Comparative Transcriptome and Metabolome Analysis of Rubber Trees (Hevea brasiliensis Muell. Arg.) Response to Aluminum Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Al Treatments
2.2. RNA Extraction, cDNA Library Construction and Sequencing
2.3. Transcriptomic Data Processing and Bioinformatic Analysis
2.4. Real-Time Quantitative PCR (qRT-PCR) Analysis
2.5. Metabolite Extraction and Detection
2.6. Statistical Analysis
3. Results
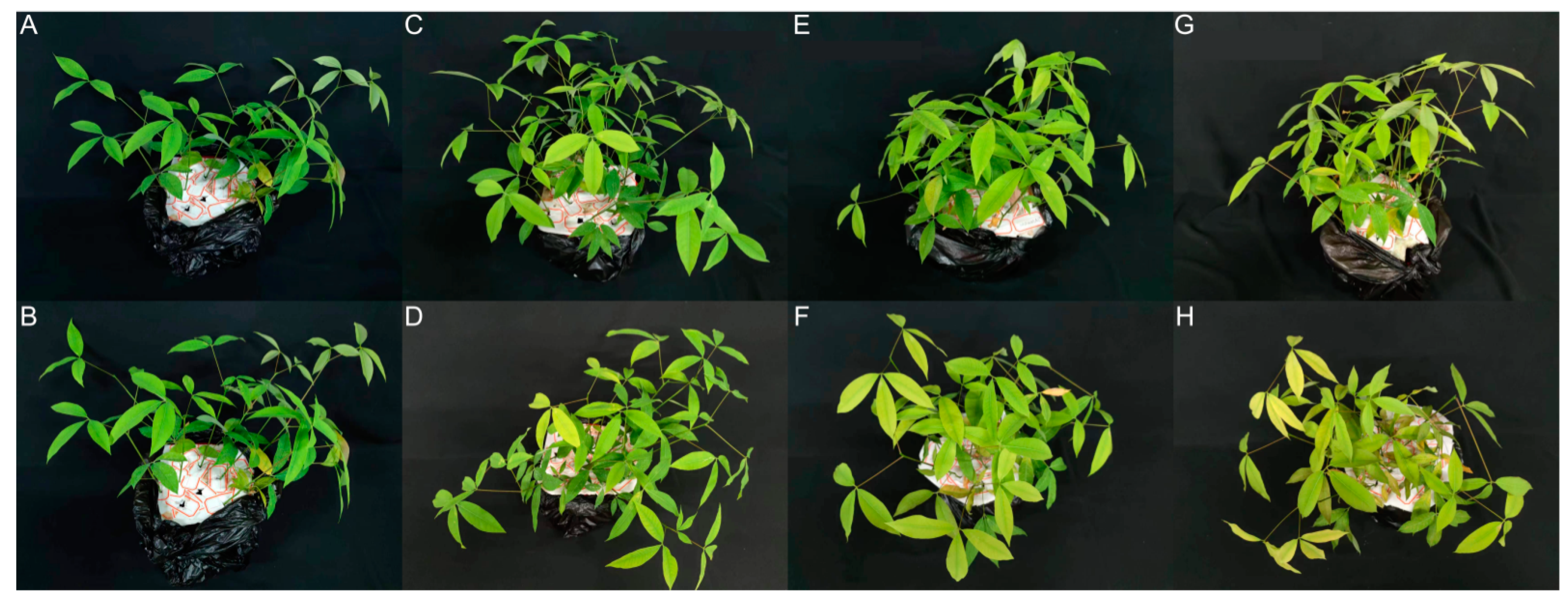
3.1. The Effects of Al Stress on Rubber Tree Saplings
3.2. Transcriptome Sequencing Data Analysis
3.3. Identification of Differentially Expressed Genes (DEGs) in Response to Al Stress
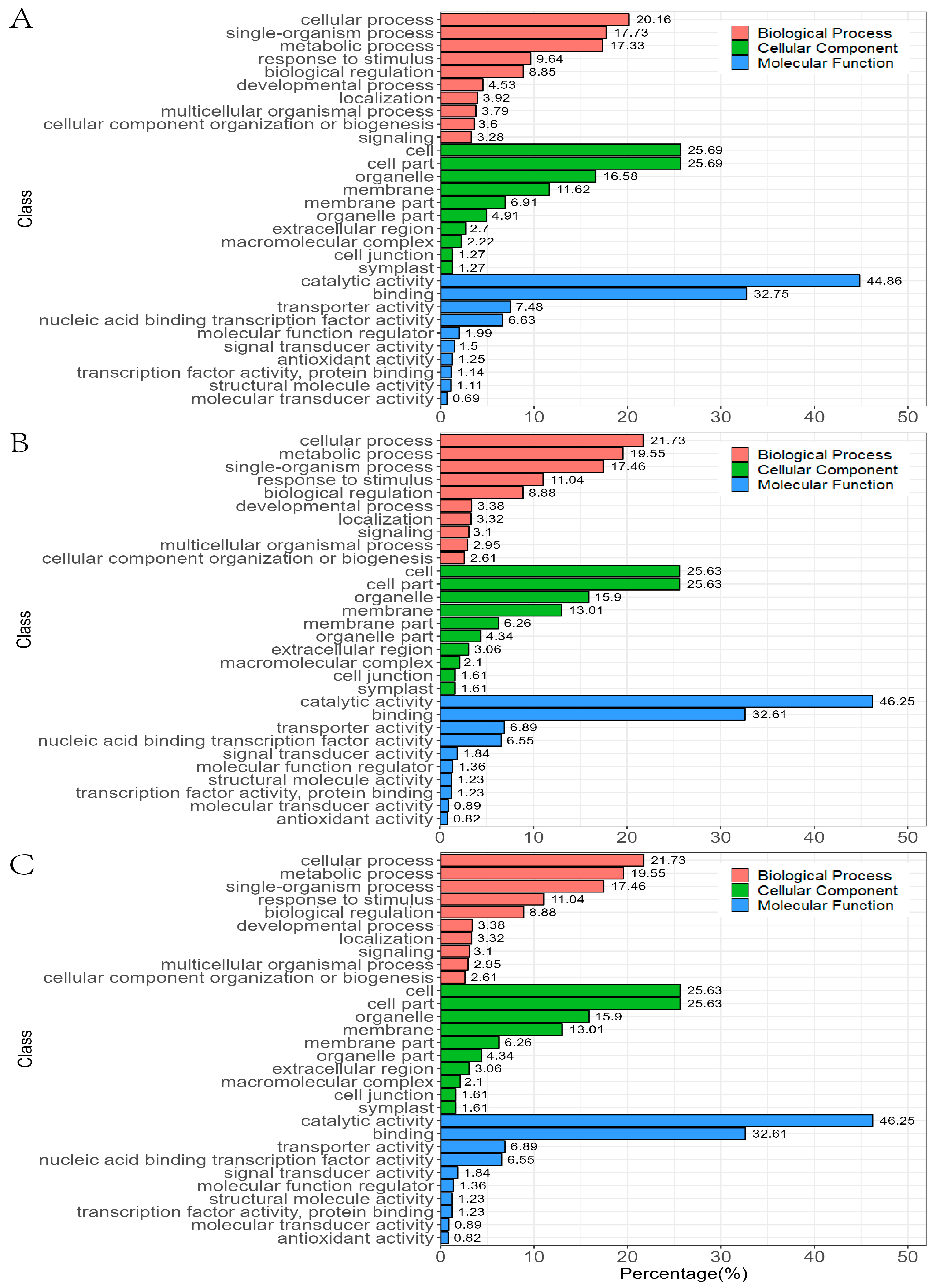
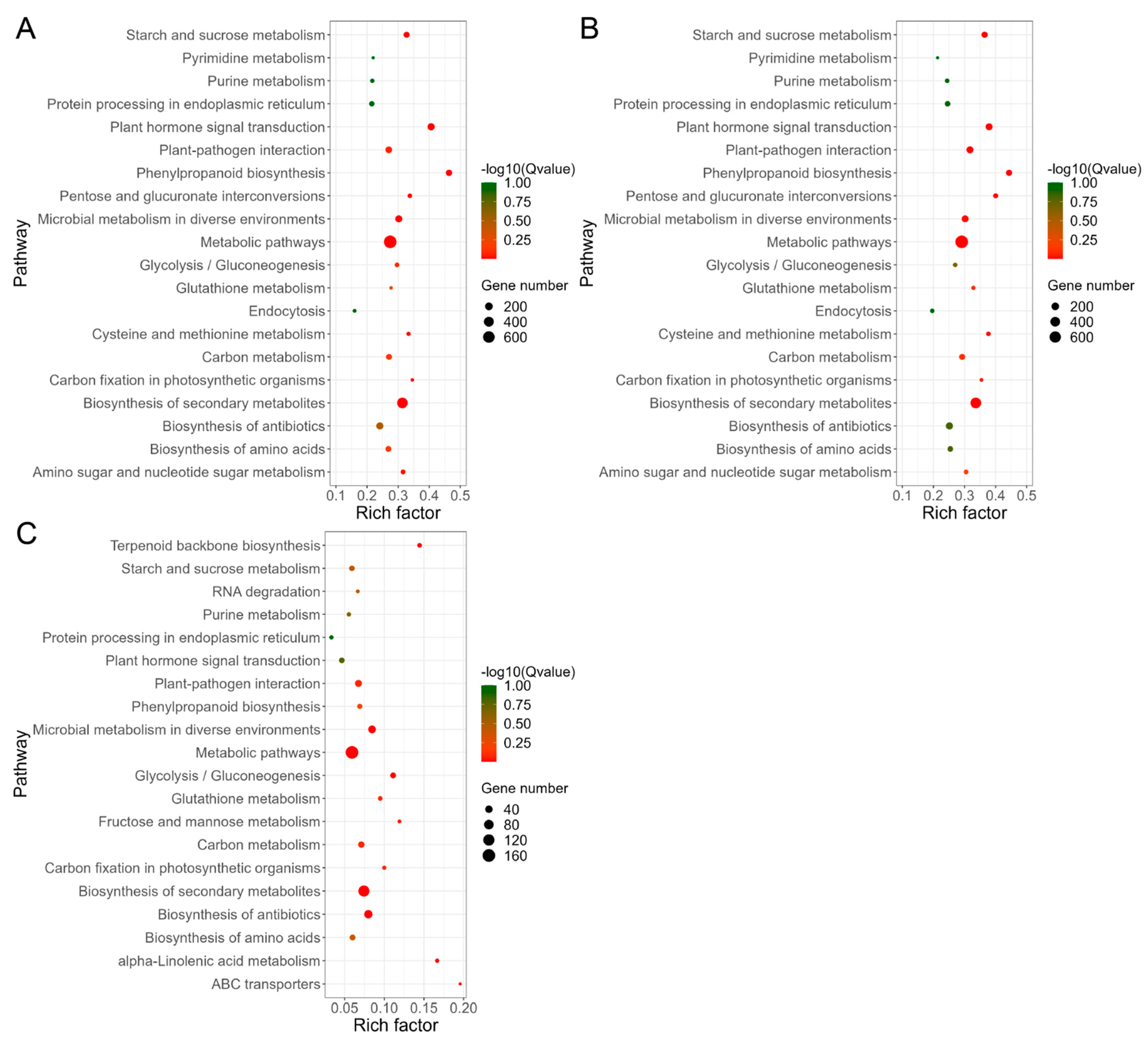
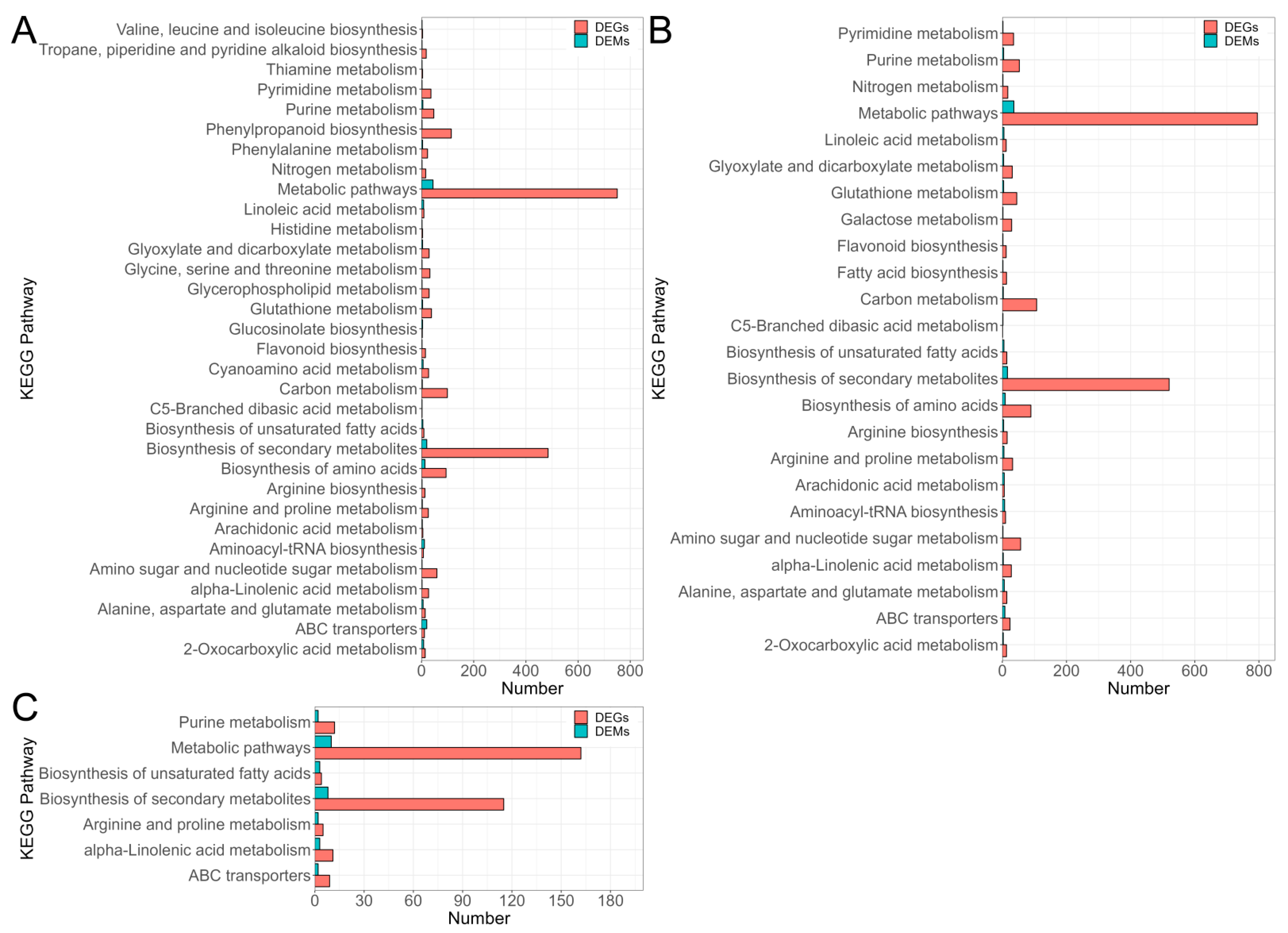
3.4. GO and KEGG Analysis of DEGs Response to Al Stress
3.5. Functional Analysis of DEGs Response to Al Stress
3.5.1. Transporter Genes
3.5.2. Transcription Factors
3.5.3. Genes Related to Cell Wall and Plasma Membrane Biosynthesis
3.5.4. Genes Associated with Oxidative Stress
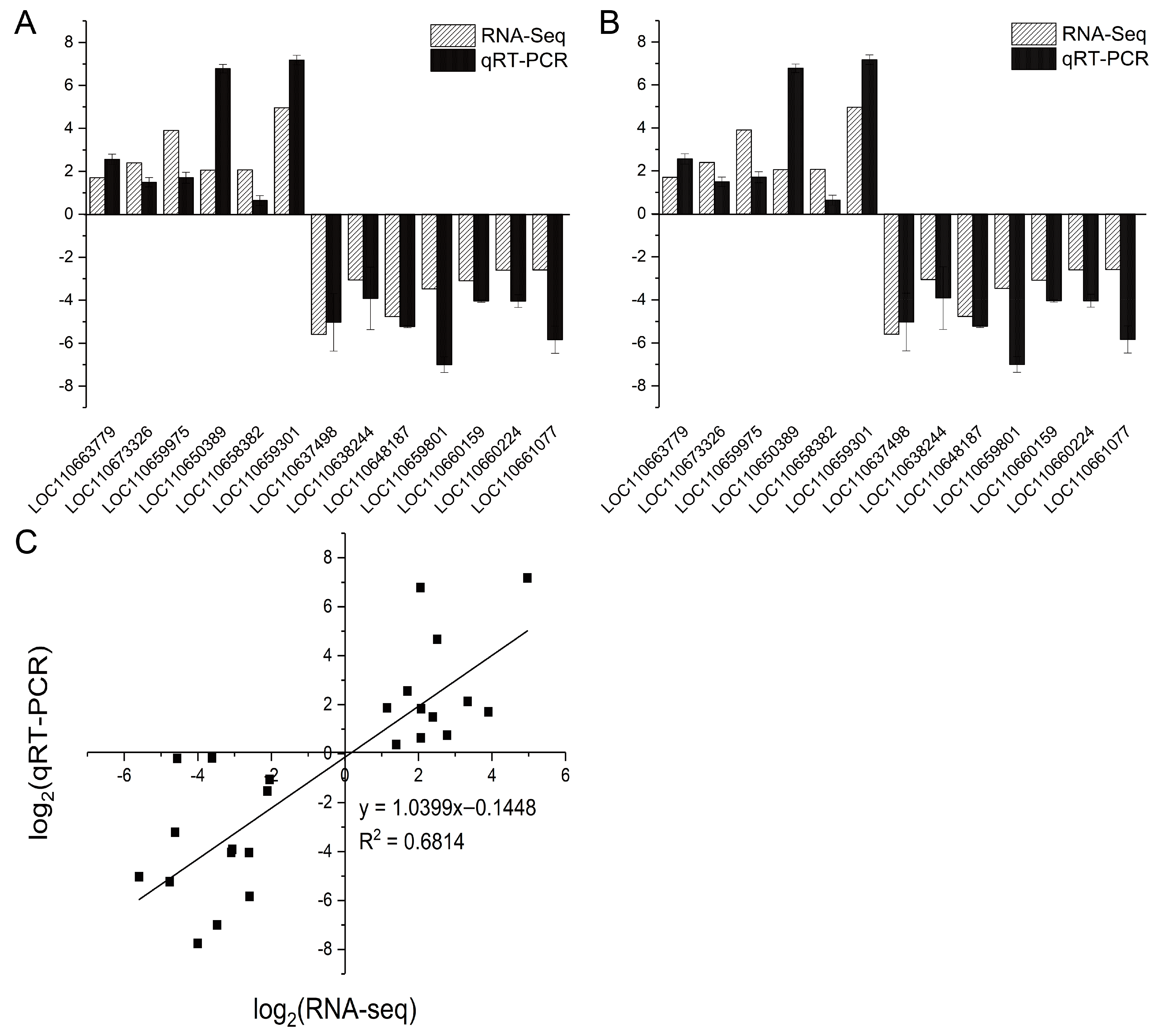
3.6. Validation of DEGs by qRT-PCR
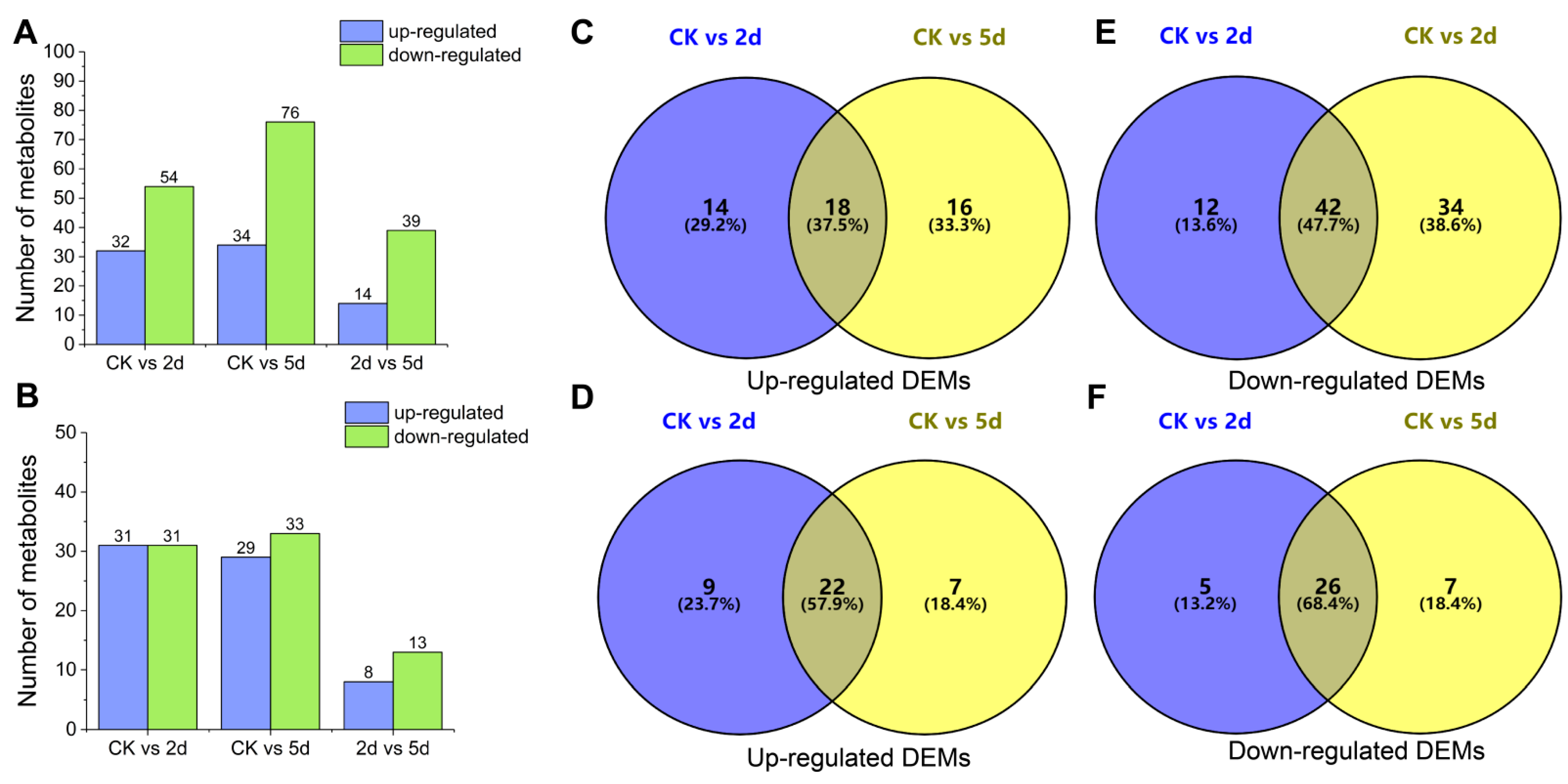
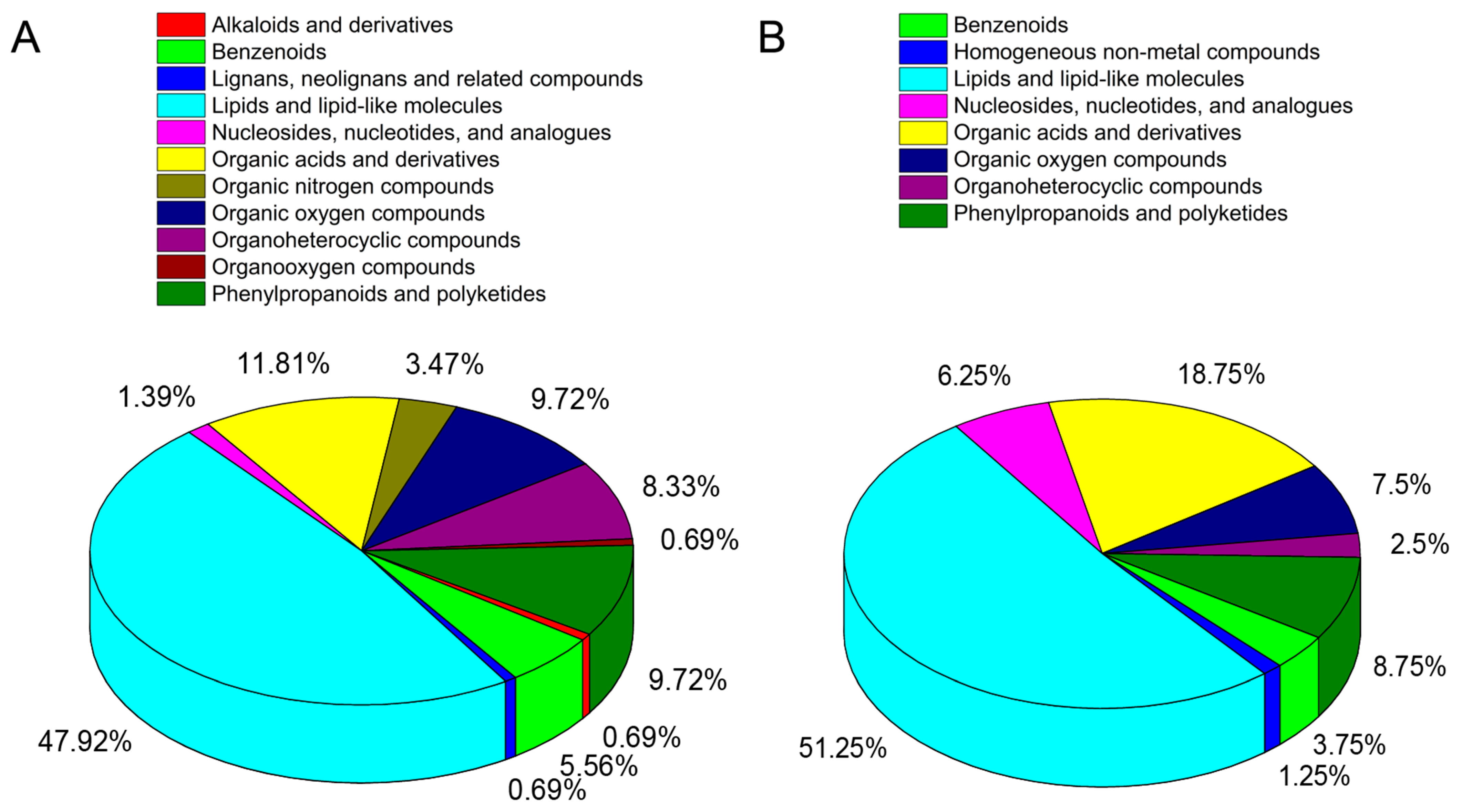
3.7. Metabolite Profiles of Rubber Tree Roots in Response to Al Stress
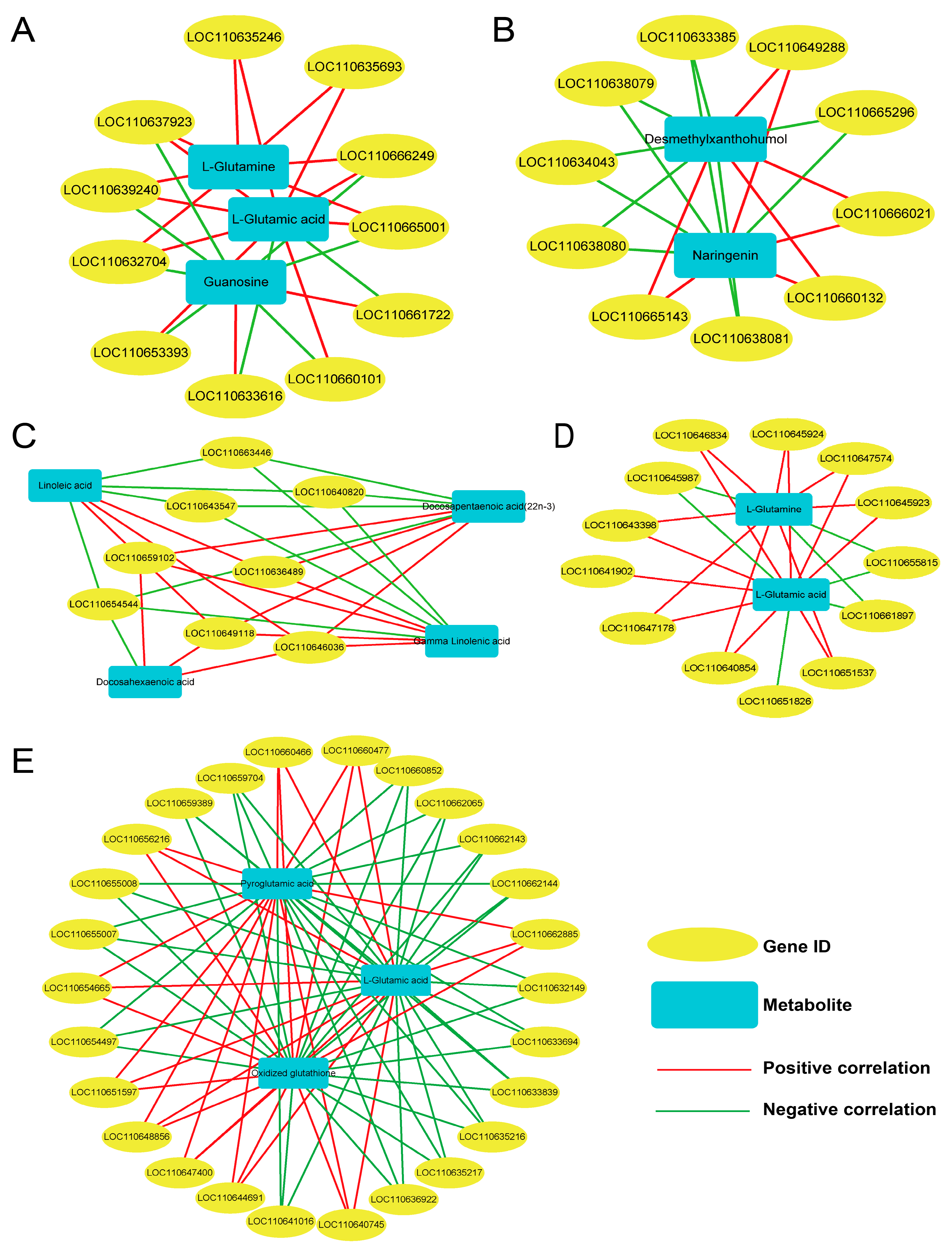
3.8. Combined Analysis of Transcriptome and Metabolome Data
4. Discussion
4.1. The Role of Organic Acids in the Al Tolerance of Rubber Trees
4.2. Accumulation of Secondary Metabolites Enhanced the Capacity of Al Tolerance in Rubber Trees
4.3. Transporter Genes Increase the Adaptation of Rubber Trees to Al Toxicity
4.4. The Cell Wall and Plasma Membrane Played a Crucial Role in Al the Tolerance of Rubber Trees
4.5. The Roles of Transcription Factor in Response to Al Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riaz, M.; Yan, L.; Wu, X.X.; Hussain, S.; Aziz, O.; Jiang, C.C. Mechanisms of organic acids and boron induced tolerance of aluminum toxicity: A review. Ecotoxicol. Environ. Saf. 2018, 165, 25–35. [Google Scholar] [CrossRef]
- Rahman, R.; Upadhyaya, H. Aluminium toxicity and its tolerance in plant: A Review. J. Plant Biol. 2021, 64, 101–121. [Google Scholar] [CrossRef]
- Ma, X.W.; An, F.; Wang, L.F.; Guo, D.; Xie, G.S.; Liu, Z.F. Genome-wide identification of aluminum-activated malate transporter (ALMT) gene family in rubber trees (Hevea brasiliensis) highlights their involvement in aluminum detoxification. Forests 2020, 11, 142. [Google Scholar] [CrossRef]
- Parra-Almuna, L.; Diaz-Cortez, A.; Ferrol, N.; Mora, M.L. Aluminium toxicity and phosphate deficiency activates antioxidant systems and up-regulates expression of phosphate transporters gene in ryegrass (Lolium perenne L.) plants. Plant Physiol. Biochem. 2018, 130, 445–454. [Google Scholar] [CrossRef]
- Liu, J.P.; Pineros, M.A.; Kochian, L.V. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 2014, 56, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F. Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 2007, 264, 225–252. [Google Scholar] [PubMed]
- Wang, Y.Q.; Yu, W.C.; Cao, Y.; Cai, Y.F.; Lyi, S.M.; Wu, W.W.; Kang, Y.; Liang, C.Y.; Liu, J.P. An exclusion mechanism is epistatic to an internal detoxification mechanism in aluminum resistance in Arabidopsis. BMC Plant Biol. 2020, 20, 122. [Google Scholar] [CrossRef]
- Famoso, A.N.; Clark, R.T.; Shaff, J.E.; Craft, E.; McCouch, S.R.; Kochian, L.V. Development of a novel aluminum tolerance phenotyping platform used for comparisons of cereal aluminum tolerance and investigations into rice aluminum tolerance mechanisms. Plant Physiol. 2010, 153, 1678–1691. [Google Scholar] [CrossRef]
- Yang, L.T.; Qi, Y.P.; Jiang, H.X.; Chen, L.S. Roles of organic acid anion secretion in aluminium tolerance of higher plants. BioMed Res. Int. 2013, 2013, 173682. [Google Scholar] [CrossRef]
- Grisel, N.; Zoller, S.; Kunzli-Gontarczyk, M.; Lampart, T.; Munsterkotter, M.; Brunner, I.; Bovet, L.; Metraux, J.P.; Sperisen, C. Transcriptome responses to aluminum stress in roots of aspen (Populus tremula). BMC Plant Biol. 2010, 10, 185. [Google Scholar] [CrossRef]
- Collins, N.C.; Shirley, N.J.; Saeed, M.; Pallotta, M.; Gustafson, J.P. An ALMT1 gene cluster controlling aluminum tolerance at the Alt4 locus of rye (Secale cereale L). Genetics 2008, 179, 669–682. [Google Scholar] [CrossRef]
- Delhaize, E.; Ryan, P.; Randall, P.J. Aluminum tolerance in wheat (Triticum aestivum L.) (II. aluminum-stimulated excretion of malic acid from root apices). Plant Physiol. 1993, 103, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Hoekenga, O.; Vision, T.; Shaff, J.; Monforte, A.; Lee, G.; Howell, S.; Kochian, L. Identification and characterization of aluminum tolerance loci in Arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping. a physiologically simple but genetically complex trait. Plant Physiol. 2003, 132, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Ligaba, A.; Katsuhara, M.; Ryan, P.R.; Shibasaka, M.; Matsumoto, H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006, 142, 1294–1303. [Google Scholar] [CrossRef]
- Liu, M.Y.; Chen, W.W.; Xu, J.M.; Fan, W.; Yang, J.L.; Zheng, S.J. The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J. Exp. Bot. 2013, 64, 1795–1804. [Google Scholar] [CrossRef]
- Magalhaes, J.V.; Liu, J.; Guimaraes, C.T.; Lana, U.G.; Alves, V.M.; Wang, Y.H.; Schaffert, R.E.; Hoekenga, O.A.; Pineros, M.A.; Shaff, J.E.; et al. A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat. Genet. 2007, 39, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Pellet, D.; Grunes, D.; Kochian, L. Organic acid exudation as an aluminum-tolerance mechanism in maize (Zea mays L.). Planta 1995, 196, 788–795. [Google Scholar] [CrossRef]
- Yang, Z.M.; Sivaguru, M.; Horst, W.J.; Matsumoto, H. Aluminium tolerance is achieved by exudation of citric acid from roots of soybean (Glycine max). Physiol. Plant. 2010, 110, 72–77. [Google Scholar] [CrossRef]
- Jian, L.Y.; Xiao, F.Z.; You, X.P.; Cheng, Z.; Feng, M.; Shao, J.Z. Aluminum regulates oxalate secretion and plasma membrane H+-ATPase activity independently in tomato roots. Planta 2011, 234, 281–291. [Google Scholar]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of al in hydrangea (identification of Al form in the leaves). Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef]
- Yang, J.L.; Zheng, S.J.; He, Y.F.; Hideaki, M. Aluminium resistance requires resistance to acid stress: A case study with spinach that exudes oxalate rapidly when exposed to Al stress. J. Exp. Bot. 2005, 56, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.J.; Ma, J.F.; Matsumoto, H. High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998, 117, 745–751. [Google Scholar] [CrossRef]
- Iuchi, S.; Koyama, H.; Iuchi, A.; Kobayashi, Y.; Kitabayashi, S.; Kobayashi, Y.; Ikka, T.; Hirayama, T.; Shinozaki, K.; Kobayashi, M. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 9900–9905. [Google Scholar] [CrossRef]
- Yamaji, N.; Huang, C.F.; Nagao, S.; Yano, M.; Sato, Y.; Nagamura, Y.; Ma, J.F. A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 2009, 21, 3339–3349. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Ma, J.F.; Ryan, P.R. Transcriptional regulation of aluminium tolerance genes. Trends Plant Sci. 2012, 17, 341–348. [Google Scholar] [CrossRef]
- Sawaki, Y.; Iuchi, S.; Kobayashi, Y.; Kobayashi, Y.; Ikka, T.; Sakurai, N.; Fujita, M.; Shinozaki, K.; Shibata, D.; Kobayashi, M.; et al. STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol. 2009, 150, 281–294. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Liu, J.; Dong, D.; Jia, X.; McCouch, S.R.; Kochian, L.V. Natural variation underlies alterations in NRAMP aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. USA 2014, 111, 6503–6508. [Google Scholar] [CrossRef]
- Huang, C.F.; Yamaji, N.; Chen, Z.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. Cell Mol. Biol. 2012, 69, 857–867. [Google Scholar] [CrossRef]
- Guo, P.; Qi, Y.P.; Yang, L.T.; Lai, N.W.; Ye, X.; Yang, Y.; Chen, L.S. Root adaptive responses to aluminum-treatment revealed by RNA-seq in two citrus species with different aluminum-tolerance. Front Plant Sci. 2017, 8, 330. [Google Scholar] [CrossRef]
- Liu, W.X.; Xiong, C.H.; Yan, L.F.; Zhang, Z.S.; Ma, L.C.; Wang, Y.R.; Liu, Y.J.; Liu, Z.P. Transcriptome analyses reveal candidate genes potentially involved in al stress response in alfalfa. Front Plant Sci. 2017, 8, 26. [Google Scholar] [CrossRef]
- Arenhart, R.A.; Bai, Y.; de Oliveira, L.F.; Neto, L.B.; Schunemann, M.; Maraschin Fdos, S.; Mariath, J.; Silverio, A.; Sachetto-Martins, G.; Margis, R.; et al. New insights into aluminum tolerance in rice: The ASR5 protein binds the STAR1 promoter and other aluminum-responsive genes. Mol. Plant 2014, 7, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.M.; Fan, W.; Jin, J.F.; Lou, H.Q.; Chen, W.W.; Yang, J.L.; Zheng, S.J. Transcriptome analysis of al-induced genes in buckwheat (Fagopyrum esculentum Moench) root apex: New insight into al toxicity and resistance mechanisms in an al accumulating species. Front Plant Sci. 2017, 8, 1141. [Google Scholar] [CrossRef]
- Mattiello, L.; Begcy, K.; da Silva, F.R.; Jorge, R.A.; Menossi, M. Transcriptome analysis highlights changes in the leaves of maize plants cultivated in acidic soil containing toxic levels of Al3+. Mol. Biol. Rep. 2014, 41, 8107–8116. [Google Scholar] [CrossRef]
- Gould, B.; McCouch, S.; Geber, M. De novo transcriptome assembly and identification of gene candidates for rapid evolution of soil al tolerance in Anthoxanthum Odoratum at the long-term park grass experiment. PLoS ONE 2015, 10, e0124424. [Google Scholar] [CrossRef]
- Chen, H.X.; Lu, C.P.; Jiang, H.; Peng, J.H. Global transcriptome analysis reveals distinct aluminum-tolerance pathways in the al-accumulating species Hydrangea Macrophylla and marker identification. PLoS ONE 2015, 10, e0144927. [Google Scholar] [CrossRef]
- Chen, L.; Wang, T.Z.; Zhao, M.G.; Tian, Q.Y.; Zhang, W.H. Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 2012, 235, 375–386. [Google Scholar] [CrossRef]
- Xie, L.H.; Li, H.J.; Zhong, Z.Z.; Guo, J.J.; Hu, G.C.; Gao, Y.; Tong, Z.H.; Liu, M.L.; Hu, S.P.; Tong, H.H.; et al. Metabolome analysis under aluminum toxicity between aluminum-tolerant and -sensitive rice (Oryza sativa L.). Plants 2022, 11, 1717. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.S.; Zhang, J.; Huang, W.L.; Yang, L.T.; Huang, Z.R.; Guo, J.; Wu, J.; Chen, L.S. Molecular mechanisms for pH-mediated amelioration of aluminum-toxicity revealed by conjoint analysis of transcriptome and metabolome in Citrus sinensis roots. Chemosphere 2022, 299, 134335. [Google Scholar] [CrossRef]
- Wang, Q.M.; Xu, Y.F.; Zhang, M.; Zhu, F.D.; Sun, M.X.; Lian, X.Y.; Zhao, G.F.; Duan, D. Transcriptome and metabolome analysis of stress tolerance to aluminium in Vitis quinquangularis. Planta 2021, 254, 105. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Peng, S.Q.; Huang, G.X.; Wu, K.X.; Fu, X.H.; Chen, Z.Q. Association of decreased expression of a Myb transcription factor with the TPD (tapping panel dryness) syndrome in Hevea brasiliensis. Plant Mol. Biol. 2003, 51, 51–58. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, G.L.; Zhao, Y.G.; Zhao, W.J.; Qi, Z.P. Chemical degradation of a Ferralsol (Oxisol) under intensive rubber (Hevea brasiliensis) farming in tropical China. Soil Tillage Res. 2007, 93, 109–116. [Google Scholar] [CrossRef]
- An, F.; Li, C.Z.; Zhang, T.T.; Wang, L.F.; Wang, J.K.; Xie, G.S. Effects of aluminum toxicity on physiological and leaf chlorophyll fluorescent characteristics of rubber tree seedlings. Ying Yong Sheng Tai Xue Bao 2018, 29, 4191–4198. [Google Scholar]
- Ma, X.W.; Liu, Z.F.; Liu, Z.; Xie, G.S.; Rookes, J.; An, F. Root secretion of oxalic and malic acids mitigates the rubber tree aluminum toxicity. J. Rubber Res. 2021, 24, 381–390. [Google Scholar] [CrossRef]
- Huang, Y.M.; Li, D.F.; Zhao, L.N.; Chen, A.G.; Li, J.J.; Tang, H.J.; Pan, G.; Chang, L.; Deng, Y.; Huang, S.Q. Comparative transcriptome combined with physiological analyses revealed key factors for differential cadmium tolerance in two contrasting hemp (Cannabis sativa L.) cultivars. Ind. Crop. Prod. 2019, 140, 111638. [Google Scholar] [CrossRef]
- Tang, C.R.; Yang, M.; Fang, Y.J.; Luo, Y.F.; Gao, S.H.; Xiao, X.H.; An, Z.W.; Zhou, B.H.; Zhang, B.; Tan, X.Y.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Lv, L.T.; Zeng, X.F.; Zhang, F.; Chen, Y.L.; Tian, W.L.; Li, J.R.; Li, X.Y.; Li, Y. Integrative analysis of metabolomics and transcriptomics reveals molecular mechanisms of anthocyanin metabolism in the zikui tea plant (Camellia sinensis cv. Zikui). Int. J. Mol. Sci. 2022, 23, 4780. [Google Scholar] [CrossRef]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.W.; An, F.; Liu, Z.F.; Xie, G.S. Screening of reference genes for quantitative real-time PCR of rubber saplings under aluminum stress. Chin. J. Trop. Crop. 2020, 41, 955–963. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Doppler, M.; Kluger, B.; Bueschl, C.; Schneider, C.; Krska, R.; Delcambre, S.; Hiller, K.; Lemmens, M.; Schuhmacher, R. Stable isotope-assisted evaluation of different extraction solvents for untargeted metabolomics of plants. Int. J. Mol. Sci. 2016, 17, 1017. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Bottcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef]
- Hu, Z.C.; Fu, Q.S.; Zheng, J.; Zhang, A.A.; Wang, H.S. Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Hortic. Res. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.H.; Lin, S.Z. Transcriptomic revelation of phenolic compounds involved in aluminum toxicity responses in roots of Cunninghamia lanceolata (Lamb.) Hook. Genes 2019, 10, 835. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.T.; Li, D.Q.; Zhu, J.J.; Shu, Z.F.; Ye, X.L.; Xing, A.Q.; Wen, B.; Ma, Y.C.; Zhu, X.J.; Fang, W.P.; et al. Aluminum relieves fluoride stress through stimulation of organic acid production in Camellia sinensis. Physiol. Mol. Biol. Plants 2020, 26, 1127–1137. [Google Scholar] [CrossRef]
- Tran, N.N.; Kazuo, N.; Julian, T.; Kounosuke, F. Role of exudation of organic acids and phosphate in aluminum tolerance of four tropical woody species. Tree Physiol. 2003, 23, 1041–1050. [Google Scholar]
- Sharma, T.; Dreyer, I.; Kochian, L.; Pineros, M.A. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front. Plant Sci. 2016, 7, 1488. [Google Scholar] [CrossRef]
- Chen, Y.; Huang, L.; Liang, X.; Dai, P.B.; Zhang, Y.X.; Li, B.H.; Lin, X.Y.; Sun, C.L. Enhancement of polyphenolic metabolism as an adaptive response of lettuce (Lactuca sativa) roots to aluminum stress. Environ. Pollut. 2020, 261, 114230. [Google Scholar] [CrossRef]
- Cong, F.; Diehl, B.G.; Hill, J.L.; Brown, N.R.; Tien, M. Covalent bond formation between amino acids and lignin: Cross-coupling between proteins and lignin. Phytochemistry 2013, 96, 449–456. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef] [PubMed]
- Su, L.t.; Lv, A.M.; Wen, W.W.; Fan, N.N.; Li, J.J.; Gao, L.; Zhou, P.; An, Y. MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis. Plant J. Cell Mol. Biol. 2022, 112, 756–771. [Google Scholar] [CrossRef]
- Kar, D.; Pradhan, A.A.; Datta, S. The role of solute transporters in aluminum toxicity and tolerance. Physiol. Plant 2021, 171, 638–652. [Google Scholar] [CrossRef] [PubMed]
- Negishi, T.; Oshima, K.; Hattori, M.; Kanai, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast and plasma membrane-localized aquaporin-family transporters in blue hydrangea sepals of aluminum hyperaccumulating plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.L.; Yao, S.C.; Xiong, W.J.; Luo, S.Z.; Wang, Y.L.; Wang, A.Q.; Xiao, D.; Zhan, J.; He, L.F. Nitric oxide inhibits Al-induced programmed cell death in root tips of peanut (Arachis hypogaea L.) by affecting physiological properties of antioxidants systems and cell wall. Front. Physiol. 2017, 8, 1037. [Google Scholar] [CrossRef]
- Wang, S.Y.; Yuan, S.L.; Su, L.T.; Lv, A.M.; Zhou, P.; An, Y. Aluminum toxicity in alfalfa (Medicago sativa) is alleviated by exogenous foliar IAA inducing reduction of Al accumulation in cell wall. Environ. Exp. Bot. 2017, 139, 1–13. [Google Scholar] [CrossRef]
- Yang, J.L.; Li, Y.Y.; Zhang, Y.J.; Zhang, S.S.; Wu, Y.R.; Wu, P.; Zheng, S.J. Cell wall polysaccharides are specifically involved in the exclusion of aluminum from the rice root apex. Plant Physiol. 2008, 146, 602–611. [Google Scholar] [CrossRef]
- Chandran, D.; Sharopova, N.; Ivashuta, S.; Gantt, J.S.; Vandenbosch, K.A.; Samac, D.A. Transcriptome profiling identified novel genes associated with aluminum toxicity, resistance and tolerance in Medicago truncatula. Planta 2008, 228, 151–166. [Google Scholar] [CrossRef]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef]
- Zhang, M.J.; Deng, X.P.; Yin, L.N.; Qi, L.Y.; Wang, X.Y.; Wang, S.W.; Li, H.B. Regulation of galactolipid biosynthesis by overexpression of the rice mgd gene contributes to enhanced aluminum tolerance in tobacco. Front. Plant Sci. 2016, 7, 337. [Google Scholar] [CrossRef]
- Maejima, E.; Watanabe, T. Proportion of phospholipids in the plasma membrane is an important factor in Al tolerance. Plant Signal. Behav. 2014, 9, e29277. [Google Scholar] [CrossRef]
- Maejima, E.; Watanabe, T.; Osaki, M.; Wagatsuma, T. Phosphorus deficiency enhances aluminum tolerance of rice (Oryza sativa) by changing the physicochemical characteristics of root plasma membranes and cell walls. J. Plant Physiol. 2014, 171, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.M.; Liu, C.; Cui, B.M.; Wang, N.; Zhao, Z.; Zhou, L.N.; Huang, K.F.; Ding, J.Z.; Du, H.M.; Jiang, W.; et al. Transcriptomic responses to aluminum (Al) stress in maize. J. Integr. Agr. 2018, 17, 1946–1958. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Xu, X.Y.; Li, G.X.; Zheng, S.J. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. Cell Mol. Biol. 2013, 76, 825–835. [Google Scholar] [CrossRef]
- Li, C.N.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Gao, L.J.; Liu, X.P.; Gao, K.K.; Cui, M.Q.; Zhu, H.H.; Li, G.X.; Yan, J.Y.; Wu, Y.R.; Ding, Z.J.; Chen, X.W.; et al. ART1 and putrescine contribute to rice aluminum resistance via OsMYB30 in cell wall modification. J. Integr. Plant Biol. 2023, 1–16. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Cheng, L.; Peng, W.; Xie, G.; Liu, Z.; Yang, Z.; Wang, Y.; An, F. Comparative Transcriptome and Metabolome Analysis of Rubber Trees (Hevea brasiliensis Muell. Arg.) Response to Aluminum Stress. Forests 2023, 14, 568. https://doi.org/10.3390/f14030568
Ma X, Cheng L, Peng W, Xie G, Liu Z, Yang Z, Wang Y, An F. Comparative Transcriptome and Metabolome Analysis of Rubber Trees (Hevea brasiliensis Muell. Arg.) Response to Aluminum Stress. Forests. 2023; 14(3):568. https://doi.org/10.3390/f14030568
Chicago/Turabian StyleMa, Xiaowei, Linlin Cheng, Wentao Peng, Guishui Xie, Zifan Liu, Zongming Yang, Ying Wang, and Feng An. 2023. "Comparative Transcriptome and Metabolome Analysis of Rubber Trees (Hevea brasiliensis Muell. Arg.) Response to Aluminum Stress" Forests 14, no. 3: 568. https://doi.org/10.3390/f14030568
APA StyleMa, X., Cheng, L., Peng, W., Xie, G., Liu, Z., Yang, Z., Wang, Y., & An, F. (2023). Comparative Transcriptome and Metabolome Analysis of Rubber Trees (Hevea brasiliensis Muell. Arg.) Response to Aluminum Stress. Forests, 14(3), 568. https://doi.org/10.3390/f14030568






