Development of an Advanced-Generation Multi-Objective Breeding Population for the 4th Cycle of Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook.)
Abstract
:1. Introduction
2. Materials and Methods
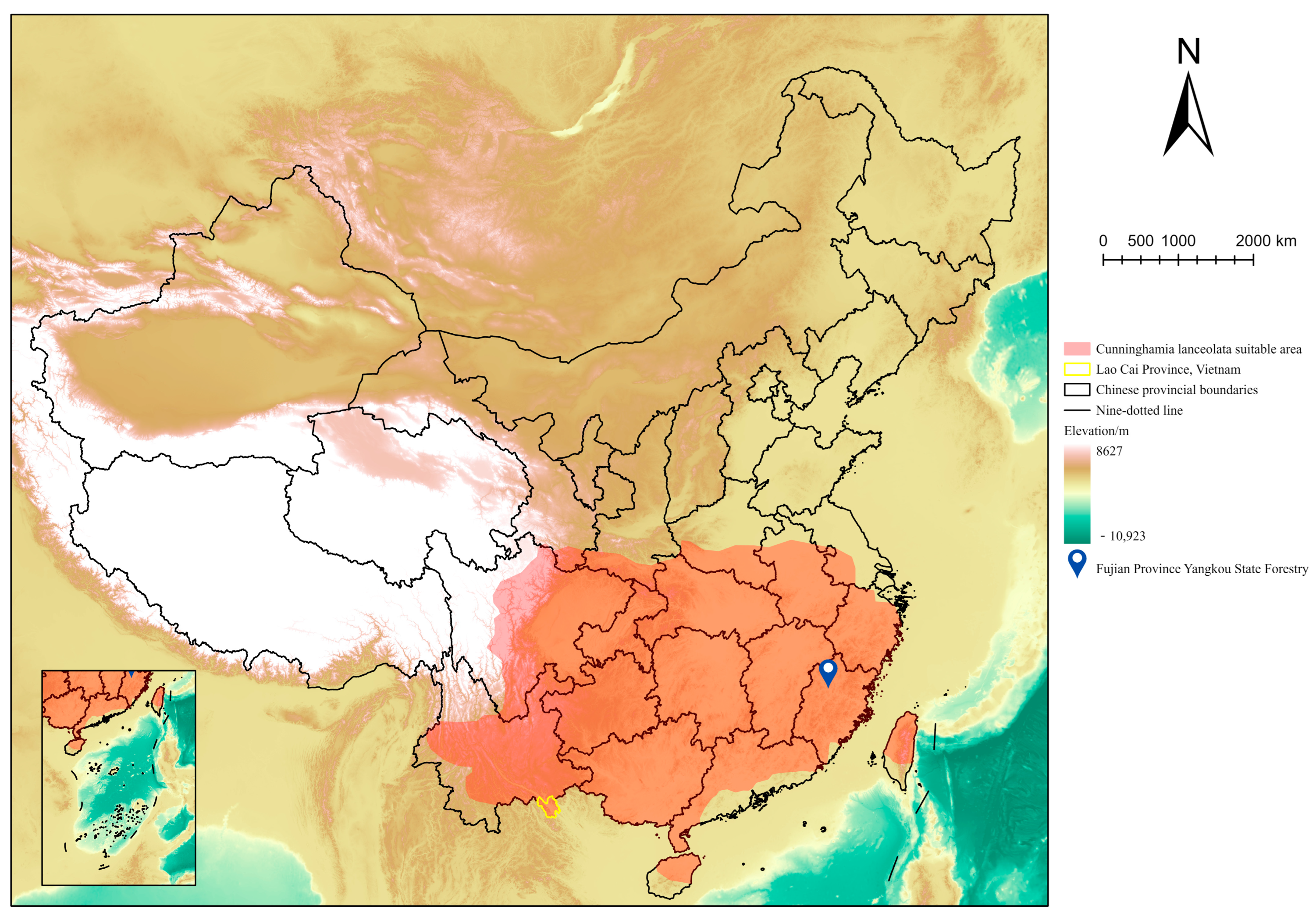
2.1. Test Material
2.2. SNP Marker Development
2.3. Trait Determination
2.4. Statistical Analysis
2.4.1. Estimates of Breeding Values
2.4.2. Genetic Diversity Analysis
2.4.3. Core Subpopulation Collection Strategy
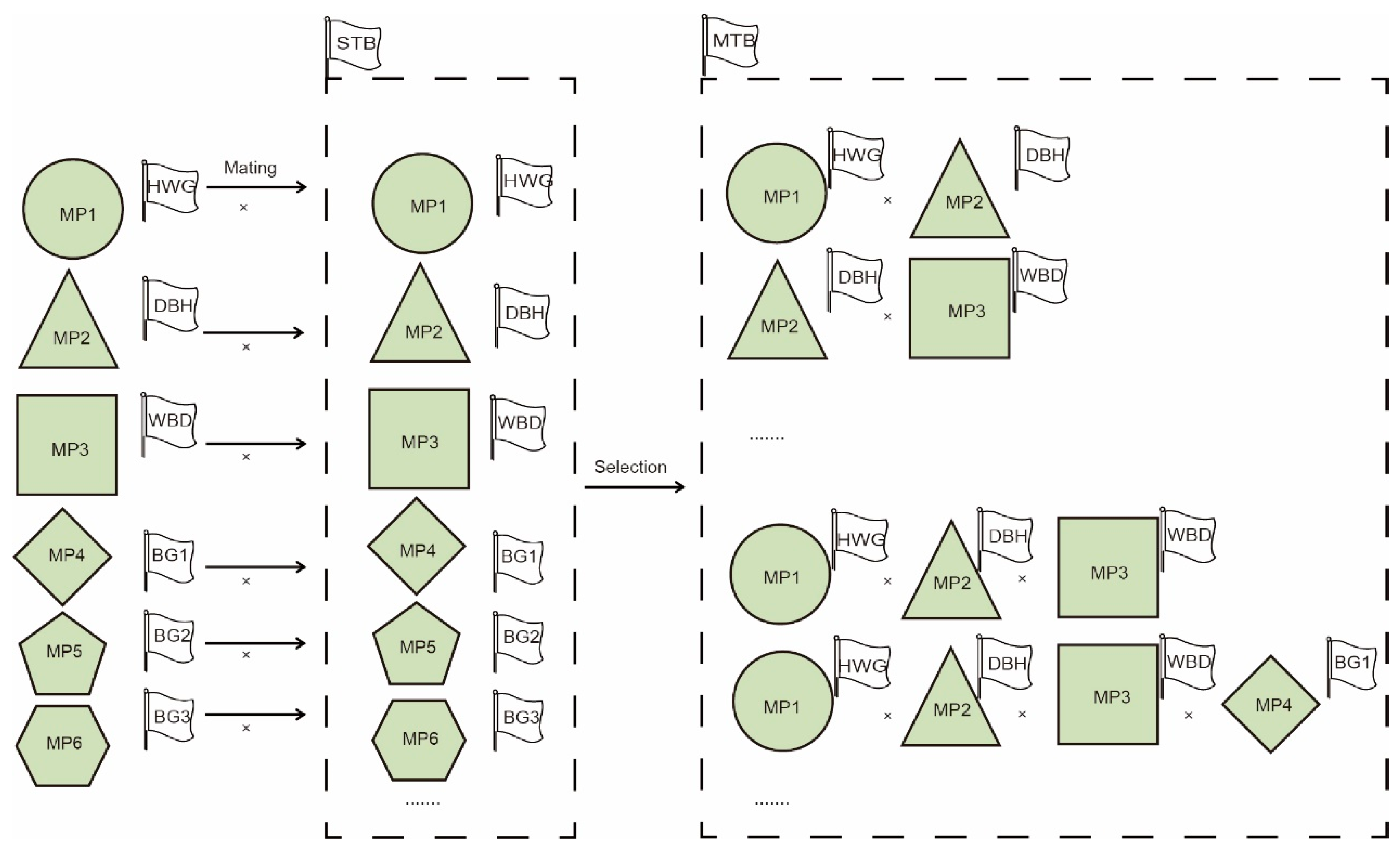
2.4.4. Structured Breeding Population Construction
3. Results
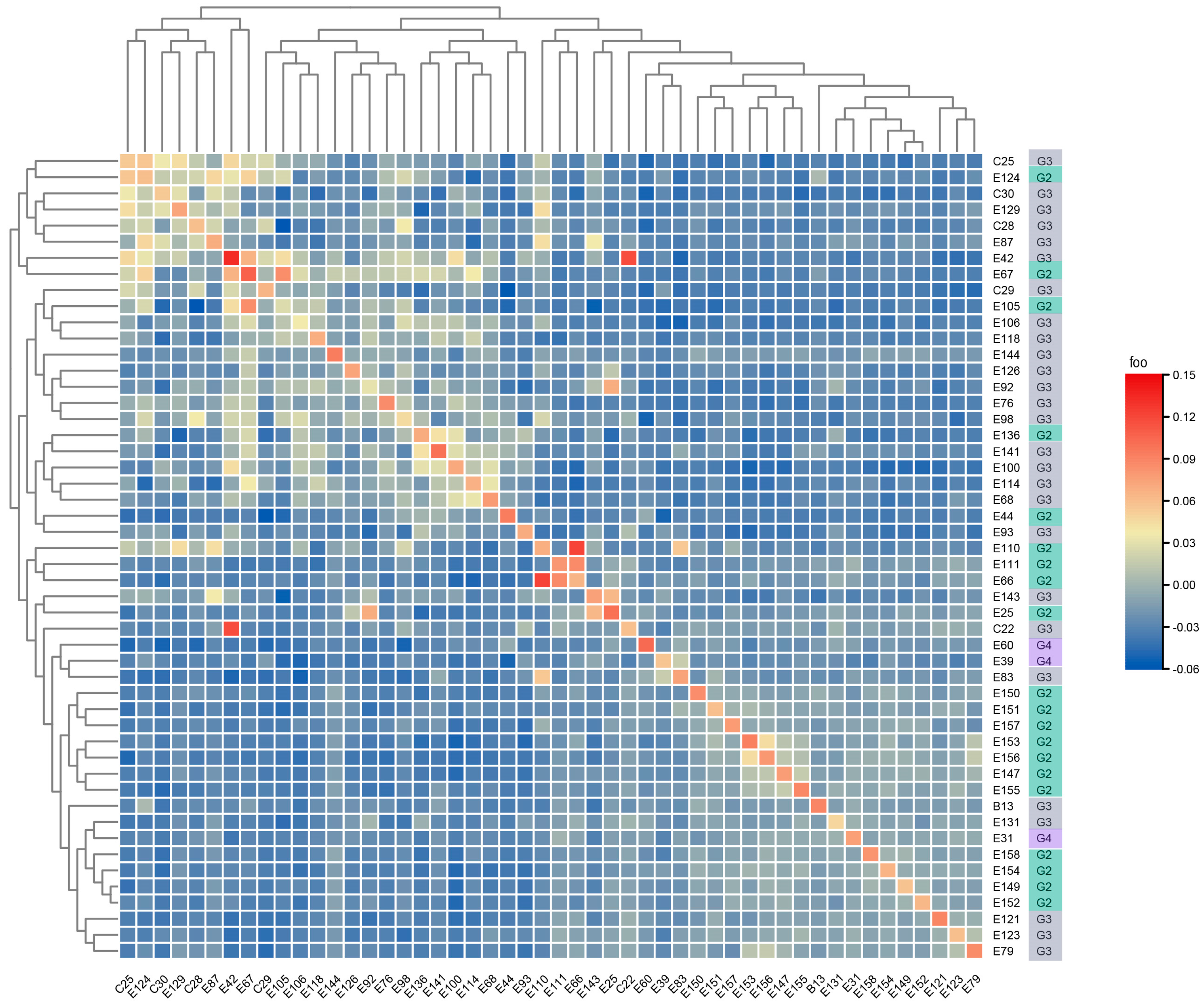
3.1. Construction of Subpopulations Based on Molecular Data
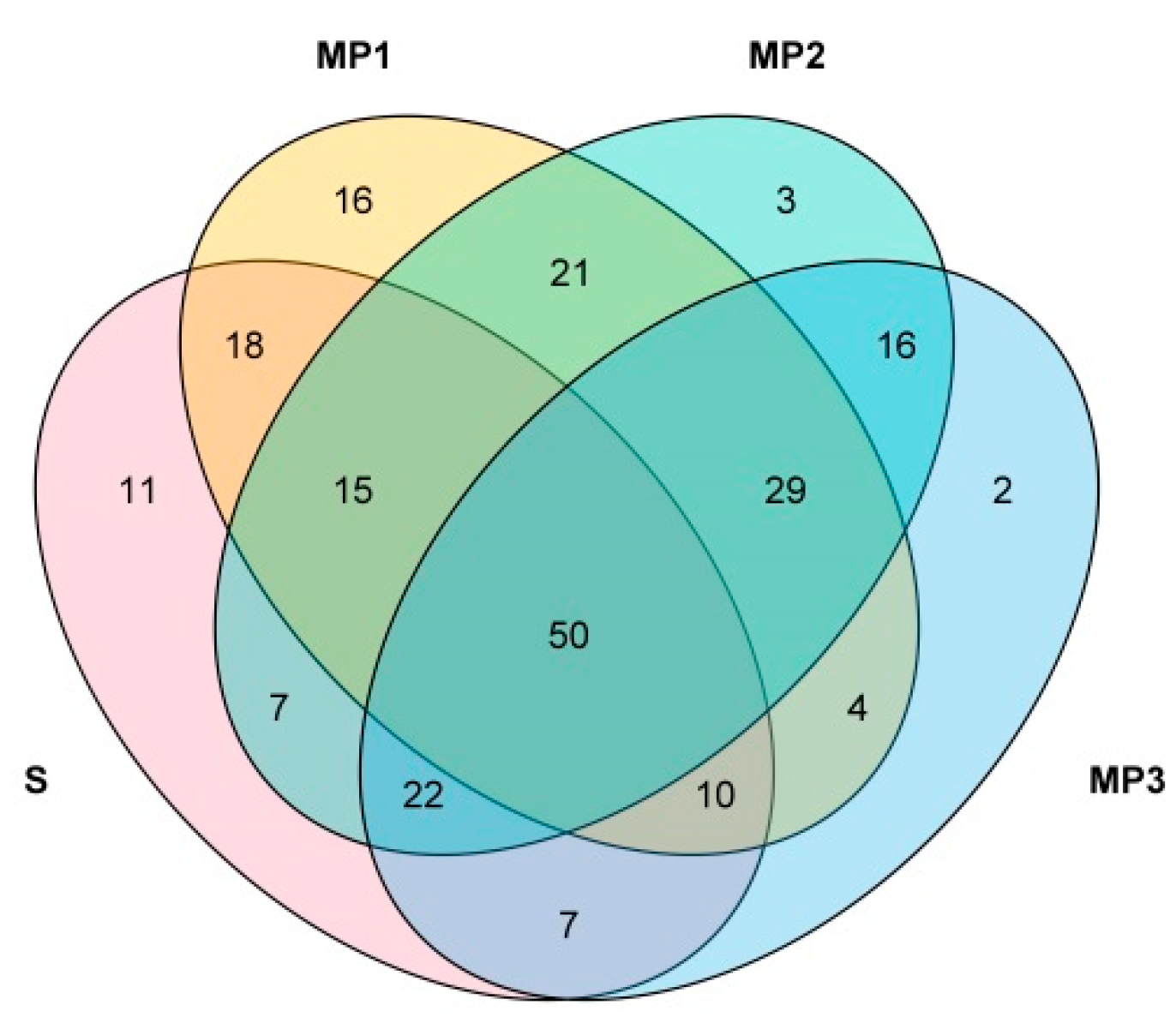
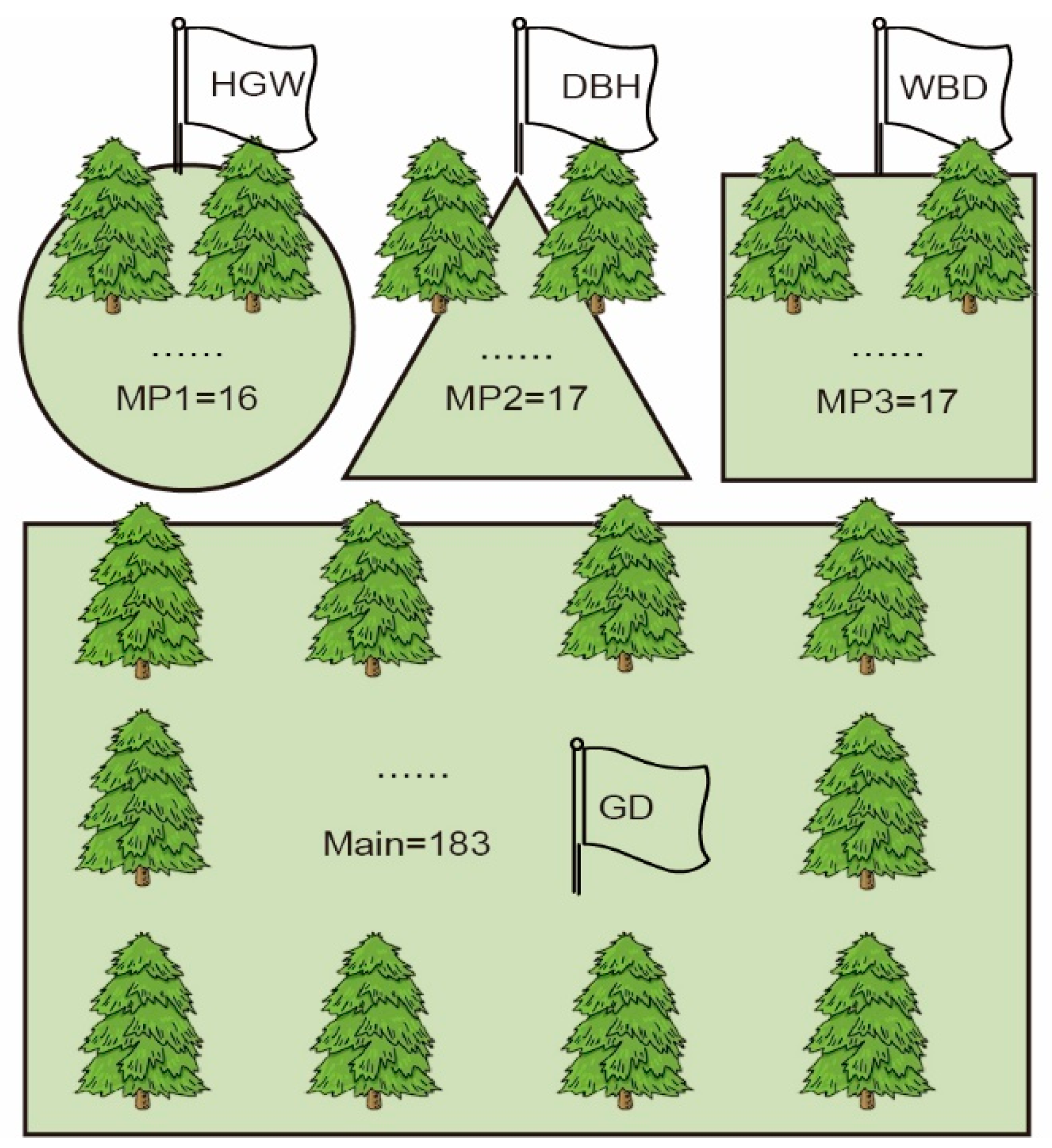
3.2. Construction of Multiple Populations of Chinese Fir Based on Phenotypic Data
3.3. Construction of a Core Breeding Chinese Fir Population
3.4. Strategies for Managing Core Populations of Chinese Fir
4. Discussion
4.1. Breeding Objectives of Chinese Fir
4.2. The Breeding Population Size
4.3. How to Maintain Genetic Diversity and Control Inbreeding
4.4. Construction and Management of the Core Breeding Population
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, Y.; Deng, X.; Xiang, W.; Chen, L.; Ouyang, S. Predicting potential suitable habitats of Chinese fir under current and future climatic scenarios based on Maxent model. Ecol. Inform. 2021, 64, 101393. [Google Scholar] [CrossRef]
- Cui, H.O.; Liu, M. Analysis on the results of the 9th national forest inventory. J. West China For. Sci. 2020, 49, 90–95. [Google Scholar]
- Kang, X. Thoughts on tree breeding strategies. J. Beijing For. Univ. 2019, 41, 15–22. [Google Scholar] [CrossRef]
- Kang, X. Research progress of forest genetics and tree breeding. J. Nanjing For. Univ. 2020, 44, 1–10. [Google Scholar] [CrossRef]
- Burdon, R.D.; Namkoong, G. Multiple populations and sublines. Silvae Genet. 1983, 32, 221–222. [Google Scholar]
- Cotterill, P.P.; Jackson, N. Gains expected from clonal orchards under alternative breeding strategies. For. Sci. 1989, 35, 183–196. [Google Scholar]
- Borralho, N.M.G.; Dutkowski, G.W. Comparison of rolling front and discrete generation breeding strategies for trees. Can. J. For. Res. 1998, 28, 987–993. [Google Scholar] [CrossRef]
- Wu, H.X.; Hallingbäck, H.R.; Sánchez, L. Performance of seven tree breeding strategies under conditions of inbreeding depression. G3 2016, 6, 529–540. [Google Scholar] [CrossRef]
- McKeand, E.; Beineke, F. Sublining for half-sib breeding populations of forest trees. Silvae Genet. 1980, 29, 14–17. [Google Scholar]
- White, T.L.; Hodge, G.R.; Powell, G.L. An advanced-generation tree improvement plan for slash pine in the southeastern United States. Silvae Genet. 1993, 42, 359–371. [Google Scholar]
- Jayawickrama, K.J.S.; Carson, M.J. A breeding strategy for the New Zealand radiata pine breeding cooperative. Silvae Genet. 2000, 49, 82–89. [Google Scholar]
- McKeand, S.E.; Bridgwater, F.E. A strategy for the third breeding cycle of loblolly pine in the Southeastern US. Silvae Genet. 1998, 47, 223–234. [Google Scholar]
- Isik, F.; McKeand, S.E. Fourth cycle breeding and testing strategy for Pinus taeda in the NC State University Cooperative Tree Improvement Program. Tree Genet. Genom. 2019, 15, 70–82. [Google Scholar] [CrossRef]
- White, T.L.; Huber, D.A.; Powell, G.L. Third-cycle breeding strategy for slash pine by the Cooperative Forest Genetics Research Program. In Proceedings of the 27th Southern Forest Tree Improvement Conference, Stillwater, OK, USA, 24–27 June 2003. [Google Scholar]
- Tóth, E.G.; Vendramin, G.G.; Bagnoli, F.; Cseke, K.; Höhn, M. High genetic diversity and distinct origin of recently fragmented Scots pine (Pinus sylvestris L.) populations along the Carpathians and the Pannonian Basin. Tree Genet. Genom. 2017, 13, 47. [Google Scholar] [CrossRef]
- Chen, Z.Q.; Zan, Y.; Milesi, P.; Zhou, L.; Chen, J.; Li, L.; Cui, B.; Niu, S.; Westin, J.; Karlsson, B. Leveraging breeding programs and genomic data in Norway spruce (Picea abies L. Karst) for GWAS analysis. Genome Biol. 2021, 22, 179. [Google Scholar] [CrossRef]
- Rosvall, O.; Ståhl, P.; Almqvist, C.; Anderson, B.; Berlin, M.; Ericsson, T.; Eriksson, M.; Gregorsson, B.; Hajek, J.; Hallander, J.J.S. Review of the Swedish Tree Breeding Programme; Skogforsk: Uppsala, Sweden, 2011. [Google Scholar]
- Bian, L.; Huang, D.; Zhang, X.; Tong, X.; Ye, D.; Shi, J. Analysis on flowering phenology and synchronization indexes of Chinese fir clonal archive. J. Nanjing For. Univ. 2020, 44, 207–212. [Google Scholar] [CrossRef]
- Ouyang, L.; Chen, J.; Zheng, R.; Xu, Y.; Lin, Y.; Huang, J.; Ye, D.; Fang, Y.; Shi, J. Genetic diversity among the germplasm collections of the Chinese fir in 1st breeding population upon SSR markers. J. Nanjing For. Univ. 2014, 38, 21–26. [Google Scholar]
- Jing, Y.; Bian, L.; Zhang, X.; Zhao, B.; Zheng, R.; Su, S.; Ye, D.; Zheng, X.; El-Kassaby, Y.A.; Shi, J. Genetic diversity and structure of the 4th cycle breeding population of Chinese fir (Cunninghamia lanceolata (lamb.) hook). Front. Plant Sci. 2023, 14, 1106615. [Google Scholar] [CrossRef]
- Chen, Y.; Shi, J. Some basic problems in genetic improvement of Chinese fir. J. Nanjing For. Univ. 1983, 7, 5–19. [Google Scholar]
- Yu, R. Study on establishing Cunninghamia lanceolata breeding poputation of the third generation. J. Fujian Coll. For. 2008, 28, 69–72. [Google Scholar] [CrossRef]
- Bian, L.; Shi, J.; Zheng, R.; Chen, J.; Wu, H.X. Genetic parameters and genotype–environment interactions of Chinese fir (Cunninghamia lanceolata) in Fujian Province. Can. J. For. Res. 2014, 44, 582–592. [Google Scholar] [CrossRef]
- Zheng, R.; Hong, Z.; Su, S.; Bian, L.; Xiao, H.; Shi, J.; Wu, H.X. Inheritance of growth and survival in two 9-year-old, open-pollinated progenies of an advanced breeding population of Chinese firs in southeastern China. J. For. Res. 2016, 27, 1067–1075. [Google Scholar] [CrossRef]
- Kerr, R.J.; Dutkowski, G.W.; Jansson, G.; Persson, T.; Westin, J. Connectedness among test series in mixed linear models of genetic evaluation for forest trees. Tree Genet. Genom. 2015, 11, 67. [Google Scholar] [CrossRef]
- Butler, D.; Cullis, B.; Gilmour, A.; Gogel, B.; Thompson, R. ASReml-R Reference Manual Version 4; VSN International Ltd.: Hemel Hempstead, UK, 2017. [Google Scholar]
- Slifer, S.H. PLINK: Key functions for data analysis. Curr. Protoc. Hum. Gen. 2018, 97, e59. [Google Scholar] [CrossRef] [PubMed]
- Ulintz, P.J.; Wu, W.; Gates, C.M. Bioinformatics analysis of whole exome sequencing data. Methods Mol. Biol. 2019, 1881, 277–318. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Thachuk, C.; Crossa, J.; Franco, J.; Dreisigacker, S.; Warburton, M.; Davenport, G.F. Core Hunter: An algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinform. 2009, 10, 243. [Google Scholar] [CrossRef]
- Jeong, N.; Kim, K.S.; Jeong, S.; Kim, J.Y.; Park, S.K.; Lee, J.S.; Jeong, S.C.; Kang, S.T.; Ha, B.K.; Kim, D.Y.; et al. Korean soybean core collection: Genotypic and phenotypic diversity population structure and genome-wide association study. PLoS ONE 2019, 14, e0224074. [Google Scholar] [CrossRef]
- Wang, N.N.; Jiang, H.W.; Zhang, Z.G.; Li, Y.Y.; Li, C.D.; Song, Y.B.; Li, Y.; Yu, Y.; Liu, C.Y.; Hu, Z.B. Gene mining of 100-grain weight and the number of four-seed pods in soybean (Glycine max). Plant Breed. 2022, 141, 143–158. [Google Scholar] [CrossRef]
- White, T.; Adams, W.; Neale, D. Forest genetics-concepts, scope, history and importance. In Forest Genetics; CABI: Wallingford, UK, 2007; pp. 1–14. [Google Scholar]
- Feng, Y.; Yang, Z.; Li, H.; Xu, H. Changes of genetic gain & genetic diversity in the breeding process of Pinus massoniana. J. Nanjing For. Univ. 2018, 61, 196–200. [Google Scholar] [CrossRef]
- López-Villalobos, A.; Eckert, C.G. Consequences of multiple mating-system shifts for population and range-wide genetic structure in a coastal dune plant. Mol. Ecol. 2018, 27, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shi, J.; Li, F.; Gan, S. Molecular characterization of elite genotypes within a second-generation Chinese fir (Cunninghamia lanceolata) breeding population using RAPD markers. Sci. Silvae Sin. 2007, 43, 50–55. [Google Scholar]
- Zheng, H.; Hu, D.; Wei, R.; Yan, S.; Wang, R. Chinese fir breeding in the high-throughput sequencing era: Insights from SNPs. Forests 2019, 10, 681. [Google Scholar] [CrossRef]
- Ye, P.; Chen, Y.; Ruan, Y.; Chen, S.; Liu, D.; Guo, M.; Lin, Q. A study of the early selection of Chinese fir. J. Nanjing For. Univ. 1981, 24, 106–116. [Google Scholar] [CrossRef]
- Wu, H.X.; Owen, J.V.; Abarquez, A.; Matheson, A.C. Inbreeding in Pinus Radiata-V. the effects of inbreeding on fecundity. Silvae Genet. 2004, 53, 80–88. [Google Scholar] [CrossRef]
- Wu, H.X.; Ker, R.; Chen, Z.; Ivkovic, M. Balancing breeding for growth and fecundity in radiata pine (Pinus radiata D. Don) breeding programme. Evol. Appl. 2021, 14, 834–846. [Google Scholar] [CrossRef]
- Kerr, R.J.; McRae, T.A.; Dutkowski, G.W.; Tier, B. Managing the rate of increase in average co-ancestry in a rolling front tree breeding strategy. J. Anim. Breed. Genet. 2015, 132, 109–120. [Google Scholar] [CrossRef]
- Wu, H.X.; Eldridge, K.G.; Matheson, A.C.; Powell, M.B.; McRae, T.A.; Butcher, T.B.; Johnson, I.G. Achievements in forest tree improvement in Australia and New Zealand 8. Successful introduction and breeding of radiata pine in Australia. Aust. For. 2007, 70, 215–225. [Google Scholar] [CrossRef]
- Hoshikawa, K.; Lin, Y.-P.; Schafleitner, R.; Shirasawa, K.; Isobe, S.; Nguyen, D.C.; Ohsawa, R.; Yoshioka, Y. Genetic diversity analysis and core collection construction for Amaranthus tricolor germplasm based on genome-wide single-nucleotide polymorphisms. Sci. Hortic. 2023, 307, 111428. [Google Scholar] [CrossRef]
- Zhu, Y.; Liang, D.; Song, Z.; Tan, Y.; Guo, X.; Wang, D. Genetic Diversity Analysis and Core Germplasm Collection Construction of Camellia oleifera Based on Fruit Phenotype and SSR Data. Genes 2022, 13, 2351. [Google Scholar] [CrossRef]
- Li, N.; Yang, Y.; Xu, F.; Chen, X.; Wei, R.; Li, Z.; Pan, W.; Zhang, W. Genetic Diversity and Population Structure Analysis of Castanopsis hystrix and Construction of a Core Collection Using Phenotypic Traits and Molecular Markers. Genes 2022, 13, 2383. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.H.D. The case for core collections. In The Use of Plant Genetic Resources; Brown, A.H.D., Frankel, O.H., Marshall, D.R., Williams, J.T., Eds.; Cambridge University Press: Cambridge, UK, 1989; pp. 136–156. [Google Scholar]
- Wang, J.; Guan, Y.; Wang, Y.; Zhu, L.; Wang, Q.; Hu, Q.; Hu, J. A strategy for finding the optimal scale of plant core collection based on Monte Carlo simulation. Sci. World J. 2014, 2014, 503473. [Google Scholar] [CrossRef] [PubMed]
- Diwan, N.; McIntosh, M.S.; Bauchan, G.R. Methods of developing a core collection of annual Medicago species. Theor. Appl. Genet. 1995, 90, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Li, T.H.; Zhang, H.L.; Qi, Y.W. Sampling strategy for a primary core collection of peach (Prunus persica (L.) Batsch.) germplasm. Eur. J. Hort. Sci. 2007, 72, 268–274. [Google Scholar]
- Roy Choudhury, D.; Singh, N.; Singh, A.K.; Kumar, S.; Srinivasan, K.; Tyagi, R.K.; Ahmad, A.; Singh, N.K.; Singh, R. Analysis of genetic diversity and population structure of rice germplasm from north-eastern region of India and development of a core germplasm set. PLoS ONE 2014, 9, e113094. [Google Scholar] [CrossRef] [PubMed]

| Species | Program | Cycle | N | Strategy | Reference |
|---|---|---|---|---|---|
| Pinus radiata | TBA | 2 (1996–2011) | 340 | Subline and multiple populations | [11] |
| 3–4 (2012–) | NA | Rolling-front | [8] | ||
| Pinus teada | NCSU | 3 (1995–2015) | 1017 | Subline within nucleus | [12] |
| 4 (2010–2024) | NA | Rolling-front | [13] | ||
| Pinus elliottii | CFGRP | 1 (1953–1986) | 2516 | Unstructured | [14] |
| 2 (1987–2002) | 1017 | Subline within nucleus | [14] | ||
| 3 (2003–2013) | NA | Subline within nucleus | [14] | ||
| Pinus sylvestris | Skogforsk | 1 | NA | Multiple populations | [15] |
| 2 | NA | Multiple populations | [15] | ||
| 3 | NA | Multiple populations and partial rolling-front | [15] | ||
| Picea abies | Skogforsk | 1 | NA | Multiple populations | [16] |
| 2 | NA | Multiple populations and partial rolling-front | [17] |
| Core Collection | Size (n) | MAF | Ne | He | Ho | PIC | Shi | Nei | Fst | PN/% | CV/% | The Most Feasible K Value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S-10% | 23 | 0.1613 | 1.3881 | 0.2428 | 0.2528 | 0.2147 | 0.4132 | 0.2538 | 0.1652 | 1.7122 | 98.2878 | 1 |
| S-15% | 35 | 0.1603 | 1.3891 | 0.2426 | 0.2487 | 0.2150 | 0.4136 | 0.2497 | 0.1650 | 0.3344 | 99.6656 | 1 |
| S-20% | 47 | 0.1583 | 1.3900 | 0.2404 | 0.2409 | 0.2152 | 0.4138 | 0.2456 | 0.1655 | 0.1018 | 99.8982 | 1 |
| S-25% | 58 | 0.1574 | 1.3911 | 0.2395 | 0.2382 | 0.2154 | 0.4140 | 0.2437 | 0.1673 | 0.0271 | 99.9729 | 1 |
| S-30% | 70 | 0.1569 | 1.3921 | 0.2392 | 0.2369 | 0.2156 | 0.4143 | 0.2427 | 0.1687 | 0.0064 | 99.9936 | 2 |
| S-35% | 82 | 0.1563 | 1.3932 | 0.2386 | 0.2348 | 0.2158 | 0.4146 | 0.2415 | 0.1697 | 0.0009 | 99.9991 | 2 |
| S-40% | 93 | 0.1567 | 1.3944 | 0.2393 | 0.2367 | 0.2160 | 0.4151 | 0.2419 | 0.1707 | 0.0003 | 99.9997 | 2 |
| S-45% | 105 | 0.1561 | 1.3955 | 0.2384 | 0.2348 | 0.2162 | 0.4152 | 0.2407 | 0.1716 | 0.0003 | 99.9997 | 3 |
| S-50% | 116 | 0.1564 | 1.3964 | 0.2390 | 0.2358 | 0.2164 | 0.4152 | 0.2411 | 0.1725 | 0 | 100 | 3 |
| S-55% | 128 | 0.1563 | 1.3973 | 0.2389 | 0.2355 | 0.2165 | 0.4158 | 0.2408 | 0.1729 | 0 | 100 | 3 |
| S-60% | 140 | 0.1560 | 1.3982 | 0.2386 | 0.2347 | 0.2168 | 0.4158 | 0.2403 | 0.1736 | 0 | 100 | 4 |
| S-65% | 151 | 0.1562 | 1.3994 | 0.2389 | 0.2353 | 0.2171 | 0.4159 | 0.2405 | 0.1750 | 0 | 100 | 4 |
| S-70% | 163 | 0.1562 | 1.4004 | 0.2388 | 0.2362 | 0.2173 | 0.4162 | 0.2403 | 0.1767 | 0 | 100 | 4 |
| MP1-10% | 23 | 0.1516 | 1.3944 | 0.2292 | 0.2234 | 0.2160 | 0.4150 | 0.2396 | 0.1849 | 3.1099 | 96.8901 | 1 |
| MP1-20% | 47 | 0.1542 | 1.3954 | 0.2348 | 0.2300 | 0.2162 | 0.4151 | 0.2399 | 0.1831 | 0.1493 | 99.8507 | 1 |
| MP1-30% | 70 | 0.1550 | 1.3964 | 0.2365 | 0.2289 | 0.2164 | 0.4152 | 0.2399 | 0.1836 | 0.0070 | 99.9930 | 2 |
| MP1-40% | 93 | 0.1552 | 1.3973 | 0.2371 | 0.2329 | 0.2167 | 0.4157 | 0.2397 | 0.1826 | 0 | 100 | 2 |
| MP1-50% | 117 | 0.1553 | 1.3985 | 0.2375 | 0.2328 | 0.2169 | 0.4159 | 0.2395 | 0.1809 | 0 | 100 | 3 |
| MP1-60% | 140 | 0.1552 | 1.3992 | 0.2375 | 0.2325 | 0.2171 | 0.4159 | 0.2392 | 0.1798 | 0 | 100 | 3 |
| MP1-70% | 163 | 0.1550 | 1.4004 | 0.2373 | 0.2319 | 0.2173 | 0.4162 | 0.2388 | 0.1792 | 0 | 100 | 4 |
| MP2-10% | 23 | 0.1525 | 1.3942 | 0.2306 | 0.2291 | 0.2162 | 0.4149 | 0.2411 | 0.1857 | 2.9859 | 97.0141 | 1 |
| MP2-20% | 47 | 0.1530 | 1.3953 | 0.2331 | 0.2287 | 0.2164 | 0.4154 | 0.2382 | 0.1828 | 0.1857 | 99.8143 | 1 |
| MP2-30% | 70 | 0.1536 | 1.3963 | 0.2345 | 0.2298 | 0.2166 | 0.4154 | 0.2379 | 0.1840 | 0.0131 | 99.9869 | 2 |
| MP2-40% | 93 | 0.1538 | 1.3973 | 0.2352 | 0.2297 | 0.2168 | 0.4158 | 0.2378 | 0.1825 | 0.0003 | 99.9997 | 3 |
| MP2-50% | 117 | 0.1543 | 1.3983 | 0.2360 | 0.2298 | 0.2169 | 0.4157 | 0.2380 | 0.1811 | 0.0003 | 99.9997 | 3 |
| MP2-60% | 140 | 0.1547 | 1.3992 | 0.2366 | 0.2318 | 0.2171 | 0.4160 | 0.2383 | 0.1786 | 0 | 100 | 4 |
| MP2-70% | 163 | 0.1547 | 1.4002 | 0.2367 | 0.2302 | 0.2172 | 0.4163 | 0.2382 | 0.1782 | 0 | 100 | 4 |
| MP3-10% | 23 | 0.1530 | 1.3943 | 0.2308 | 0.2305 | 0.2160 | 0.4149 | 0.2413 | 0.1860 | 3.6031 | 96.3969 | 1 |
| MP3-20% | 47 | 0.1540 | 1.3953 | 0.2342 | 0.2320 | 0.2163 | 0.4153 | 0.2393 | 0.1844 | 0.2654 | 99.7346 | 1 |
| MP3-30% | 70 | 0.1542 | 1.3964 | 0.2350 | 0.2327 | 0.2165 | 0.4155 | 0.2384 | 0.1829 | 0.0422 | 99.9578 | 2 |
| MP3-40% | 93 | 0.1549 | 1.3973 | 0.2363 | 0.2346 | 0.2166 | 0.4157 | 0.2389 | 0.1826 | 0.0081 | 99.9919 | 2 |
| MP3-50% | 117 | 0.1548 | 1.3984 | 0.2363 | 0.2322 | 0.2168 | 0.4159 | 0.2383 | 0.1815 | 0.0009 | 99.9991 | 3 |
| MP3-60% | 140 | 0.1549 | 1.3993 | 0.2368 | 0.2331 | 0.2170 | 0.4161 | 0.2385 | 0.1801 | 0 | 100 | 3 |
| MP3-70% | 163 | 0.1551 | 1.4003 | 0.2372 | 0.2327 | 0.2172 | 0.4163 | 0.2387 | 0.1778 | 0 | 100 | 4 |
| Entire collection | 233 | 0.1552 | 1.4013 | 0.2377 | 0.2323 | 0.2175 | 0.4165 | 0.2387 | 0.1773 | 0 | 100 | 4 |
| Core Collection | Size (n) | HGW/g | DBH/cm | WD/(kg/m3) | ΔGHGW | ΔGDBH | ΔGWBD |
|---|---|---|---|---|---|---|---|
| S-10% | 23 | 0.575 | 21.92 | 306.57 | 1.05% | −5.94% | −6.21% |
| S-15% | 35 | 0.570 | 23.30 | 316.21 | 0.17% | −0.02% | −3.27% |
| S-20% | 47 | 0.570 | 23.32 | 326.23 | 0.17% | 0.07% | −0.20% |
| S-25% | 58 | 0.566 | 23.39 | 327.15 | −0.53% | 0.37% | 0.08% |
| S-30% | 70 | 0.563 | 23.48 | 325.39 | −1.06% | 0.76% | −0.46% |
| S-35% | 82 | 0.556 | 24.10 | 327.18 | −2.29% | 3.42% | 0.09% |
| S-40% | 93 | 0.568 | 24.15 | 326.88 | −0.18% | 3.63% | 0.00% |
| S-45% | 105 | 0.559 | 24.35 | 327.32 | −1.76% | 4.49% | 0.13% |
| S-50% | 116 | 0.559 | 24.17 | 326.73 | −1.76% | 3.72% | −0.05% |
| S-55% | 128 | 0.567 | 23.97 | 324.92 | −0.36% | 2.86% | −0.60% |
| S-60% | 140 | 0.560 | 23.96 | 324.12 | −1.59% | 2.82% | −0.85% |
| S-65% | 151 | 0.556 | 23.73 | 325.71 | −2.29% | 1.83% | −0.36% |
| S-70% | 163 | 0.573 | 24.00 | 326.90 | 0.70% | 2.99% | 0.00% |
| MP1-10% | 23 | 0.873 | 21.60 | 324.91 | 53.42% | −7.31% | −0.60% |
| MP1-20% | 47 | 0.792 | 22.52 | 327.71 | 39.18% | −3.36% | 0.25% |
| MP1-30% | 70 | 0.748 | 23.03 | 328.33 | 31.45% | −1.17% | 0.44% |
| MP1-40% | 93 | 0.715 | 22.79 | 322.22 | 25.65% | −2.20% | −1.43% |
| MP1-50% | 117 | 0.686 | 22.98 | 322.33 | 20.55% | −1.39% | −1.39% |
| MP1-60% | 140 | 0.661 | 22.94 | 322.33 | 16.16% | −1.56% | −1.39% |
| MP1-70% | 163 | 0.638 | 23.10 | 324.21 | 12.12% | −0.87% | −0.82% |
| MP2-10% | 23 | 0.587 | 33.13 | 361.14 | 3.16% | 42.17% | 10.48% |
| MP2-20% | 47 | 0.558 | 30.54 | 351.94 | −1.94% | 31.05% | 7.66% |
| MP2-30% | 70 | 0.530 | 29.33 | 345.33 | −6.86% | 25.86% | 5.64% |
| MP2-40% | 93 | 0.535 | 28.40 | 344.13 | −5.98% | 21.87% | 5.28% |
| MP2-50% | 117 | 0.540 | 27.45 | 342.00 | −5.10% | 17.79% | 4.62% |
| MP2-60% | 140 | 0.556 | 25.57 | 337.35 | −2.29% | 9.73% | 3.20% |
| MP2-70% | 163 | 0.558 | 25.73 | 332.34 | −1.94% | 10.41% | 1.67% |
| MP3-10% | 23 | 0.511 | 26.76 | 405.17 | −10.20% | 14.83% | 23.95% |
| MP3-20% | 47 | 0.524 | 25.82 | 387.76 | −7.91% | 10.80% | 18.62% |
| MP3-30% | 70 | 0.543 | 25.57 | 376.87 | −4.58% | 9.73% | 15.29% |
| MP3-40% | 93 | 0.546 | 25.03 | 367.95 | −4.05% | 7.41% | 12.56% |
| MP3-50% | 117 | 0.550 | 24.77 | 359.84 | −3.35% | 6.29% | 10.08% |
| MP3-60% | 140 | 0.557 | 24.48 | 325.69 | −2.12% | 5.05% | −0.37% |
| MP3-70% | 163 | 0.561 | 24.31 | 346.40 | −1.41% | 4.32% | 5.97% |
| Entire Collection | 233 | 0.569 | 23.30 | 326.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, B.; Bian, L.; Feng, Q.; Wu, J.; Zhang, X.; Zheng, R.; Zheng, X.; Yang, Z.; Chen, Z.; Wu, H.X.; et al. Development of an Advanced-Generation Multi-Objective Breeding Population for the 4th Cycle of Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook.). Forests 2023, 14, 1658. https://doi.org/10.3390/f14081658
Zhao B, Bian L, Feng Q, Wu J, Zhang X, Zheng R, Zheng X, Yang Z, Chen Z, Wu HX, et al. Development of an Advanced-Generation Multi-Objective Breeding Population for the 4th Cycle of Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook.). Forests. 2023; 14(8):1658. https://doi.org/10.3390/f14081658
Chicago/Turabian StyleZhao, Benwen, Liming Bian, Qihang Feng, Jinzhang Wu, Xuefeng Zhang, Renhua Zheng, Xueyan Zheng, Zhiyuan Yang, Zhiqiang Chen, Harry X. Wu, and et al. 2023. "Development of an Advanced-Generation Multi-Objective Breeding Population for the 4th Cycle of Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook.)" Forests 14, no. 8: 1658. https://doi.org/10.3390/f14081658
APA StyleZhao, B., Bian, L., Feng, Q., Wu, J., Zhang, X., Zheng, R., Zheng, X., Yang, Z., Chen, Z., Wu, H. X., & Shi, J. (2023). Development of an Advanced-Generation Multi-Objective Breeding Population for the 4th Cycle of Chinese Fir (Cunninghamia lanceolata (Lamb.) Hook.). Forests, 14(8), 1658. https://doi.org/10.3390/f14081658







