Characterization of 33 HbbZIP Gene Family Members and Analysis of Their Expression Profiles in Rubber Tree in Response to ABA, Glyphosate and Powdery Mildew Treatment
Abstract
1. Introduction
2. Methods
2.1. Plant Material
2.2. Gene Sequence Acquisition
2.3. Structural and Functional Analyses
2.4. Sequence Alignment and Phylogenetic Analysis
2.5. Expression Analysis of 33 HbbZIPs via qRT-PCR
3. Results
3.1. Identification and Analysis of bZIP Gene Family Members in Rubber Tree
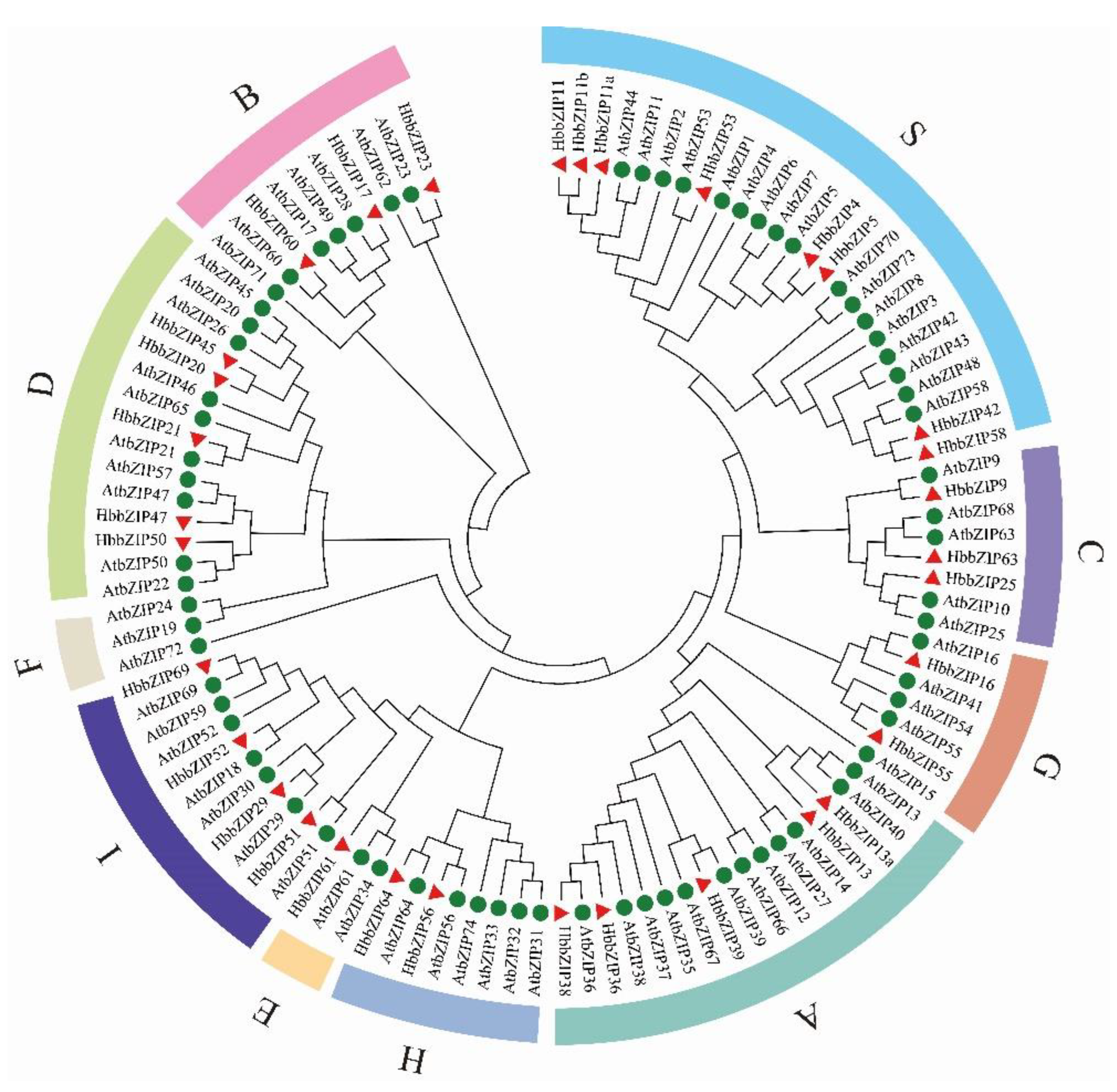
3.2. Phylogenetic Tree of 33 HbbZIP Family Genes and AtbZIP Family Genes
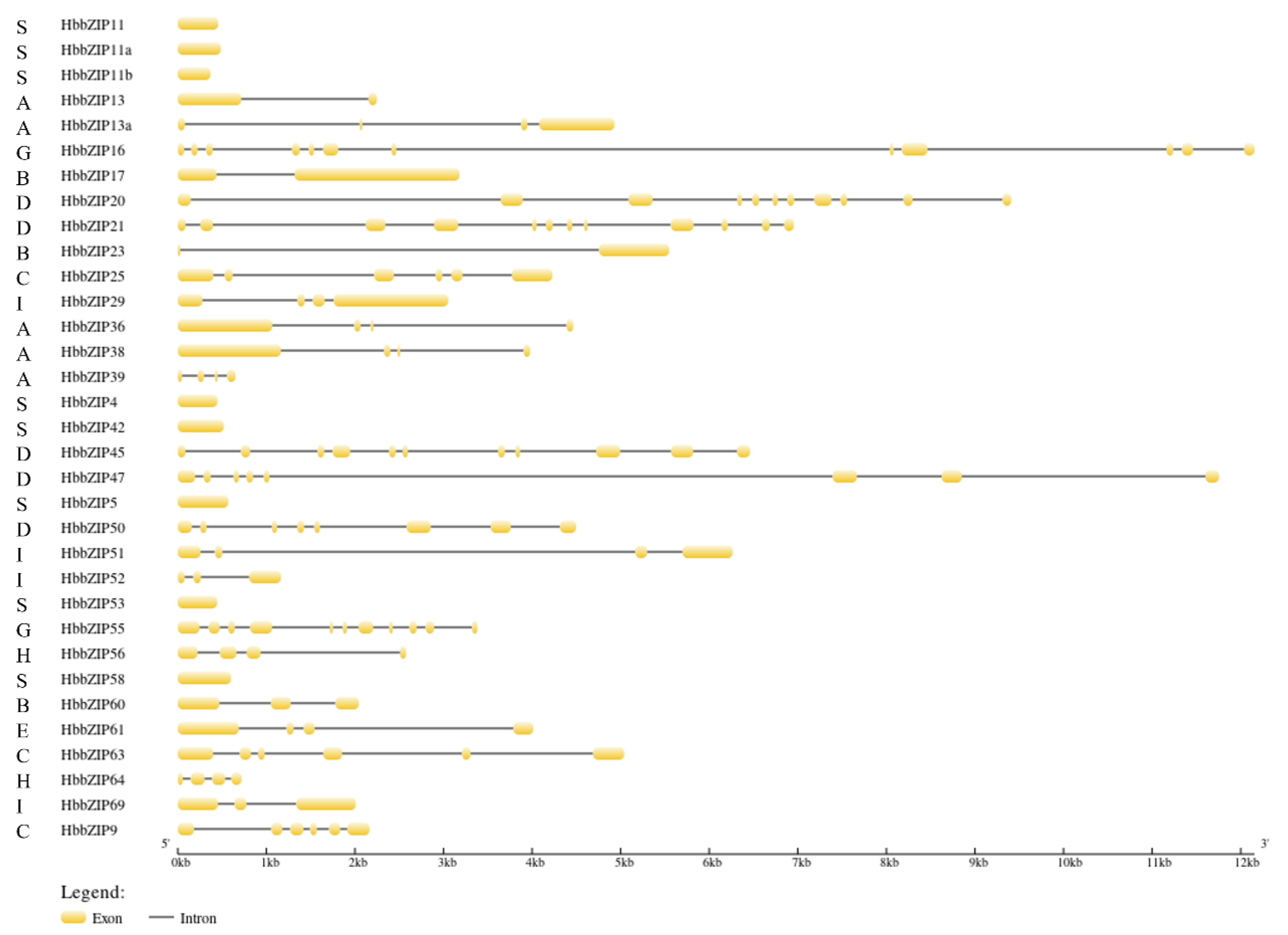
3.3. Analysis of the Gene Structure and Function of the bZIP Family Members in Rubber Tree
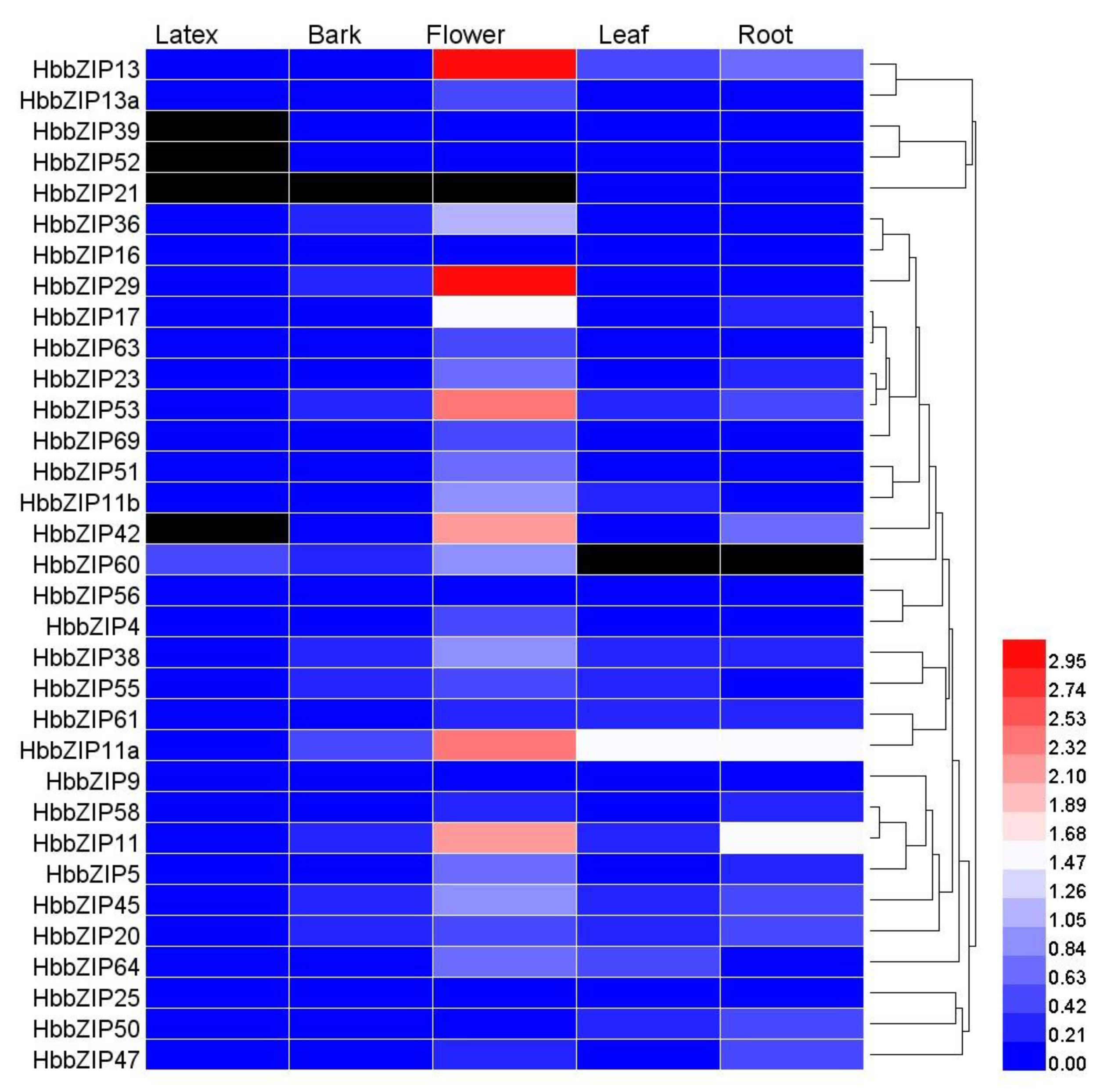
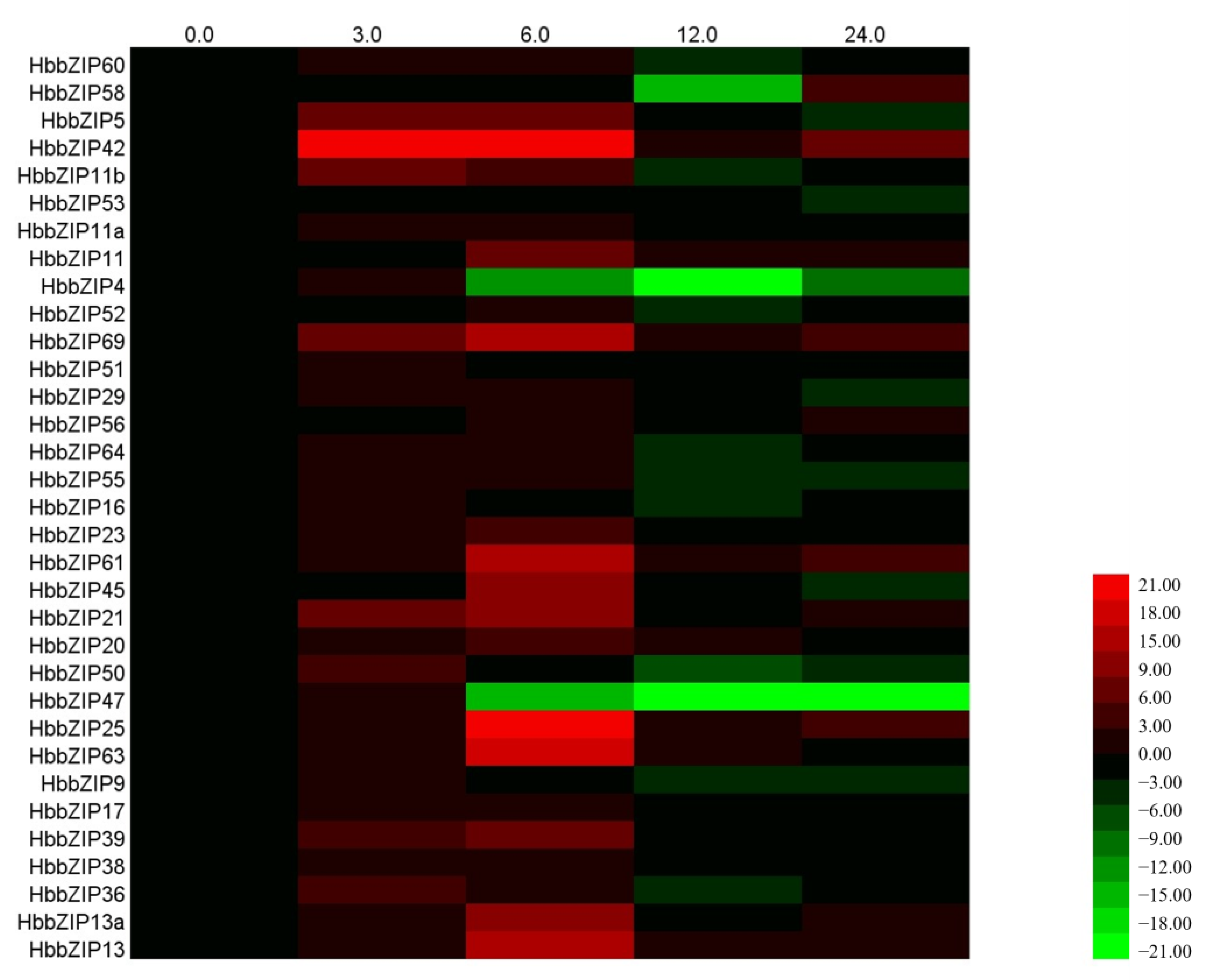
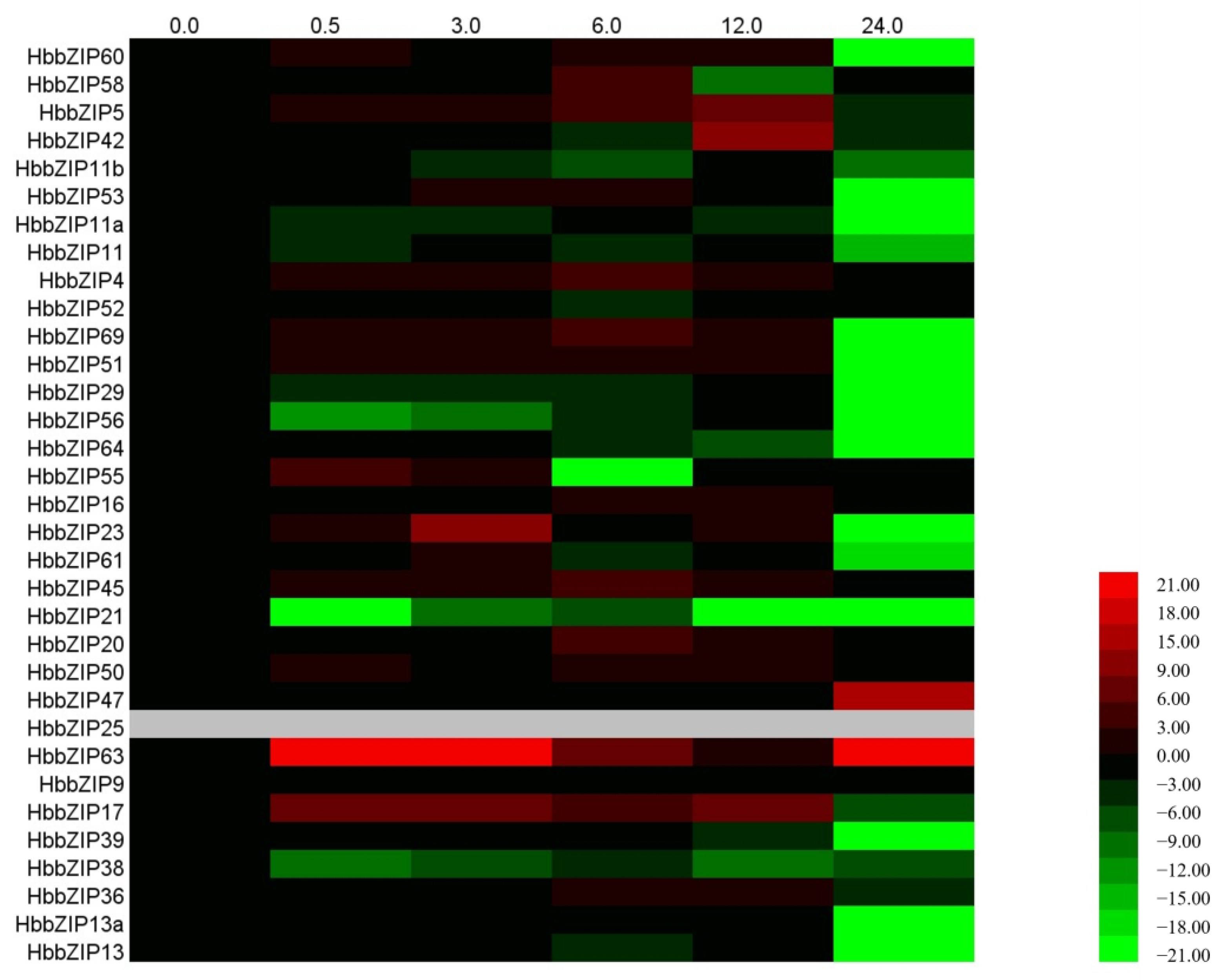
3.4. Analysis of HbbZIP Expression in Different Tissues in Response to ABA, Powdery Mildew and Glyphosate Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bai, Y.; Zhu, W.; Hu, X.; Sun, C.; Li, Y.; Wang, D.; Wang, Q.; Pei, G.; Zhang, Y.; Guo, A.; et al. Genome-Wide Analysis of the bZIP Gene Family Identifies Two ABI5-Like bZIP Transcription Factors, BrABI5a and BrABI5b, as Positive Modulators of ABA Signalling in Chinese Cabbage. PLoS ONE 2016, 11, e0158966. [Google Scholar] [CrossRef]
- Baloglu, M.C.; Eldem, V.; Hajyzadeh, M.; Unver, T. Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 2014, 9, e96014. [Google Scholar] [CrossRef]
- Correa, L.G.; Riano-Pachon, D.M.; Schrago, C.G.; dos Santos, R.V.; Mueller-Roeber, B.; Vincentz, M. The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 2008, 3, e2944. [Google Scholar] [CrossRef]
- Dai, S.; Zhang, Z.; Chen, S.; Beachy, R.N. RF2b, a rice bZIP transcription activator, interacts with RF2a and is involved in symptom development of rice tungro disease. Proc. Natl. Acad. Sci. USA 2004, 101, 687–692. [Google Scholar] [CrossRef]
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237. [Google Scholar] [CrossRef] [PubMed]
- Firdous, S.; Iqbal, S.; Anwar, S.; Jabeen, H. Identification and analysis of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene from glyphosate-resistant Ochrobactrum intermedium Sq20. Pest Manag. Sci. 2018, 74, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Xu, S.; Lu, Z.; Su, L.; Zhang, L.; Sun, W.; Zhang, Y.; Jiang, C.; Liu, Z.; Duan, L.; et al. Genomic characterization of bZIP transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Int. J. Biol. Macromol. 2022, 223, 1619–1631. [Google Scholar] [CrossRef]
- Heinekamp, T.; Strathmann, A.; Kuhlmann, M.; Froissard, M.; Muller, A.; Perrot-Rechenmann, C.; Droge-Laser, W. The tobacco bZIP transcription factor BZI-1 binds the GH3 promoter in vivo and modulates auxin-induced transcription. Plant J. 2004, 38, 298–309. [Google Scholar] [CrossRef]
- Hu, W.; Wang, L.; Tie, W.; Yan, Y.; Ding, Z.; Liu, J.; Li, M.; Peng, M.; Xu, B.; Jin, Z. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci. Rep. 2016, 6, 30203. [Google Scholar] [CrossRef]
- Hu, W.; Yang, H.; Yan, Y.; Wei, Y.; Tie, W.; Ding, Z.; Zuo, J.; Peng, M.; Li, K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci. Rep. 2016, 6, 22783. [Google Scholar] [CrossRef] [PubMed]
- Jakoby, M.; Weisshaar, B.; Droge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Xu, W.; Liu, A. Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 2014, 239, 299–312. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; He, Q.; Li, S.; Liu, W.; Lin, C.; Miao, W. A Candidate Secreted Effector Protein of Rubber Tree Powdery Mildew Fungus Contributes to Infection by Regulating Plant ABA Biosynthesis. Front. Microbiol. 2020, 11, 591387. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Lim, S.; Han, S.W.; Lee, S.C. Expression and Functional Roles of the Pepper Pathogen-Induced bZIP Transcription Factor CabZIP2 in Enhanced Disease Resistance to Bacterial Pathogen Infection. Mol. Plant Microbe Interact. 2015, 28, 825–833. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.M. Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef]
- Liu, J.; Shu, D.; Tan, Z.; Ma, M.; Guo, N.; Gao, S.; Duan, G.; Kuai, B.; Hu, Y.; Li, S.; et al. The Arabidopsis IDD14 transcription factor interacts with bZIP-type ABFs/AREBs and cooperatively regulates ABA-mediated drought tolerance. New Phytol. 2022, 236, 929–942. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Nazri, A.Z.; Griffin, J.H.C.; Peaston, K.A.; Alexander-Webber, D.G.A.; Williams, L.E. F-group bZIPs in barley-a role in Zn deficiency. Plant Cell Environ. 2017, 40, 2754–2770. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Kohls, S.; Landsberg, E.; Stock, K.; Rmheld, V. Relevance of glyphosate transfer to non-target plants via the rhizosphere. Qual. Quant. 2006, 20, 330–350. [Google Scholar]
- Newman, M.M.; Hoilett, N.; Lorenz, N.; Dick, R.P.; Liles, M.R.; Ramsier, C.; Kloepper, J.W. Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci. Total Environ. 2016, 543, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 2008, 146, 333–350. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Pontes, L.C.G.; Cardoso, C.M.Y.; Callegari, D.M.; Dos Reis, S.P.; do Socorro Alves Namias, É.; da Cunha Ferreira, S.; de Souza, C.R.B. A Cassava CPRF-2-like bZIP Transcription Factor Showed Increased Transcript Levels during Light Treatment. Protein Pept. Lett. 2020, 27, 904–914. [Google Scholar] [CrossRef]
- Qin, B.; Wang, M.; He, H.X.; Xiao, H.X.; Zhang, Y.; Wang, L.F. Identification and Characterization of a Potential Candidate Mlo Gene Conferring Susceptibility to Powdery Mildew in Rubber Tree. Phytopathology 2019, 109, 1236–1245. [Google Scholar] [CrossRef]
- Qin, B.; Zheng, F.; Zhang, Y. Molecular cloning and characterization of a Mlo gene in rubber tree (Hevea brasiliensis). J. Plant Physiol. 2015, 175, 78–85. [Google Scholar] [CrossRef]
- Sant’Anna, I.C.; Gouvea, L.R.L.; Martins, M.A.; Scaloppi Junior, E.J.; de Freitas, R.S.; Goncalves, P.S. Genetic diversity associated with natural rubber quality in elite genotypes of the rubber tree. Sci. Rep. 2021, 11, 1081. [Google Scholar] [CrossRef]
- Santos, J.O.D.; de Oliveira, L.E.M.; de Souza, T.; Lopes, G.M.; Coelho, V.T.; Gomes, M.P. Physiological mechanisms responsible for tolerance to, and recuperation from, drought conditions in four different rubber clones. Ind. Crops Prod. 2019, 141, 111714. [Google Scholar] [CrossRef]
- Shaw, D.E. Powdery mildew of rubber in Papua. Papua New Guinea. Agric. J. 1967, 19, 140–146. [Google Scholar]
- Sornaraj, P.; Luang, S.; Lopato, S.; Hrmova, M. Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta 2016, 1860, 46–56. [Google Scholar] [CrossRef]
- Spaunhorst, D.J.; Nie, H.; Todd, J.R.; Young, J.M.; Young, B.G.; Johnson, W.G. Confirmation of herbicide resistance mutations Trp574Leu, DeltaG210, and EPSPS gene amplification and control of multiple herbicide-resistant Palmer amaranth (Amaranthus palmeri) with chlorimuron-ethyl, fomesafen, and glyphosate. PLoS ONE 2019, 14, e0214458. [Google Scholar] [CrossRef] [PubMed]
- Sutjit, C.; Nualsri, C.; Duangpan, S.; Nakkanong, K. Characterization of 9-cis-epoxycarotenoid dioxygenase3 gene from Hevea brasiliensis and its expression responses by tissue type during drought stress. Pak. J. Biotechnol. 2019, 16, 175–182. [Google Scholar]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Uno, Y.; Furihata, T.; Abe, H.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 2000, 97, 11632–11637. [Google Scholar] [CrossRef] [PubMed]
- Van Stempvoort, D.R.; Spoelstra, J.; Senger, N.D.; Brown, S.J.; Post, R.; Struger, J. Glyphosate residues in rural groundwater, Nottawasaga River Watershed, Ontario, Canada. Pest Manag. Sci. 2016, 72, 1862–1872. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, J.; Zhang, B.; Vanitha, J.; Ramachandran, S.; Jiang, S.Y. Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on sorghum. J. Integr. Plant Biol. 2011, 53, 212–231. [Google Scholar] [CrossRef]
- Wang, L.-F.; Wang, M.; Zhang, Y. Effects of powdery mildew infection on chloroplast and mitochondrial functions in rubber tree. Trop. Plant Pathol. 2014, 39, 242–250. [Google Scholar] [CrossRef]
- Wang, Y.H.; Que, F.; Li, T.; Zhang, R.R.; Khadr, A.; Xu, Z.S.; Tian, Y.S.; Xiong, A.S. DcABF3, an ABF transcription factor from carrot, alters stomatal density and reduces ABA sensitivity in transgenic Arabidopsis. Plant Sci. 2021, 302, 110699. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yan, G.; Duan, Z.; Wang, Z.; Kang, C.; Guo, L.; Liu, K.; Tu, J.; Shen, J.; Yi, B.; et al. Roles of the Brassica napus DELLA Protein BnaA6.RGA, in Modulating Drought Tolerance by Interacting with the ABA Signaling Component BnaA10.ABF2. Front. Plant Sci. 2020, 11, 577. [Google Scholar] [CrossRef]
- Xiao, S.; Liu, Y.; Wang, A.; Liu, Y.; Li, X.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. The response of tartary buckwheat and 19 bZIP genes to abscisic acid (ABA). Mol. Biol. Rep. 2021, 48, 4341–4350. [Google Scholar] [CrossRef]
- Yamashita, S.; Takahashi, S. Molecular Mechanisms of Natural Rubber Biosynthesis. Annu. Rev. Biochem. 2020, 89, 821–851. [Google Scholar] [CrossRef]
- Yao, L.; Hao, X.; Cao, H.; Ding, C.; Yang, Y.; Wang, L.; Wang, X. ABA-dependent bZIP transcription factor, CsbZIP18, from Camellia sinensis negatively regulates freezing tolerance in Arabidopsis. Plant Cell Rep. 2020, 39, 553–565. [Google Scholar] [CrossRef]
- Yong, X.; Zheng, T.; Zhuo, X.; Ahmad, S.; Li, L.; Li, P.; Yu, J.; Wang, J.; Cheng, T.; Zhang, Q. Genome-wide identification, characterisation, and evolution of ABF/AREB subfamily in nine Rosaceae species and expression analysis in mei (Prunus mume). PeerJ 2021, 9, e10785. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Zancanela, D.C.; Funari, C.S.; Herculano, R.D.; Mello, V.M.; Rodrigues, C.M.; Borges, F.A.; de Barros, N.R.; Marcos, C.M.; Almeida, A.M.F.; Guastaldi, A.C. Natural rubber latex membranes incorporated with three different types of propolis: Physical-chemistry and antimicrobial behaviours. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 97, 576–582. [Google Scholar] [CrossRef]
- Zhang, B.; Feng, C.; Chen, L.; Li, B.; Zhang, X.; Yang, X. Identification and Functional Analysis of bZIP Genes in Cotton Response to Drought Stress. Int. J. Mol. Sci. 2022, 23, 14894. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Wang, M.; Zheng, F.C.; Yang, Y. Research Progress in the Effects of Glyphosate on Plant Physiology. Chin. J. Trop. Agric. 2016, 36, 55–61. [Google Scholar]
- Zhang, Y.; Gao, W.; Li, H.; Wang, Y.; Li, D.; Xue, C.; Liu, Z.; Liu, M.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef] [PubMed]


| Name | Website |
|---|---|
| NCBI ORF Finder | https://www.ncbi.nlm.nih.gov/orffinder/ (1 January 2020) |
| ExPASy ProtParam | http://web.expasy.org/protparam/ (1 January 2020) |
| NCBI Conserved Domains Database | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (1 January 2020) |
| Program SCANPROSITE | http://prosite.expasy.org/scanprosite/ (1 January 2020) |
| SWISS-MODEL TMHMM Server 2.0 | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 (1 January 2020) |
| SignalP 5.0 Server | https://services.healthtech.dtu.dk/service.php?SignalP-5.0 (1 January 2020) |
| MEME | http://meme-suite.org/tools/meme (1 January 2020) |
| WoLF PSORT prediction | http://psort.hgc.jp/form.html (1 January 2020) |
| PlantCARE | http://bioinformatics.psb.ugent.be/ (1 January 2020) |
| ClustalW | http://www.clustal.org/ (1 January 2020) |
| GSDS | gsds.gao-lab.org (1 January 2020) |
| NCBI database | http://www.ncbi.nlm.nih.gov/guide/ (1 January 2020) |
| TAIR | http://www.arabidopsis.org/ (1 January 2020) |
| Rice Genome Annotation Project | http://rice.plantbiology.msu.edu/analyses_search_locus.shtml (1 January 2020) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wang, Y.; Zhang, D.; Liu, Q.; Liu, Y.; Qin, B.; Liang, X.; Wang, L.; Zhang, Y. Characterization of 33 HbbZIP Gene Family Members and Analysis of Their Expression Profiles in Rubber Tree in Response to ABA, Glyphosate and Powdery Mildew Treatment. Forests 2023, 14, 556. https://doi.org/10.3390/f14030556
Wang M, Wang Y, Zhang D, Liu Q, Liu Y, Qin B, Liang X, Wang L, Zhang Y. Characterization of 33 HbbZIP Gene Family Members and Analysis of Their Expression Profiles in Rubber Tree in Response to ABA, Glyphosate and Powdery Mildew Treatment. Forests. 2023; 14(3):556. https://doi.org/10.3390/f14030556
Chicago/Turabian StyleWang, Meng, Yan Wang, Dong Zhang, Qifeng Liu, Yanchao Liu, Bi Qin, Xiaoyu Liang, Lifeng Wang, and Yu Zhang. 2023. "Characterization of 33 HbbZIP Gene Family Members and Analysis of Their Expression Profiles in Rubber Tree in Response to ABA, Glyphosate and Powdery Mildew Treatment" Forests 14, no. 3: 556. https://doi.org/10.3390/f14030556
APA StyleWang, M., Wang, Y., Zhang, D., Liu, Q., Liu, Y., Qin, B., Liang, X., Wang, L., & Zhang, Y. (2023). Characterization of 33 HbbZIP Gene Family Members and Analysis of Their Expression Profiles in Rubber Tree in Response to ABA, Glyphosate and Powdery Mildew Treatment. Forests, 14(3), 556. https://doi.org/10.3390/f14030556





