | 60% transformation efficiency | [58] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | pHEV2.1 | pCAMBIA1381Z | gus; | 14 independent transgenic callus lines | [59] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | CaMV 35s | pCAMBIA2300 | GFP; EcGSH1 | 23 independent transgenic callus lines | [60] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | CaMV 35s | pCAMBIA2300 | HbCuZnSOD-GFP | 72 independent transgenic callus lines; 8.4–9.5 regenerated transgenic plantlets per gram of callus | [47] |
| CATAS 7-33-97 | Anther-derived callus | EHA105; | CaMV 35s | pCAMBIA2301 | gus; | 11 transgenic plantlets from 2.2 million anther callus | [9] |
| CATAS 7-33-97 | Anther-derived callus | EHA105; LBA4404 | CaMV 35s | pBLGC | Bean Chitinase; Tobaccoβ-1,3-Glucase gene | 0.33% transformation efficiency | [52] |
| CATAS 87-6-62 | Cotyledonary somatic embryos | EHA105 | CaMV 35s | pCAMBIA2301 | gus | 0.5% transformation efficiency | [10] |
| RRII 105 | Inner integument tissues of immature fruit-derived friable callus | - | Super promoter | pBIB | HMGR1 | 44% regeneration efficiency | [61] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | CaMV 35s; HEV2.1 | pCamway | HbERF-IX c5 | 16 transgenic callus obtained from 300 callus (5.3% transformation efficiency); 2–223 resistant embryos per gram of callus; 1–6 regenerated transgenic plantlets per gram of callus | [45] |
| PB260 | Inner integument tissues of immature fruit-derived friable callus | EHA105 | CaMV 35s; HEV2.1 | pCAMBIA2300-GFP-EcGSH1 | EcGSH1 | 13 transgenic callus lines; 77–1039 resistant embryos per gram of callus; 0–35 regenerated transgenic plantlets per gram of callus | [46] |
| Haiken 2 | Anther-derived callus | Particle bombardment | CaMV 35s | pBI121 | GAI-GUS | 149 resistant embryoids obtained from 8771 callus (1.7% transformation efficiency); 8 regenerated transgenic plantlets obtained from 149 resistant embryoids (5.4% regeneration efficiency) | [49] |
| CATAS 7-33-97 | Cotyledonary somatic embryos | EHA105; | CaMV 35s | pCAMBIA2300 | HbAN2; gus | - | [53] |
| CATAS 7-33-97 | Cotyledonary somatic embryos | EHA105 | CaMV 35s | pCAMBIA2301 | gus | - | [54] |
5. Genotype Dependence Is a Major Problem in the Application of Somatic Embryogenesis Technologies in H. brasiliensis
6. The Somatic Embryogenesis Mechanism in Plants
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhou, Y.J.; Li, J.H.; Ross Friedman, C.; Wang, H.F. Variation of Soil Bacterial Communities in a Chronosequence of Rubber Tree (Hevea brasiliensis) Plantations. Front. Plant Sci. 2017, 8, 849. [Google Scholar] [CrossRef]
- Metcalfe, C.R. Distribution of latex in the plant kingdom. Econ. Bot. 1967, 21, 115–127. [Google Scholar] [CrossRef]
- Cherian, S.; Ryu, S.B.; Cornish, K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019, 17, 2041–2061. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Xu, X.; Ruan, J.; Liu, S.; Wu, S.; Shao, X.; Wang, X.; Gan, L.; Qin, B.; Yang, Y.; et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2017, 5, 78–87. [Google Scholar] [CrossRef]
- Tokumoto, Y.; Uefuji, H.; Yamamoto, N.; Kajiura, H.; Nakazawa, Y. Gene coexpression network for trans-1,4-polyisoprene biosynthesis involving mevalonate and methylerythritol phosphate pathways in Eucommia ulmoides Oliver. Plant Biotechnol. 2017, 34, 165–172. [Google Scholar] [CrossRef]
- Premadasa, R.B. ANRPC Releases Natural Rubber Trends November 2021. Available online: www.anrpc.org. (accessed on 24 December 2021).
- Priyadarshan, P.M.; Goncalves, P.D.S. Hevea gene pool for breeding. Genet. Resour. Crop Evol. 2003, 50, 101–114. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Huang, T.D.; Li, Z.; Sun, A.H.; Zhou, Q.N.; Hua, Y.W.; Huang, H.S. Establishment of Agrobacterium tumefaciens-mediated Anther Calli Transformation System in Hevea brasiliensis ACTA Agron. Sin. 2010, 36, 1691–1697. [Google Scholar]
- Huang, T.D.; Li, J.; Li, Y.T.; Huang, H.S.; Hua, Y.W. Somatic Embryo, an Alternative Target Tissue for Agrobacterium-mediated Transformation in Hevea brasiliensis. J. Rubber Res. 2015, 18, 171–188. [Google Scholar]
- Montoro, P.; Etienne, H.; Michaux-Ferrière, N.; Carron, M.-P. Callus friability and somatic embryogenesis in Hevea brasiliensis. Plant Cell Tissue Organ Cult. 1993, 33, 331–338. [Google Scholar] [CrossRef]
- Wang, Z.; Zeng, X.; Chen, C.; Wu, H.; Li, Q.; Fan, G.A.; Lu, W. Induction of rubber plantlets from anther of Hevea brasiliensis Muell-Arg in vitro. Chin. J. Trop. Crops 1980, 1, 25–26. [Google Scholar]
- Carron, M.P.; Enjalric, F. Studies on vegetative micropropagation of Hevea brasiliensis by somatic embryogenesis and in vitro microcutting. Plant Tissue Cult. 1982, 1982, 751–752. [Google Scholar]
- Hua, Y.W.; Huang, T.D.; Huang, H.S. Micropropagation of self-rooting juvenile clones by secondary somatic embryogenesis in Hevea brasiliensis. Plant Breed. 2010, 129, 202–207. [Google Scholar] [CrossRef]
- Carron, M.P.; Granent, F.; Keli, J. Budding from rejuvenated clones: A good compromise between micropropagation and conventional budding. In Proceedings of the International Rubber Conference, Siem Reap, Cambodia, 12–13 November 2007; pp. 367–373. [Google Scholar]
- Xu, Z.H.; Zhang, X.S.; Su, Y.H.; Hu, Y.X.; Xu, L.; Wang, J.W. Plant cell totipotency and regeneration. Sci. China 2019, 49, 1282–1300. [Google Scholar]
- Zimmerman, J.L. Somatic Embryogenesis: A Model for Early Development in Higher Plants. Plant Cell 1993, 5, 1411–1423. [Google Scholar] [CrossRef]
- Arnold, S.V.; Sabala, I.; Bozhkov, P.; Dyachok, J.; Filonova, L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult. 2002, 69, 233–249. [Google Scholar] [CrossRef]
- Smertenko, A.; Bozhkov, P.V. Somatic embryogenesis: Life and death processes during apical-basal patterning. J. Exp. Bot. 2014, 65, 1343–1360. [Google Scholar] [CrossRef]
- Wang, T.D.; Huang, T.D.; Huang, H.S.; Hua, Y.W. Origin of Secondary Somatic Embryos and Genetic Stability of the Regenerated Plants in Hevea brasiliensis. J. Rubber Res. 2017, 20, 101–116. [Google Scholar] [CrossRef]
- Kala, R.G.; Gimicha, G.C.; Jayasree, P.K. Somatic Embryogenesis in Leaf Cultures of Hevea Brasiliensis: Effect of Source Plant. Nat. Rubber Res. 2009, 22, 117–126. [Google Scholar]
- Sushamakumari, S.; Asokan, M.P.; Anthony, P.; Lowe, K.C.; Power, J.B.; Davey, M.R. Plant regeneration from embryogenic cell suspension-derived protoplasts of rubber. Plant Cell Tissue Organ Cult. 2000, 61, 81–85. [Google Scholar] [CrossRef]
- Dai, X.M.; Huang, T.D.; Li, J.; Yang, X.F.; Huang, H.S. Effects of Different Explants on Isolation and Regeneration of Protoplast in Rubber Tree. Mol. Plant Breed. 2014, 12, 1259–1264. [Google Scholar]
- Pascal, M. Somatic embryogenesis in Hevea brasiliensis: Advances and limitations. In Proceedings of the IRRDB Biotechnology Workshop, Bogor, Indonésie, 7–8 October 2014; p. 10. [Google Scholar]
- Jin, H.F.; Tian, W.M.; Shi, M.J. Current Situation and Industrial Development of Natural Rubber in China. Chin. J. Trop. Agric. 2017, 37, 98–104. [Google Scholar]
- Van Helten, W.M. Het Oculeeren Van Hevea; Ned.: Washington, DC, USA, 1918; Volume 2, p. 87. [Google Scholar]
- Hao, B.Z.; Wu, J.L. High yield characteristics of young clones of hevea brasiliensis. Chin. J. Trop. Agric. 1996, 2, 1–9. [Google Scholar]
- Veisseire, P.; Linossier, L.; Coudret, A. Effect of abscisic acid and cytokinins on the development of somatic embryos in Hevea brasiliensis. Plant Cell Tissue Organ Cult. 1994, 39, 219–223. [Google Scholar] [CrossRef]
- Carron, M.P.; Etienne, H.; Michaux-Ferriere, N.; Montoro, P. Somatic Embryogenesis in Rubber Tree (Hevea brasiliensis Müll. Arg.). Biotechnol. Agric. For. 1995, 30, 353–369. [Google Scholar]
- Blanc, G.; Lardet, L.; Martin, A.; Jacob, J.; Carron, M. Differential carbohydrate metabolism conducts morphogenesis in embryogenic callus of Hevea brasiliensis (Müll. Arg.). J. Exp. Bot. 2002, 53, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Carron, M.P.; Lardet, L.; Leconte, A.; Dea, B.G.; Keli, J.; Granet, F.; Julien, J.; Teerawatanasuk, K.; Montoro, P. Field trials network emphasizes the improvement of growth and yield through micropropagation in rubber tree (Hevea brasiliensis, Muell.-Arg.). Acta Hortic. 2009, 812, 485–492. [Google Scholar] [CrossRef]
- Lardet, L.; Martin, F.; Dessailly, F.; Carron, M.-P.; Montoro, P. Effect of exogenous calcium on post-thaw growth recovery and subsequent plant regeneration of cryopreserved embryogenic calli of Hevea brasiliensis (Müll. Arg.). Plant Cell Rep. 2007, 26, 559–569. [Google Scholar] [CrossRef]
- Chen, X.T. Cultivation and trial planting of new rubber planting materials with fast growth and high yield. In Proceedings of the Symposium on the Construction and Planning of Tropical Crop Industrial, Haikou, China, 14 June 2006; pp. 105–108. [Google Scholar]
- Zhou, Q.N.; Sun, A.; Hua, Y.W.; Jiang, Z.H. Cryopreservation and plant regeneration of anther callus in Hevea by vitrification. Afr. J. Biotechnol. 2012, 11, 7212–7217. [Google Scholar]
- Zhou, G.Z.; Luan, L.L.; Zhang, S.M.; Song, Y.F.; He, C.Z.; Chen, J.M. Response of embryogenic calli to low-temperature storage with exogenous antioxidants in rubber tree. Mol. Plant Breed. 2018, 16, 2677–2685. [Google Scholar]
- Hua, Y.W. Self-root juvenile clones micropropagation technique of Hevea brasiliensis. In World Tropical Agriculture Information; Natinal Press Publications: Haikou, China, 2019; pp. 49–50. [Google Scholar]
- Yang, J.W.; Huang, T.D.; Hua, Y.W.; Huang, H.S. The low temperature resistance analysis of the self-rooting juvenile clones of the rubber tree. Chin. J. Trop. Crops 2012, 33, 1235–1238. [Google Scholar]
- Chen, X.T.; Wang, Z.Y.; Wu, H.D.; Zhang, X.J. A New Planting Material of Hevea brasiliensis-Self-rooting Juvenile-type Clone. Chin. J. Trop. Crops 2002, 23, 19–23. [Google Scholar]
- Li, H.L.; Guo, D.; Zhu, J.H.; Wang, Y.; Chen, X.T.; Peng, S.Q. Comparative Transcriptome Analysis of Latex Reveals Molecular Mechanisms Underlying Increased Rubber Yield in Hevea brasiliensis Self-Rooting Juvenile Clones. Front. Plant Sci. 2016, 8, e75307. [Google Scholar] [CrossRef] [PubMed]
- Paardekooper, E. Exploitation of the Rubber Tree; Longman Scientific and Technical: New York, NY, USA, 1989. [Google Scholar]
- Cheng, H.; Song, X.; Hu, Y.; Wu, T.; Yang, Q.; An, Z.; Feng, S.; Deng, Z.; Wu, W.; Zeng, X.; et al. Chromosome-level wild Hevea brasiliensis genome provides new tools for genomic-assisted breeding and valuable loci to elevate rubber yield. Plant Biotechnol. J. 2023, 21, 1058–1072. [Google Scholar] [CrossRef]
- Liu, J.; Shi, C.; Shi, C.-C.; Li, W.; Zhang, Q.-J.; Zhang, Y.; Li, K.; Lu, H.-F.; Shi, C.; Zhu, S.-T.; et al. The Chromosome-Based Rubber Tree Genome Provides New Insights into Spurge Genome Evolution and Rubber Biosynthesis. Mol. Plant 2020, 13, 336–350. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Baptiste, C.; Oliver, G.; Martin, F.; Montoro, P. Efficient Agrobacterium tumefaciens-mediated transformation of embryogenic calli and regeneration of Hevea brasiliensis Müll Arg. plants. Plant Cell Rep. 2006, 24, 724–733. [Google Scholar] [CrossRef] [PubMed]
- Etienne, H.; Lartaud, M.; Michaux-Ferriére, N.; Carron, M.P.; Berthouly, M.; Teisson, C. Improvement of somatic embryogenesis in Hevea brasiliensis (Müll. Arg.) using the temporary immersion technique. Vitr. Cell. Dev. Biol.-Plant 1997, 33, 81–87. [Google Scholar] [CrossRef]
- Lestari, R.; Rio, M.; Martin, F.; Leclercq, J.; Woraathasin, N.; Roques, S.; Dessailly, F.; Clément-Vidal, A.; Sanier, C.; Fabre, D.; et al. Overexpression of Hevea brasiliensis ethylene response factor HbERF-IXc5 enhances growth and tolerance to abiotic stress and affects laticifer differentiation. Plant Biotechnol. J. 2018, 16, 322–336. [Google Scholar] [CrossRef]
- Martin, F.; Abati, V.; Burel, A.; Clément-Vidal, A.; Sanier, C.; Fabre, D.; Woraathasin, N.; Rio, M.; Besret, P.; Farinas, B.; et al. Overexpression of EcGSH1 induces glutathione production and alters somatic embryogenesis and plant development in Hevea brasiliensis. Ind. Crops Prod. 2018, 112, 803–814. [Google Scholar] [CrossRef]
- Leclercq, J.; Martin, F.; Sanier, C.; Clément-Vidal, A.; Fabre, D.; Oliver, G.; Lardet, L.; Ayar, A.; Peyramard, M.; Montoro, P. Over-expression of a cytosolic isoform of the HbCuZnSOD gene in Hevea brasiliensis changes its response to a water deficit. Plant Mol. Biol. 2012, 80, 255–272. [Google Scholar] [CrossRef]
- Jayashree, R.; Rekha, K.; Venkatachalam, P.; Uratsu, S.L.; Dandekar, A.M.; Jayasree, P.K.; Kala, R.G.; Priya, P.; Sobha, S.; Ashokan, M.P. Genetic transformation and regeneration of rubber tree ( Hevea brasiliensis Muell. Arg) transgenic plants with a constitutive version of an anti-oxidative stress superoxide dismutase gene. Plant Cell Rep. 2003, 22, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Wang, Y.; Chen, X.T.; Zhang, X.J. Studies on GAI transgenic plants of Hevea brasiliensis by particle bombardment. J. Trop. Subtrop. Bot. 2020, 18, 165–169. [Google Scholar]
- Arokiaraj, P.; Jones, H.; Cheong, K.F.; Coomber, S.; Charlwood, B.V. Gene insertion into Hevea brasiliensis. Plant Cell Rep. 1994, 13, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Arokiaraj, P.; Florian, R.; Eva, O.; Samsul, A.; Hafsah, J.; Carter, D.; Yeang, H.Y. Expression of human serum albumin in transgenic Hevea brasiliensis. J. Rubber Res. 2002, 5, 157–166. [Google Scholar]
- Huang, T.D.; Sun, A.H.; Yang, J.W.; Hua, Y.W.; Huang, H.S. Genetic transformation of chitinase and β-1,3-glucase gene to rubber tree. Chin. Agric. Sci. Bull. 2012, 28, 28–33. [Google Scholar] [CrossRef]
- Huang, T.; Xin, S.; Fang, Y.; Chen, T.; Chang, J.; Ko, N.C.K.; Huang, H.; Hua, Y. Use of a novel R2R3-MYB transcriptional activator of anthocyanin biosynthesis as visual selection marker for rubber tree (Hevea brasiliensis) transformation. Ind. Crops Prod. 2021, 174, 114–225. [Google Scholar] [CrossRef]
- Udayabhanu, J.; Huang, T.; Xin, S.; Cheng, J.; Hua, Y.; Huang, H. Optimization of the Transformation Protocol for Increased Efficiency of Genetic Transformation in Hevea brasiliensis. Plants 2022, 11, 1067. [Google Scholar] [CrossRef]
- Gu, X.; Peng, S.; Dai, X.; Zhou, Q.; Sun, X.; Gui, M.; Huang, h.; Hua, Y.; Huang, T.; Zhang, Y. Differential Analysis of Anther Embryogenesis between Different Genotypes of Hevea brasiliensis. For. Res. 2022, 35, 143–152. [Google Scholar]
- Arokiaraj, P.; Yeang, H.Y.; Cheong, K.F.; Hamezh, S.; Jones, H.; Coomber, S.; Charlwood, B.V. CaMV 35S promoter directs β-glucuronidase expression in the laticiferous system of transgenic Hevea brasiliensis (rubber tree). Plant Cell Rep. 1998, 17, 621–625. [Google Scholar] [CrossRef]
- Montoro, P.; Rattana, W.; Pujade-Renaud, V.; Michaux-Ferrière, N.; Monkolsook, Y.; Kanthapura, R.; Adunsadthapong, S. Production of Hevea brasiliensis transgenic embryogenic callus lines by Agrobacterium tumefaciens: Roles of calcium. Plant Cell Rep. 2003, 21, 1095–1102. [Google Scholar] [CrossRef]
- Kala, R.G.; Anu, K.S.; Manesh, K.; Saleena, A.; Jayasree, P.K.; Narayanan, P.R.; Thomas, G.A.; Thulaseedharan, A. Agrobacterium mediated genetic transformation in Hevea brasiliensis for recombinant protein production. J. Plant. Crops 2006, 34, 582–586. [Google Scholar]
- Montoro, P.; Lagier, S.; Baptiste, C.; Marteaux, B.; Pujade-Renaud, V.; Alemanno, L.L. Expression of the HEV2.1 gene promoter in transgenic Hevea brasiliensis. Plant Cell TissueOrgan Cult. 2008, 94, 55–63. [Google Scholar] [CrossRef]
- Leclercq, J.; Lardet, L.; Martin, F.; Chapuset, T.; Oliver, G.; Montoro, P. The green fluorescent protein as an efficient selection marker for Agrobacterium tumefaciens-mediated transformation in Hevea brasiliensis (Müll. Arg). Plant Cell Rep. 2010, 29, 513–522. [Google Scholar] [CrossRef]
- Jayashree, R.; Nazeem, P.A.; Rekha, K.; Sreelatha, S.; Thulaseedharan, A.; Krishnakumar, R.; Kala, R.G.; Vineetha, M.; Leda, P.; Jinu, U.; et al. Over-expression of 3-hydroxy-3- methylglutaryl-coenzyme A reductase 1 (hmgr1) gene under super-promoter for enhanced latex biosynthesis in rubber tree (Hevea brasiliensis Muell. Arg.). Plant Physiol. Biochem. 2018, 127, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.Y.; Zhang, B.T.; Ding, W.; Liu, X.D.; Yang, D.L.; Wei, P.L.; Cao, F.Q.; Zhu, S.H.; Zhang, F.; Mao, Y.F.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Wang, J.J.; Chen, H.D. A novel CRISPR/Cas9 system for efficiently generating Cas9-free multiplex mutants in Arabidopsis. Abiotech 2020, 1, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Yang, X.; Wang, C.; Fan, Y.; Xin, S.; Hua, Y.; Wang, K.; Huang, H. CRISPR/Cas9-mediated genome editing in Hevea brasiliensis. Ind. Crops Prod. 2021, 164, 113418. [Google Scholar] [CrossRef]
- Fan, Y.; Xin, S.; Dai, X.; Yang, X.; Huang, H.; Hua, Y. Efficient genome editing of rubber tree (hevea brasiliensis) protoplasts using CRISPR/Cas9 ribonucleoproteins. Ind. Crops Prod. 2020, 146, 112–146. [Google Scholar] [CrossRef]
- Carron, M.P.; Lardet, L.; Dea, B.G. Hevea micropropagation by somatic embryogenesis, technology. Plant. Res. Dev. 1998, 5, 187–192. [Google Scholar]
- Montoro, P.; Etienne, H.; Carron, M.P. Effect of calcium on callus friability and somatic embryogenesis in Hevea brasiliensis Müll. Arg.: Relations with callus mineral nutrition, nitrogen metabolism and water parameters. J. Exp. Bot. 1995, 46, 255–261. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.; Ohto, M.A.; Yee, K.M.; West, M.A.L.; Harada, J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 1998, 93, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.; Kwong, L.; Matsudaira, K.; Pelletier, J.; Lepiniec, L.; Fischer, R.; Goldberg, R.; Harada, J. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. United States Am. 2001, 98, 11806–11811. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Campagne, M.M.V.L. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muino, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM Transcription Factor Activates the LEC1-ABI3-FUS3-LEC2 Network to Induce Somatic Embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar] [CrossRef]
- Levine, M. Differentiation of carrot root tissue grown in vitro. Bull. Torrey Bot. Club 1947, 74, 321–328. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhang, X.S. The hormonal control of regeneration in plants. Curr. Top. Dev. Biol. 2014, 108, 35–69. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Bai, B.; Zhang, X.S. Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front. Plant Sci. 2015, 5, 792. [Google Scholar] [CrossRef]
- Bai, B.; Su, Y.H.; Yuan, J.; Zhang, X.S. Induction of somatic embryos in Arabidopsis requires local YUCCA expression mediated by the down-regulation of ethylene biosynthesis. Mol. Plant 2013, 6, 1247–1260. [Google Scholar] [CrossRef]
- Su, Y. Molecular Basis of ABA Function in the Arabidopsis Somatic Embryogenesis. Master’s Thesis, Shandong Agricultural University, Taian, China, 2011. [Google Scholar]
- Xu, M.; Du, Q.; Tian, C.; Wang, Y.; Jiao, Y. Stochastic gene expression drives mesophyll protoplast regeneration. Sci. Adv. 2021, 7, eabg8466. [Google Scholar] [CrossRef]
- Wang, F.X.; Shang, G.D.; Wu, L.Y.; Xu, Z.G.; Zhao, X.Y.; Wang, J.W. Chromatin Accessibility Dynamics and a Hierarchical Transcriptional Regulatory Network Structure for Plant Somatic Embryogenesis. Dev. Cell 2020, 54, 742–757. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef]
- Nelson-Vasilchik, K.; Hague, J.; Mookkan, M.; Zhang, Z.J.; Kausch, A. Transformation of Recalcitrant Sorghum Varieties Facilitated by Baby Boom and Wuschel2. Curr. Protoc. Plant Biol. 2018, 3, e20076. [Google Scholar] [CrossRef]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhang, Y.; Lu, M.H.; Chai, Y.P.; Jiang, Y.Y.; Zhou, Y.; Wang, X.C.; Chen, Q.J. A Novel Ternary Vector System United with Morphogenic Genes Enhances CRISPR/Cas Delivery in Maize. Plant Physiol 2019, 181, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Martin-Ortigosa, S.; Finer, J.; Semsang, N.; Gunadi, A.; Batts, L.; Takhare, D.; Rush, B.; Schmitz, O.; Stuiver, M.; et al. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 2020, 11, 572319. [Google Scholar] [CrossRef]
- Lian, Z.; Nguyen, C.D.; Liu, L.; Wang, G.; Chen, J.; Wang, S.; Yi, G.; Wilson, S.; Ozias-Akins, P.; Gong, H.; et al. Application of Developmental Regulators to Improve In-Planta or In Vitro Transformation in Plants. Plant Biotechnol. J. 2022, 20, 1622–1635. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, M.; Li, Y.; Zhang, Q.; Keith, L.; Henry, D.; Jin, S.; Zhang, X. Multimics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation (SRA) process. Plant Biotechnol. J. 2018, 17, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Barkess, G. Chromatin remodeling and genome stability. Genome Biol. 2006, 7, 319. [Google Scholar] [CrossRef]
- Görisch, S.M.; Wachsmuth, M.; Tóth, K.F.; Lichter, P.; Rippe, K. Histone acetylation increases chromatin accessibility. J. Cell Sci. 2005, 118, 5825–5834. [Google Scholar] [CrossRef]
- Chuang, D.M.; Leng, Y.; Marinova, Z.; Kim, H.J.; Chiu, C.T. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009, 32, 591–601. [Google Scholar] [CrossRef]
- Wójcikowska, B.; Wójcik, A.M.; Gaj, M.D. Epigenetic Regulation of Auxin-Induced Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2020, 21, 2307. [Google Scholar] [CrossRef]
- Tanaka, M.; Kikuchi, A.; Kamada, H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008, 146, 149–161. [Google Scholar] [CrossRef]
- Charbit, E.; Legavre, T.; Lardet, L.; Bourgeois, E.; Ferriere, N.; Carron, M.P. Identification of differentially expressed cDNA sequences and histological characteristics of Hevea brasiliensis calli in relation to their embryogenic and regenerative capacities. Plant Cell Rep. 2004, 22, 539–548. [Google Scholar] [CrossRef]
- Li, H.L.; Wang, Y.; Guo, D.; Tian, W.M.; Peng, S.Q. Three MADS-box genes of Hevea brasiliensis expressed during somatic embryogenesis and in the laticifer cells. Mol. Biol. Rep. 2011, 38, 4045–4052. [Google Scholar] [CrossRef]
- Piyatrakul, P.; Putranto, R.A.; Martin, F.; Rio, M.; Dessailly, F.; Leclercq, J.; Dufayard, J.F.; Lardet, L.; Montoro, P. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol. 2012, 12, 244. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.L.; Zhou, Y.K.; Guo, D.; Zhu, J.H.; Peng, S.Q. Transcriptomes analysis reveals novel insight into the molecular mechanisms of somatic embryogenesis in Hevea brasiliensis. BMC Genom. 2021, 22, 183. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 2013, 343, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wei, W.; Tang, J.; Tan, L.; Zhu, J.-K.; Chu, C.; Qian, Y. Genome-wide targeted mutagenesis in rice using CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Simmons, S.K.; Guo, A.; Shetty, A.S.; Arlotta, P. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 2020, 370, eaaz6063. [Google Scholar] [CrossRef]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. Vitr. Cell. Dev. Biol. Plant J. Tissue Cult. Assoc. 2018, 54, 240–252. [Google Scholar] [CrossRef]
- Pan, C.; Li, G.; Malzahn, A.A.; Cheng, Y.; Leyson, B.; Sretenovic, S.; Gurel, F.; Coleman, G.D.; Qi, Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants 2022, 8, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Raman, V.; Rojas, C.M.; Vasudevan, B.; Dunning, K.; Kolape, J.; Oh, S.; Yun, J.; Yang, L.; Li, G.; Pant, B.D.; et al. Agrobacterium expressing a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nat. Commun. 2022, 13, 2581. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2022, 4, 100345. [Google Scholar] [CrossRef]
- Zhao, X.; Meng, Z.; Wang, Y.; Chen, W.; Sun, C.; Cui, B.; Cui, J.; Yu, M.; Zeng, Z.; Guo, S.; et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat. Plants 2017, 3, 956–964. [Google Scholar] [CrossRef]
- Wang, Z.P.; Zhang, Z.B.; Zheng, D.Y.; Zhang, T.T.; Li, X.L.; Zhang, C.; Yu, R.; Wei, J.H.; Wu, Z.Y. Efficient and genotype independent maize transformation using pollen transfected by DNA-coated magnetic nanoparticles. J. Integr. Plant Biol. 2022, 64, 1145–1156. [Google Scholar] [CrossRef]
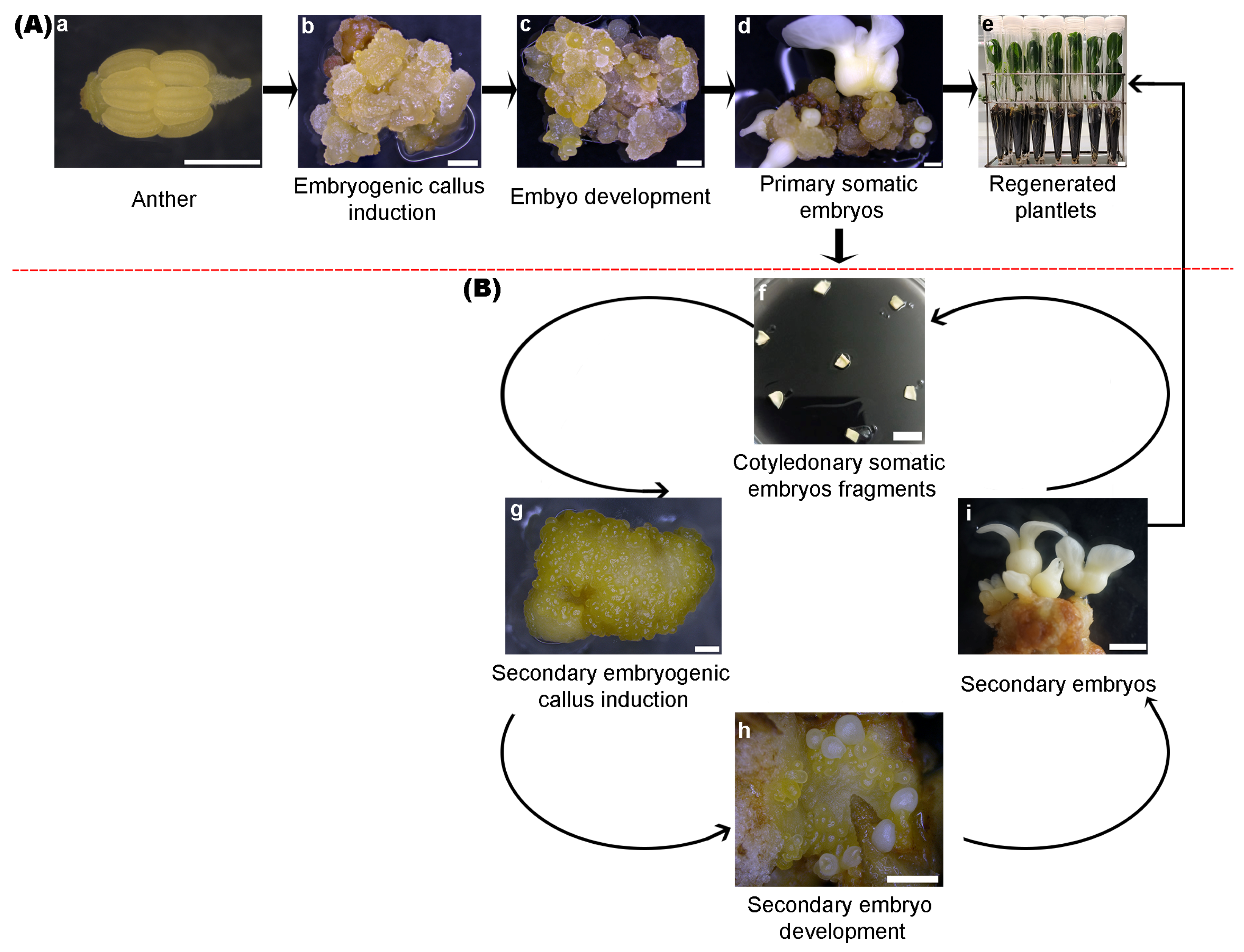
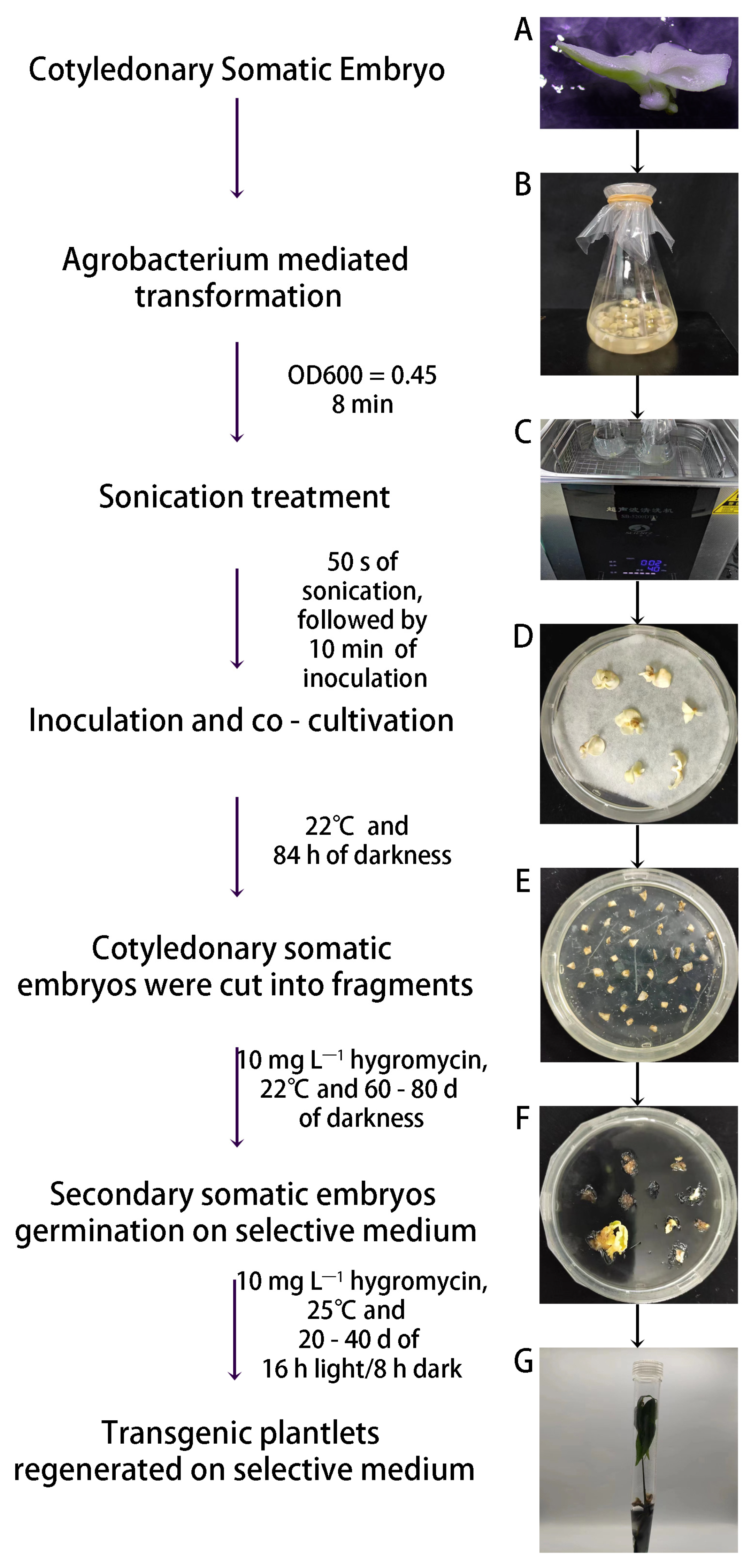
| Primary Somatic Embryogenesis | ||||
|---|---|---|---|---|
| Explant | Callus Induction Rate | Embryogenic Callus Induction Rate | Somatic Embryo Induction Rate | Clones with High Somatic Embryogenesis Ability |
| Immature seed inner integument | 87%–99% | 36%–51% | 43%–209% (number of somatic embryos/total number of explants × 100%) | PB217, PB260, PB280, PB310, PR107, RRIM600 |
| Anther | 53.7%–96.5% | 26.8%–95% | 0%–23.1% (number of callus-forming cotyledonary somatic embryos/total number of callus inoculated × 100%) | PB86, RRIM600, CATAS88-13, Haiken1, Dafeng95, Haiken2, CATAS917, CATAS73397, CATAS879, CATAS918, Xuyu3, Xuyu141-2 |
| Secondary somatic embryogenesis | ||||
| Explant | Annual multiplication coefficient | Clones | ||
| Cotyledonary somatic embryo | 696–2525 | CATAS73397, CATAS88-13 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Gu, X.; Xu, Z.; Yin, Z.; Yang, X.; Lin, R.; Zhou, Q.; Huang, H.; Huang, T. Current Achievements and Future Challenges of Genotype-Dependent Somatic Embryogenesis Techniques in Hevea brasiliensis. Forests 2023, 14, 1891. https://doi.org/10.3390/f14091891
Wang X, Gu X, Xu Z, Yin Z, Yang X, Lin R, Zhou Q, Huang H, Huang T. Current Achievements and Future Challenges of Genotype-Dependent Somatic Embryogenesis Techniques in Hevea brasiliensis. Forests. 2023; 14(9):1891. https://doi.org/10.3390/f14091891
Chicago/Turabian StyleWang, Xiaoyi, Xiaochuan Gu, Zhengwei Xu, Zhaochen Yin, Xianfeng Yang, Rong Lin, Quannan Zhou, Huasun Huang, and Tiandai Huang. 2023. "Current Achievements and Future Challenges of Genotype-Dependent Somatic Embryogenesis Techniques in Hevea brasiliensis" Forests 14, no. 9: 1891. https://doi.org/10.3390/f14091891
APA StyleWang, X., Gu, X., Xu, Z., Yin, Z., Yang, X., Lin, R., Zhou, Q., Huang, H., & Huang, T. (2023). Current Achievements and Future Challenges of Genotype-Dependent Somatic Embryogenesis Techniques in Hevea brasiliensis. Forests, 14(9), 1891. https://doi.org/10.3390/f14091891





