Foliar Application of dsRNA to Induce Gene Silencing in Emerald Ash Borer: Systemic Distribution, Persistence, and Bioactivity †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ash Seedlings
2.2. Insects
2.3. Gene Selection
2.4. dsRNA Synthesis
2.5. dsRNA Exposure
2.6. Plant Processing and RNA Isolation
2.7. Recovery of Exogenously Applied dsRNA
2.7.1. Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and Gel Electrophoresis
2.7.2. Sanger Sequencing
2.8. Biological Activity of Exogenous dsRNA
2.8.1. Bioassay
2.8.2. Gene Expression
3. Results
3.1. Seedling Measurements
3.2. Recovery of Exogenous dsRNA
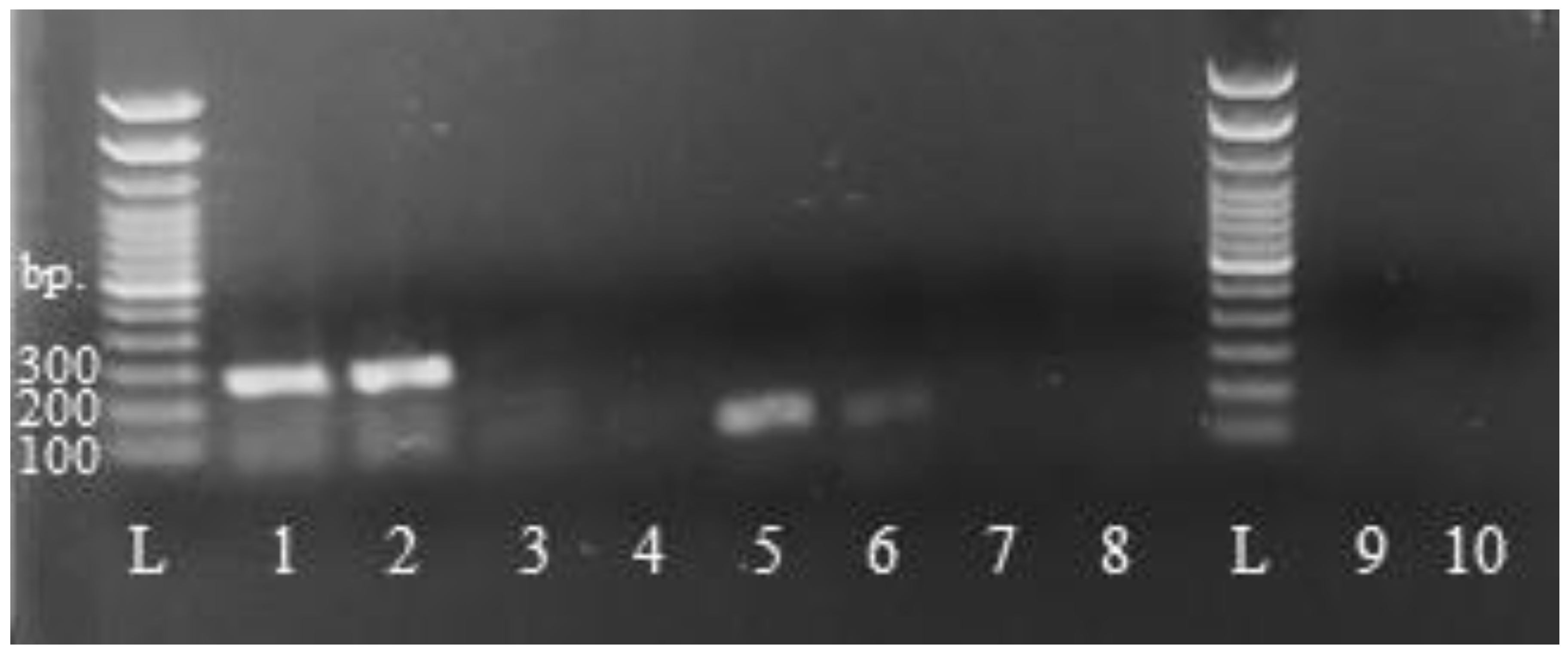
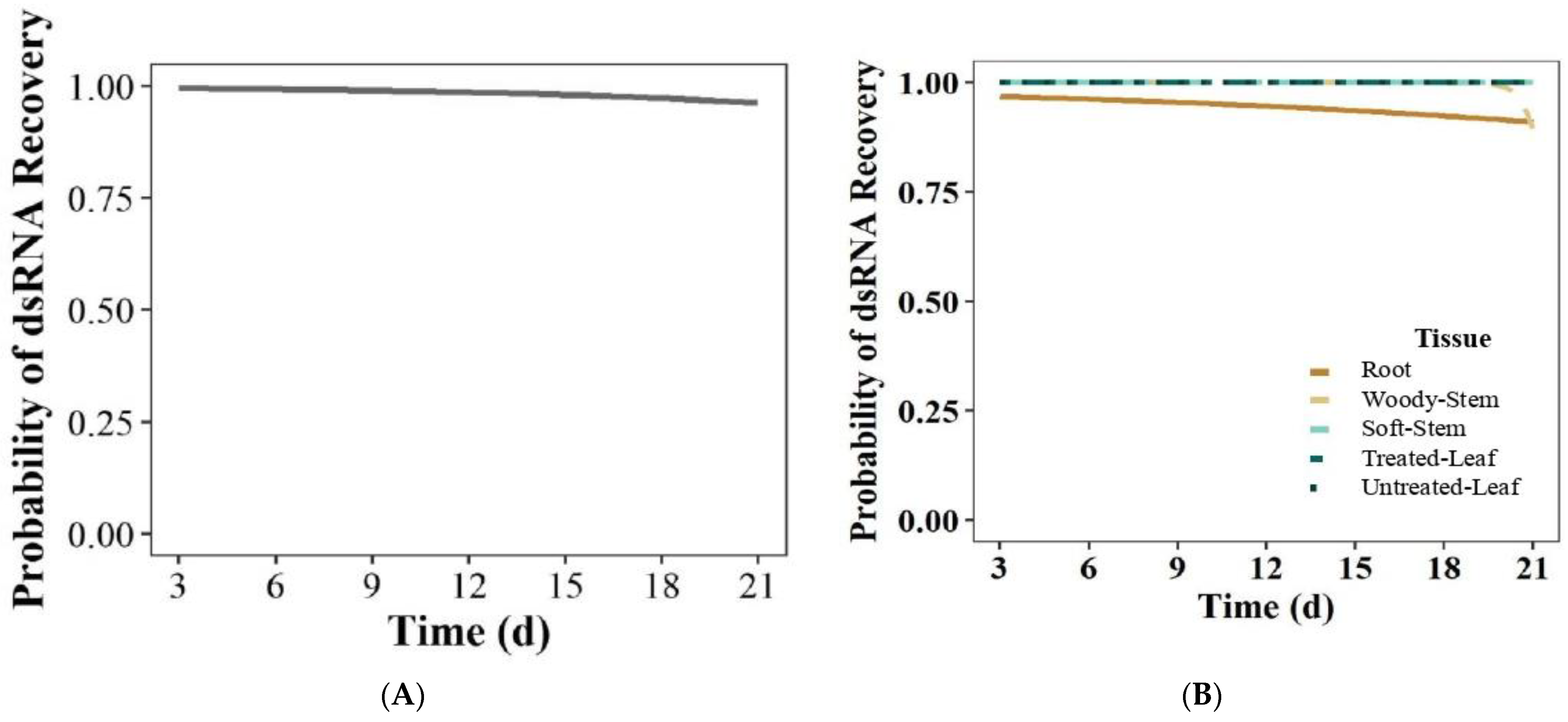
3.2.1. Gel Imaging and Logistic Regression Modeling
3.2.2. Sanger Sequencing
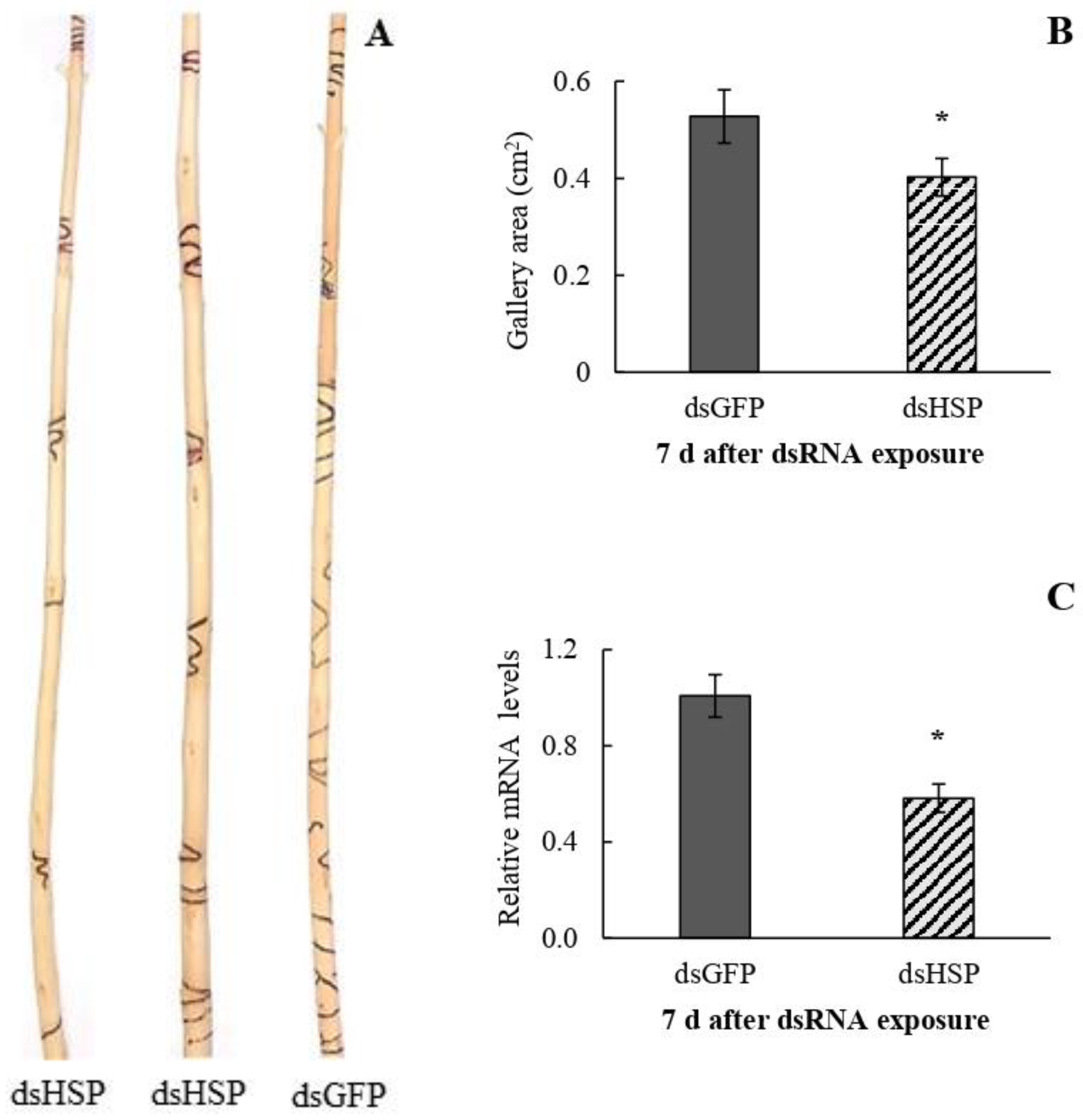
3.3. dsRNA Bioactivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- USDA APHIS. Emerald Ash Borer. 2022. Available online: https://www.aphis.usda.gov/aphis/ourfocus/planthealth/plant-pest-and-disease-programs/pests-and-diseases/emerald-ash-borer (accessed on 15 October 2022).
- Herms, D.A.; McCullough, D.G. Emerald ash borer invasion of North America: History, biology, ecology, impacts, and management. Annu. Rev. Entomol. 2014, 59, 13–30. [Google Scholar] [CrossRef]
- Aukema, J.E.; Leung, B.; Kovacs, K.; Chivers, C.; Britton, K.O.; Englin, J.; Frankel, S.J.; Haight, R.G.; Holmes, T.P.; Liebhold, A.M.; et al. Economic impacts of non-native forest insects in the continental United States. PLoS ONE 2011, 6, e24587. [Google Scholar] [CrossRef] [PubMed]
- McCullough, D.G. Challenges, tactics and integrated management of emerald ash borer in North America. Forestry Int. J. For. Res. 2020, 93, 197–211. [Google Scholar] [CrossRef]
- Donovan, G.H.; Butry, D.T.; Michael, Y.L.; Prestemon, J.P.; Liebhold, A.M.; Gatziolis, D.; Mao, M.Y. The relationship between trees and human health: Evidence from the spread of the emerald ash borer. Am. J. Prev. Med. 2013, 44, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, K.F.; Mercader, R.J.; Haight, R.G.; Siegert, N.W.; McCullough, D.G.; Liebhold, A.M. The influence of satellite populations of emerald ash borer on projected economic costs in U.S. communities, 2010–2020. J. Environ. Manag. 2011, 92, 2170–2181. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Bauer, L.S.; Miller, D.L.; Zhao, T.; Gao, R.; Song, L.; Luan, Q.; Jin, R.; Gao, C. Seasonal abundance of Agrilus planipennis (Coleoptera: Buprestidae) and its natural enemies Oobius agrili (Hymenoptera: Encyrtidae) and Tetrastichus planipennisi (Hymenoptera: Eulophidae) in China. Biol. Control. 2007, 42, 61–71. [Google Scholar] [CrossRef]
- Wharton, M.E.; Barbour, R.W. Trees and Shrubs of Kentucky; University Press of Kentucky: Lexington, KY, USA, 1993; 582p. [Google Scholar]
- Sadof, C.S.; Hughes, G.P.; Witte, A.R.; Peterson, D.J.; Ginzel, M.D. Tools for staging and managing emerald ash borer in the urban forest. Arboric. Urban For. 2017, 43, 15–26. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect. Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef]
- Zhang, J.; Khan, S.A.; Heckel, D.G.; Bock, R. Next-generation insect-resistant plants: RNAi-mediated crop protection. Trends Biotechnol. 2017, 35, 871–882. [Google Scholar] [CrossRef]
- Hunter, W.B.; Glick, E.; Paldi, N.; Bextine, B.R. Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest suppression. Southwest Entomol. 2012, 37, 85–87. [Google Scholar] [CrossRef]
- Dalakouras, A.; Jarausch, W.; Buchholz, G.; Bassler, A.; Braun, M.; Manthey, T.; Krczal, G.; Wassenegger, M. Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petiole absorption. Front. Plant Sci. 2018, 9, 1253. [Google Scholar] [CrossRef]
- Ghosh, S.K.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double-stranded RNA oral delivery methods to induce RNA interference in phloem and plant-sap-feeding hemipteran insects. J. Vis. Exp. 2018, 135, e57390. [Google Scholar] [CrossRef]
- Bragg, Z.; Rieske, L.K. Feasibility of systemically applied dsRNAs for pest-specific RNAi-induced gene silencing in white oak. Front. Plant Sci. 2022, 13, 830226. [Google Scholar] [CrossRef] [PubMed]
- Bragg, Z.; Rieske, L.K. Spatial distribution and retention in loblolly pine seedlings of exogenous dsRNAs applied through roots. Int. J. Mol. Sci. 2022, 23, 9167. [Google Scholar] [CrossRef]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Baum, J.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Mishra, S.K.; Sridharan, K.; Barnes, E.R.; Alyokhin, A.; Tuttle, R.; Kokulapalan, W.; Garby, D.; Skizim, N.J.; Tang, Y.-W.; et al. First sprayable double-stranded RNA-based biopesticide product targets Proteasome subunit beta type-5 in Colorado potato beetle (Leptinotarsa decemlineata). Front. Plant Sci. 2021, 12, 728652. [Google Scholar] [CrossRef]
- Zhao, C.; Alvarez-Gonzales, M.A.; Poland, T.M.; Mittapalli, O. Core RNAi machinery and gene knockdown in the emerald ash borer (Agrilus planipennis). J. Insect Physiol. 2015, 72, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, T.B.; Duan, J.J.; Palli, S.R.; Rieske, L.K. Identification of highly effective target genes for RNAi-mediated control of emerald ash borer, Agrilus planipennis. Sci. Rep. 2018, 8, 5020. [Google Scholar] [CrossRef]
- Kyre, B.R.; Rodrigues, T.B.; Rieske, L.K. RNA interference and validation of reference genes for gene expression analyses using qPCR in southern pine beetle, Dendroctonus frontalis. Sci. Rep. 2019, 9, 5640. [Google Scholar] [CrossRef] [PubMed]
- Kyre, B.R.; Bentz, B.J.; Rieske, L.K. Susceptibility of mountain pine beetle (Dendroctonus ponderosae Hopkins) to gene silencing through RNAi provides potential as a novel management tool. For. Ecol. Manag. 2020, 473, 118322. [Google Scholar] [CrossRef]
- Kyre, B.R.; Rieske, L.K. Using RNAi to silence heat shock protein has congeneric effects in North America’s Dendroctonus bark beetles. For. Ecol. Manag. 2022, 520, 120367. [Google Scholar] [CrossRef]
- Rodrigues, T.B.; Dhandapani, R.K.; Duan, J.J.; Palli, S.R. RNA interference in the Asian longhorned beetle: Identification of key RNAi genes and reference genes for RT-qPCR. Sci. Rep. 2017, 7, 8913. [Google Scholar] [CrossRef] [PubMed]
- Dhandapani, R.K.; Gurusamy, D.; Duan, J.J.; Palli, S.R. RNAi for management of Asian long-horned beetle, Anoplophora glabripennis: Identification of target genes. J. Pest Sci. 2020, 93, 823–832. [Google Scholar] [CrossRef]
- Pampolini, F.; Rieske, L.K. Emerald ash borer specific gene silencing has no effect on non-target organisms. Front. Agron. 2020, 2, 608827. [Google Scholar] [CrossRef]
- Pampolini, F.; Rodrigues, T.B.; Leelesh, R.S.; Kawashima, T.; Rieske, L.K. Confocal microscopy provides visual evidence and confirms the feasibility of dsRNA delivery to emerald ash borer through plant tissues. J. Pest Sci. 2020, 93, 1143–1153. [Google Scholar] [CrossRef]
- Yu, N.; Christiaens, O.; Liu, J.; Niu, J.; Cappelle, K.; Caccia, S.; Huvenne, H.; Smagghe, G. Delivery of dsRNA for RNAi in insects: An overview and future directions. Insect Sci. 2013, 20, 4–14. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Miao, X. Feasibility, limitation, and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 2013, 20, 15–30. [Google Scholar] [CrossRef]
- Lelito, J.P.; Watt, T.J.; Duan, J.J. Mass-rearing of emerald ash borer and its parasitoids. In Biology and Control of Emerald Ash Borer; Van Driesche, R.G., Reardon, R.C., Eds.; FHTET-2014-09; Department of Agriculture, Forest Service, Forest Health Technology Enterprise Team: Morgantown, WV, USA, 2015; Chapter 8; pp. 129–137. Available online: https://www.fs.fed.us/foresthealth/technology/pdfs/FHTET-2014-09_Biology_Control_EAB.pdf (accessed on 27 April 2015).
- Rivera-Vega, L.; Mamidala, P.; Koch, J.L.; Mason, M.; Mittapalli, O. Evaluation of reference genes for expression studies in ash (Fraxinus spp.). Plant Mol. Biol. Rep. 2012, 30, 242–245. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Olson, D.; Rieske, L.K. Host range expansion may provide enemy free space for the highly invasive emerald ash borer. Biol. Invasions. 2018, 21, 625–635. [Google Scholar] [CrossRef]
- Rajarapu, S.P.; Mamidala, P.; Mittapalli, O. Validation of reference genes for gene expression studies in the emerald ash borer (Agrilus planipennis). Insect Sci. 2012, 19, 41–46. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Willow, J.; Cook, S.M.; Veromann, E.; Smagghe, G. Uniting RNAi technology and conservation biocontrol to promote global food security and agrobiodiversity. Front. Bioeng. Biotechnol. 2022, 10, 871651. [Google Scholar] [CrossRef]
- Hoang, B.T.L.; Fletcher, S.J.; Brosnan, C.A.; Ghodke, A.B.; Manzie, N.; Mitter, N. RNAi as a foliar spray: Efficiency and challenges to field applications. Int. J. Mol. Sci. 2022, 23, 6639. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Dubrovina, A.S. Physiological conditions and dsRNA application approaches for exogenously induced RNA interference in Arabidopsis thaliana. Plants 2021, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Dean, R. Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 2020, 21, 589–601. [Google Scholar] [CrossRef]
- de Andrade, E.C.; Hunter, W.B. RNA interference–natural gene-based technology for highly specific pest control (HiSPeC). In RNA Interference; InTech: Rijeka, Croatia, 2016. [Google Scholar] [CrossRef]
- Biedenkopf, D.; Will, T.; Knauer, T.; Jelonek, L.; Furch, A.; Busche, T.; Koch, A. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2020, 2, 12. [Google Scholar] [CrossRef]
- Doering-Saad, C.; Newbury, H.J.; Bale, J.S.; Pritchard, J. Use of aphid stylectomy and RT-PCR for the detection of transporter mRNAs in sieve elements. J. Exp. Bot. 2002, 53, 631–637. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Andrade, E.C.; Hunter, W.B.; Christiaens, O.; Smagghe, G. Asian citrus psyllid RNAi pathway RNAi evidence. Sci. Rep. 2016, 6, 38082. [Google Scholar] [CrossRef]
- Brosnan, C.A.; Sawyer, A.; de Felippes, F.F.; Carroll, B.J.; Waterhouse, P.M.; Mitter, N. Intact double stranded RNA is mobile and triggers RNAi against viral and fungal plant pathogens. bioRxiv. 2021; preprint. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.M.; Thomma, B.; Huang, H.D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 16151. [Google Scholar] [CrossRef]
- Cagliari, D.; Dias, N.P.; Galdeano, D.M.; dos Santos, E.Á.; Smagghe, G.; Zotti, M.J. Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 2019, 10, 1319. [Google Scholar] [CrossRef]
| Day 3 | Day 7 | Day 14 | Day 21 | All Time Points | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Root | 100% | (9/9) | 100% | (9/9) | 77.7% | (7/9) | 100% | (9/9) | 94.4 | (34/36) |
| Woody stem | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 88.8% | (8/9) | 97.2% | (35/36) |
| Soft stem | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (36/36) |
| Treated leaf | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (36/36) |
| Untreated leaf | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (9/9) | 100% | (36/36) |
| All tissues | 100% | (45/45) | 100% | (45/45) | 93.3% | (42/45) | 95.5% | (43/45) | 98.3% | (177/180) |
| Characteristic | Odds Ratio | Confidence Interval | p-Value |
|---|---|---|---|
| Time (d) | 0.88 | (0.68, 1.06) | 0.18 |
| RCD (mm) | 0.05 | (0.00, 4637) | 0.59 |
| Height (cm) | 0.97 | (0.82, 1.13) | 0.69 |
| Tissue | N/A | N/A | 0.19 |
| Replicate | N/A | N/A | 0.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pampolini, F.; Rieske, L.K. Foliar Application of dsRNA to Induce Gene Silencing in Emerald Ash Borer: Systemic Distribution, Persistence, and Bioactivity. Forests 2023, 14, 1853. https://doi.org/10.3390/f14091853
Pampolini F, Rieske LK. Foliar Application of dsRNA to Induce Gene Silencing in Emerald Ash Borer: Systemic Distribution, Persistence, and Bioactivity. Forests. 2023; 14(9):1853. https://doi.org/10.3390/f14091853
Chicago/Turabian StylePampolini, Flávia, and Lynne K. Rieske. 2023. "Foliar Application of dsRNA to Induce Gene Silencing in Emerald Ash Borer: Systemic Distribution, Persistence, and Bioactivity" Forests 14, no. 9: 1853. https://doi.org/10.3390/f14091853
APA StylePampolini, F., & Rieske, L. K. (2023). Foliar Application of dsRNA to Induce Gene Silencing in Emerald Ash Borer: Systemic Distribution, Persistence, and Bioactivity. Forests, 14(9), 1853. https://doi.org/10.3390/f14091853







