Prediction of Genetic Gains from Selection in Tree Breeding
Abstract
1. Introduction
2. Genetic Gains from Selection at Different Levels
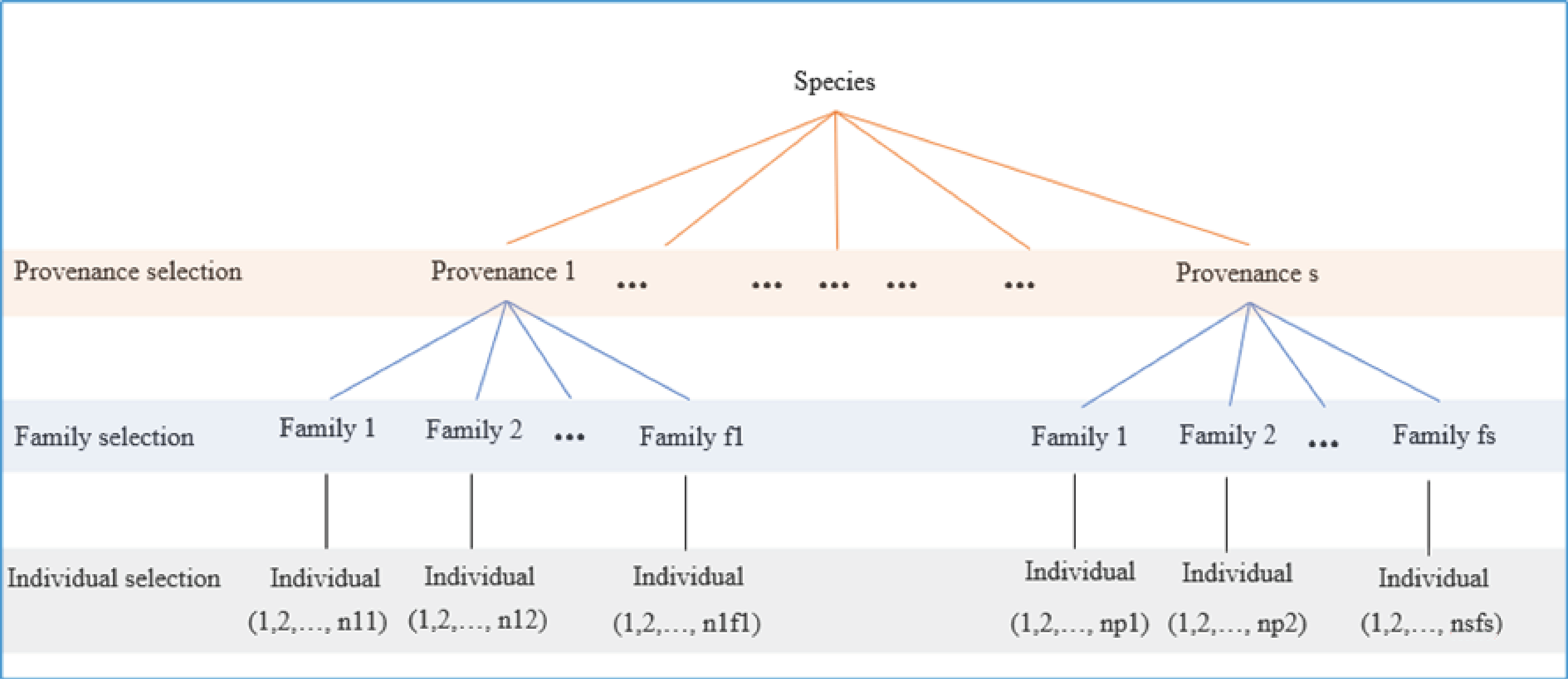
2.1. Selection with Populations of Hierarchical Structure
| Source of Variation | Degree of Freedom | Expected Mean Square |
|---|---|---|
| Block | ||
| Provenance | ||
| Family within provenance | ||
| Provenance × Block | ||
| Family × Block | ||
| Error |
| Level of Selection | Selection Differential S | Genetic Gains |
|---|---|---|
| Provenance | Difference in trait mean between the selected provenances and the whole provenances investigated | |
| Family | Difference in trait mean between the selected families and the whole families investigated | |
| Individual | Difference in trait mean between the selected individuals and the whole individuals investigated | (half-sib family) |
2.2. Factors Causing Biased Prediction of Genetic Gains
3. Improving Prediction of Genetic Gains
3.1. Conventional Breeding Approach
3.2. Marker-Assisted Selection
3.3. Genome-Wide Association Study
3.4. Genomic Selection
4. Advancing Prediction of Genetic Gains Using Multi-Omics Traits
4.1. Gene Expression Traits
4.2. Epigenetic Traits
5. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C. On the Origin of Species by Means of Natural Selection, or Preservation of Favored Races in the Struggle for Life, 1st ed.; J. Murray: London, UK, 1859. [Google Scholar]
- Lush, J.L. Animal Breeding Plans; Iowa State College Press: Ames, IA, USA, 1937. [Google Scholar]
- Griffing, B. Theoretical consequences of truncation selection based on the individual phenotype. Aust. J. Biol. Sci. 1960, 13, 307. [Google Scholar] [CrossRef]
- Griffing, B. Selection in reference to biological groups. I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 1967, 20, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics; Pearson: London, UK, 1996. [Google Scholar]
- Fisher, R.A. The Genetical Theory of Natural Selection; Oxford University Press: Oxford, UK, 1930. [Google Scholar]
- Robertson, A. A mathematical theory of the culling process in dairy cattle. Anim. Sci. 1966, 8, 95–108. [Google Scholar] [CrossRef]
- Robertson, A. The spectrum of genetic variation. In Population Biology and Evolution; Lewontin, R., Ed.; Syracuse University Press: Syracuse, NY, USA, 1968; pp. 5–16. [Google Scholar]
- Price, G.R. Selection and covariance. Nature 1970, 227, 520–521. [Google Scholar] [CrossRef]
- Price, G.R. Extension of covariance selection mathematics. Ann. Hum. Genet. 1972, 35, 485–490. [Google Scholar] [CrossRef]
- Morrissey, M.B.; Parker, D.J.; Korsten, P.; Pemberton, J.M.; Kruuk, L.E.B.; Wilson, A.J. The prediction of adaptive evolution: Em pirical application of the secondary theorem of selection and comparison to the breeder’s equation. Evolution 2012, 66, 2399–2410. [Google Scholar] [CrossRef] [PubMed]
- Queller, D.C. Fundamental theorems of evolution. Am. Nat. 2017, 189, 345–353. [Google Scholar] [CrossRef]
- Lande, R. Quantitative genetic-analysis of multivariate evolution, applied to brain–body size allometry. Evolution 1979, 33, 402–416. [Google Scholar] [CrossRef] [PubMed]
- Roff, D.A. A centennial celebration for quantitative genetics. Evolution 2007, 61, 1017–1032. [Google Scholar] [CrossRef]
- Namkoong, G. Introduction to Quantitative Genetics in Forestry; U.S. Department of Agriculture: Washington, DC, USA, 1979.
- Wright, S. Evolution and the Genetics of Populations, Volume 2: Theory of Gene Frequencies; University of Chicago Press: Chicago, IL, USA, 1969. [Google Scholar]
- Yang, R.C.; Yeh, F.C.; Yanchuk, A.D. A comparison of isozyme and quantitative genetic variation in Pinus contorta ssp. latifolia by FST. Genetics 1996, 142, 1045–1052. [Google Scholar] [CrossRef]
- Wei, R.-P. Response to Selection with restrictions while considering effective family number. Hereditas 1995, 123, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.-P.; Lindgren, D. Selection effects on diversity and genetic gain. Silva Fenn. 1991, 25, 229–234. [Google Scholar] [CrossRef]
- Lindgren, D.; Mullin, T.J. Balancing gain and relatedness in selection. Silvae Genet. 1997, 46, 124–129. [Google Scholar]
- Wei, R.-P.; Yeh, F.C.; Lindgren, D. Expected gain and status number following restricted individual and combined-index selection. Genome 1997, 40, 1–8. [Google Scholar] [CrossRef]
- Hill, W.G.; Robertson, A. The effect of linkage on limits to artificial selection. Genet. Res. 1966, 8, 269–294. [Google Scholar] [CrossRef]
- Lynch, M.; Walsh, B. Genetics and Analysis of Quantitative Traits; Sinauer Associates Inc.: Sunderland, MA, USA, 1998. [Google Scholar]
- Wright, S. The genetic structure of populations. Ann. Eugen. 1951, 15, 323–354. [Google Scholar] [CrossRef]
- Walsh, B.; Lynch, M. Evolution and Selection of Quantitative Traits; Oxford University Press: Oxford, UK, 2018. [Google Scholar] [CrossRef]
- Kimura, M. On the fixation probability of mutant genes in a subdivided population. Genetics 1962, 47, 713–719. [Google Scholar] [CrossRef]
- Kimura, M.; Crow, J.F. Effect of overall phenotypic selection on genetic change at individual loci. Proc. Natl. Acad. Sci. USA 1978, 75, 6168–6171. [Google Scholar] [CrossRef]
- Robertson, A. A theory of limits in artificial selection. Proc. R. Soc. London. Ser. B Biol. Sci. 1960, 153, 234–259. [Google Scholar] [CrossRef]
- Hill, W.G. Predictions of response to artificial selection from new mutations. Genet. Res. 1982, 40, 255–278. [Google Scholar] [CrossRef]
- Hu, X.S.; Zeng, W.; Li, B.L. Impacts of one-way gene flow on genetic variance components in a natural population. Silvae Genet. 2003, 52, 18–24. [Google Scholar]
- Hu, X.S.; Chen, X.Y.; Yeh, F.C. Forest Population Genetics; China Forestry Publishers Education Branch: Beijing, China, 2021. [Google Scholar]
- Xu, Y.B.; Li, P.; Zou, C.; Lu, Y.; Xie, C.; Zhang, X.; Prasanna, B.M.; Olsen, M.S. Article navigation enhancing genetic gain in the era of molecular breeding. J. Exp. Bot. 2017, 68, 2641–2666. [Google Scholar] [CrossRef]
- Hu, X.S.; Wu, R.L.; Han, Y.F. Exploring the sustainable management of genetic resources of forest tree populations II. Analysis of natural and artificial population management of important native tree species in China. For. Res. 2001, 14, 1–7. [Google Scholar]
- Mulder, H.A.; Bijma, P. Effects of genotype × environment interaction on genetic gain in breeding programs. J. Anim. Sci. 2005, 83, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Allwright, M.R.; Taylor, G. Molecular breeding for improved second generation bioenergy crops. Trends Plant Sci. 2016, 21, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Endelman, J.B.; Atlin, G.N.; Beyene, Y.; Semagn, K.; Zhang, X.; Sorrells, M.E.; Jannink, J. Optimal design of preliminary yield trials with genome-wide markers. Crop Sci. 2014, 54, 48–59. [Google Scholar] [CrossRef]
- Lande, R.; Thompson, R. Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 1990, 124, 743–756. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, J.; Lou, X.; Lu, Y. A method for marker-assisted selection based on QTLs with epistatic effects. Genetica 2003, 119, 75–86. [Google Scholar] [CrossRef]
- Hu, X.-S. A general framework for marker-assisted selection. Theor. Popul. Biol. 2007, 71, 524–542. [Google Scholar] [CrossRef]
- Wu, H.X. Study of early selection in tree breeding 4. efficiency of marker-aided early selection (MAES). Silvae Genet. 2002, 51, 261–269. [Google Scholar]
- Wang, X.; Ma, P.; Liu, J.; Zhang, Q.; Zhang, Y.; Ding, X.; Jiang, L.; Wang, Y.; Zhang, Y.; Sun, D.; et al. Genome-wide association study in Chinese Holstein cows reveal two candidate genes for somatic cell score as an indicator for mastitis susceptibility. BMC Genet. 2015, 16, 111. [Google Scholar] [CrossRef]
- Li, Y.-H.; Reif, J.C.; Hong, H.-L.; Li, H.-H.; Liu, Z.-X.; Ma, Y.-S.; Li, J.; Tian, Y.; Li, Y.-F.; Li, W.-B.; et al. Genome-wide association mapping of QTL underlying seed oil and protein contents of a diverse panel of soybean accessions. Plant Sci. 2018, 266, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Koochakpouk, Z.; Solouki, M.; Fakheri, B.A.; Aghnoum, R.; Nezhad, N.M. Genome wide association analysis of plant height, spike and awn length in barley (Hordeum vulgare L.) exposed to Mn stress. J. Anim. Plant Sci. 2020, 30, 384–390. [Google Scholar] [CrossRef]
- Feng, J.L.; Jiang, Y.; Yang, Z.J.; Chen, S.P.; EI-Kassaby, Y.A.; Chen, H. Marker-assisted selection in C. oleifera hybrid population. Silvae Genet. 2020, 69, 63–72. [Google Scholar] [CrossRef]
- Vijayan, K.; Srivastava, P.P.; Nair, C.V.; Awasthi, A.K.; Tikader, A.; Sreenivasa, B.; Urs, S.R. Molecular characterization and identification of markers associated with leaf yield traits in mulberry using ISSR markers. Plant Breed. 2006, 125, 298–301. [Google Scholar] [CrossRef]
- Kar, P.K.; Srivastava, P.P.; Awasthi, A.K.; Urs, S.R. Genetic variability and association of ISSR markers with some biochemical traits in mulberry (Morus spp.) genetic resources available in India. Tree Genet. Genomes 2008, 4, 75–83. [Google Scholar] [CrossRef]
- Vijayan, K.; Nair, C.V.; Chatterjee, S.N. Diversification of mulberry (Morus indica var. S36)—A vegetatively propagated tree species. Casp. J. Env. Sci. 2009, 7, 23–30. [Google Scholar]
- Ahmar, S.; Ballesta, P.; Ali, M.; Mora-Poblete, F. Achievements and challenges of genomics-assisted breeding in forest trees: From marker-assisted selection to genome editing. Int. J. Mol. Sci. 2021, 22, 10583. [Google Scholar] [CrossRef]
- Nagano, S.; Hirao, T.; Takashima, Y.; Matsushita, M.; Mishima, K.; Takahashi, M.; Iki, T.; Ishiguri, F.; Hiraoka, Y. SNP Genotyping with target amplicon sequencing using a multiplexed primer panel and its application to genomic prediction in Japanese Cedar, Cryptomeria japonica (L.f.) D.Don. Forests 2020, 11, 898. [Google Scholar] [CrossRef]
- McKown, A.D.; Guy, R.D.; Quamme, L.; Klápště, J.; La Mantia, J.; Constabel, C.P.; El-Kassaby, Y.A.; Hamelin, R.C.; Zifkin, M.; Azam, M.S. Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol. Ecol. 2014, 23, 5771–5790. [Google Scholar] [CrossRef]
- Du, Q.; Gong, C.; Wang, Q.; Zhou, D.; Yang, H.; Pan, W.; Li, B.; Zhang, D. Genetic architecture of growth traits in Populus revealed by integrated quantitative trait locus (QTL) analysis and association studies. N. Phytol. 2016, 209, 1067–1082. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, A.M.; Neves, L.G.; Resende, M.F.R.; Vazquez, A.I.; Campos, G.; Dervinis, C.; Sykes, R.; Davis, M.; Davenport, R.; Barbazuk, W.B.; et al. Genome-wide association study reveals putative regulators of bioenergy traits in Populus deltoides. New Phytol. 2017, 213, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.P.; Suren, H.; Holliday, J.; Richards, J.H.; Fiehn, O.; Famula, R.; Stanton, B.J.; Shuren, R.; Sykes, R.; Davis, M.F.; et al. Exome resequencing and GWAS for growth, ecophysiology, and chemical and metabolomic composition of wood of Populus trichocarpa. BMC Genom. 2019, 20, 875. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, C.; Ballesta, P.; Ahmar, S.; Fiaz, S.; Heidari, P.; Maldonado, C.; Mora-Poblete, F. Haplotype- and SNP-based GWAS for growth and wood quality traits in Eucalyptus cladocalyx trees under arid conditions. Plants 2021, 10, 148. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Gmitter, F.G. QTL mapping of mandarin (Citrus reticulata) fruit characters using high-throughput SNP markers. Tree Genet. Genomes 2016, 12, 77. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-Kanegae, H.; Onogi, A.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T.; et al. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7, 4721. [Google Scholar] [CrossRef]
- Duan, N.; Bai, Y.; Sun, H.; Wang, N.; Ma, Y.; Li, M.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 429. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Lamara, M.; Raherison, E.; Lenz, P.; Beaulieu, J.; Bousquet, J.; MacKay, J. Genetic architecture of wood properties based on association analysis and co-expression networks in white spruce. N. Phytol. 2016, 210, 240–255. [Google Scholar] [CrossRef]
- Tan, B.; Ingvarsson, P.K. Integrating genome-wide association mapping of additive and dominance genetic effects to improve genomic prediction accuracy in Eucalyptus. Plant Genome 2022, 15, e20208. [Google Scholar] [CrossRef]
- Fraser, H.B.; Hirsh, A.E.; Steinmetz, L.M.; Scharfe, C.; Feldman, M.W. Evolutionary rate in the protein interaction network. Science 2002, 296, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M.E.; Hayes, B.J. Genomic selection. J. Anim. Breed. Genet. 2007, 124, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.L.; Ma, Y.L.; Xing, T.; Zhu, M.J.; Yu, M.; Li, X.Y.; Liu, X.L.; Zhao, S.H. Progress and perspectives on genome-wide selection models. J. Anim. Husb. Vet. Med. 2019, 50, 233–242. [Google Scholar] [CrossRef]
- Zhu, S.; Huang, M.R. Progress and prospects of genomic selection in genetic breeding research of forest trees. Sci. Silvae Sin. 2020, 56, 176–185. [Google Scholar]
- McGowan, M.; Wang, J.; Dong, H.; Liu, X.; Jia, Y.; Wang, X.; Iwata, H.; Li, Y.; Lipka, A.E.; Zhang, Z. Ideas in genomic selection with the potential to transform plant molecular breeding: A review. Plant Breed Rev. 2021, 45, 273–319. [Google Scholar] [CrossRef]
- Calleja-Rodriguez, A.; Chen, Z.; Suontama, M.; Pan, J.; Wu, H.X. Genomic predictions with nonadditive effects improved estimates of additive effects and oredictions of total genetic values in Pinus sylvestris. Front. Plant Sci. 2021, 12, 666820. [Google Scholar] [CrossRef] [PubMed]
- Denis, M.; Favreau, B.; Ueno, S.; Camus-Kulandaivelu, L.; Chaix, G.; Gion, J.-M.; Nourrisier-Mountou, S.; Polidori, J.; Bouvet, J.-M. Genetic variation of wood chemical traits and association with underlying genes in Eucalyptus urophylla. Tree Genet. Genomes 2013, 9, 927–942. [Google Scholar] [CrossRef]
- VanRaden, P. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Sun, W.; Hu, Y. eQTL Mapping Using RNA-seq Data. Stat. Biosci. 2012, 5, 198–219. [Google Scholar] [CrossRef]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Gao, J. Integration of multi-omics data for expression quantitative trait loci (eQTL) analysis and eQTL epistasis. Methods Mol. Biol. 2020, 2082, 157–171. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Claringbould, A.; Westra, H.J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300. [Google Scholar] [CrossRef] [PubMed]
- Petretto, E.; Mangion, J.; Dickens, N.J.; Cook, S.A.; Kumaran, M.K.; Lu, H.; Fischer, J.; Maatz, H.; Kren, V.; Pravenec, M.; et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006, 2, e172. [Google Scholar] [CrossRef]
- Lloyd-Jones, L.R.; Holloway, A.; McRae, A.; Yang, J.; Small, K.; Zhao, J.; Zeng, B.; Bakshi, A.; Metspalu, A.; Dermitzakis, M.; et al. The genetic architecture of gene expression in peripheral blood. Am. J. Hum. Genet. 2017, 100, 228–237. [Google Scholar] [CrossRef]
- Yao, D.W.; O’Connor, L.J.; Price, A.L.; Gusev, A. Quantifying genetic effects on disease mediated by assayed gene expression levels. Nat. Genet. 2020, 52, 626–633. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Oster, M.; Reyer, H.; Hadlich, F.; Trakooljul, N.; Rodehutscord, M.; Camarinha-Silva, A.; Bennewitz, J.; Wimmers, K. Genetic regulation and heritability of miRNA and mRNA expression link to phosphorus utilization and gut microbiome. Open Biol. 2021, 11, 200182. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Bei, X.; Shahid, M.Q.; Wu, J.; Chen, Z.; Wang, L.; Liu, X. Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice. PLoS ONE 2019, 14, e0214953. [Google Scholar] [CrossRef]
- Hussey, S.G.; Mizrachi, E.; Groover, A.; Berger, D.K.; Myburg, A.A. Genome-wide mapping of histone H3 lysine 4 trimethylation in Eucalyptus grandis developing xylem. BMC Plant Biol. 2015, 15, 117. [Google Scholar] [CrossRef]
- Kawashima, T.; Lorković, Z.J.; Nishihama, R.; Ishizaki, K.; Axelsson, E.; Yelagandula, R.; Kohchi, T.; Berger, F. Diversification of histone H2A variants during plant evolution. Trends Plant Sci. 2015, 20, 419–425. [Google Scholar] [CrossRef]
- Hou, Q.; Wan, X. Epigenome and epitranscriptome: Potential resources for crop improvement. Int. J. Mol. Sci. 2021, 22, 12912. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.-Y.; Huang, H.-W.; Shu, C.-W.; Hou, M.-F.; Yuan, S.-S.F.; Wang, H.-R.; Chang, Y.-T.; Farooqi, A.A.; Tang, J.-Y.; Chang, H.-W. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett. 2016, 373, 185–192. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Zhao, X. Epigenetic marks: Regulators of livestock phenotypes and conceivable provenances of missing variation in livestock improvement programs. Front. Genet. 2015, 6, 302. [Google Scholar] [CrossRef] [PubMed]
- Granada, L.; Lemos, M.F.; Cabral, H.N.; Bossier, P.; Novais, S.C. Epigenetics in aquaculture—The last frontier. Rev. Aquac. 2018, 10, 994–1013. [Google Scholar] [CrossRef]
- Yu, J.; Xu, F.; Wei, Z.; Zhang, X.; Chen, T.; Pu, L. Epigenomic landscape and epigenetic regulation in maize. Theor. Appl. Genet. 2020, 133, 1467–1489. [Google Scholar] [CrossRef]
- Yakovlev, I.A.; Carneros, E.; Lee, Y.; Olsen, J.E.; Fossdal, C.G. Transcriptional profiling of epigenetic regulators in somatic embryos during temperature induced formation of an epigenetic memory in Norway spruce. Planta 2016, 243, 1237–1249. [Google Scholar] [CrossRef]
- Sáez-Laguna, E.; Guevara, M.; Díaz, L.-M.; Sánchez-Gómez, D.; Collada, C.; Aranda, I.; Cervera, M.-T. Epigenetic variability in the genetically uniform forest tree species Pinus pinea L. PLoS ONE 2014, 9, e103145. [Google Scholar] [CrossRef]
- Bräutigam, K.; Vining, K.J.; Lafon-Placette, C.; Fossdal, C.G.; Mirouze, M.; Marcos, J.G.; Fluch, S.; Fraga, M.F.; Guevara, M.Á.; Abarca, D.; et al. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol. Evol. 2013, 3, 399–415. [Google Scholar] [CrossRef]
- Sow, M.D.; Allona, I.; Ambroise, C.; Conde, D.; Fichot, R.; Gribkova, S.; Jorge, V.; Le-Provost, G.; Pâques, L.; Plomion, C.; et al. Epigenetics in forest trees: State of the art and potential implications for breeding and management in a context of climate change. Adv. Bot. Res. 2018, 88, 387–453. [Google Scholar] [CrossRef]
- Amaral, J.; Ribeyre, Z.; Vigneaud, J.; Sow, M.D.; Fichot, R.; Messier, C.; Pinto, G.; Nolet, P.; Maury, S. Advances and promises of epigenetics for forest trees. Forests 2020, 11, 976. [Google Scholar] [CrossRef]
- Klupczyńska, E.A.; Ratajczak, E. Can forest trees cope with climate change?—Effects of DNA methylation on gene expression and adaptation to environmental change. Int. J. Mol. Sci. 2021, 22, 13524. [Google Scholar] [CrossRef] [PubMed]
- Baldanzi, S.; Watson, R.; McQuaid, C.; Gouws, G.; Porri, F. Epigenetic variation among natural populations of the South African sandhopper Talorchestia capensis. Evol. Ecol. 2017, 31, 77–91. [Google Scholar] [CrossRef]
- Lande, R.; Arnold, S.J. The measurement of selection on correlated characters. Evolution 1983, 37, 1210–1226. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.-H.; Xiao, Y.; Lv, Y.-W.; Yeh, F.C.; Wang, X.; Hu, X.-S. Prediction of Genetic Gains from Selection in Tree Breeding. Forests 2023, 14, 520. https://doi.org/10.3390/f14030520
He Z-H, Xiao Y, Lv Y-W, Yeh FC, Wang X, Hu X-S. Prediction of Genetic Gains from Selection in Tree Breeding. Forests. 2023; 14(3):520. https://doi.org/10.3390/f14030520
Chicago/Turabian StyleHe, Zi-Han, Yu Xiao, Yan-Wen Lv, Francis C. Yeh, Xi Wang, and Xin-Sheng Hu. 2023. "Prediction of Genetic Gains from Selection in Tree Breeding" Forests 14, no. 3: 520. https://doi.org/10.3390/f14030520
APA StyleHe, Z.-H., Xiao, Y., Lv, Y.-W., Yeh, F. C., Wang, X., & Hu, X.-S. (2023). Prediction of Genetic Gains from Selection in Tree Breeding. Forests, 14(3), 520. https://doi.org/10.3390/f14030520




