Male-Specific Sequence in Populus simonii Provides Insights into Gender Determination of Poplar
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction, PCR Amplification, and Gel Electrophoresis
2.3. Sanger Sequencing and SNP Analysis
2.4. Chromosome Preparation
2.5. Fluorescence In Situ Hybridization (FISH) Analysis
3. Results and Discussion
3.1. Poplar Gender Verification and Other Sex Markers in Populus
3.2. SNP Detection of the Male-Specific Sequence
3.3. Cytogenetic Location of the Male-Specific Sequence in P. simonii
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Taylor, G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 2002, 90, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Wu, H.; Chen, Y.; Li, X.; Hou, J.; Lu, J.; Wei, S.; Dai, X.; Olson, M.S.; Liu, J.; et al. Evidences for a role of two Y-specific genes in sex determination in Populus deltoides. Nat. Commun. 2020, 11, 5893. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Zhang, T.; Han, Y.; Wu, Y.; Shi, J.; Xi, M.; Jiang, J. Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides. Chromosoma 2018, 127, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.Q.; Ferguson, D.K.; Bera, S.; Li, C.S. Seed hairs of poplar trees as natural airborne pollen trap for allergenic pollen grains. Grana 2008, 47, 241–245. [Google Scholar] [CrossRef]
- Ortega, M.A.; Zhou, R.; Chen, M.S.S.; Bewg, W.P.; Simon, B.; Tsai, C.J. In vitro floral development in poplar: Insights into seed trichome regulation and trimonoecy. New Phytol. 2023, 237, 1078–1081. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, T.; Rao, P.; Yang, N.; Li, G.; Jia, L.; An, X.; Chen, Z. Morphology, sucrose metabolism and gene network reveal the molecular mechanism of seed fiber development in poplar. Int. J. Biol. Macromol. 2023, 246, 125633. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Yang, M. Isozymes analysis of three poplar species with different sex. J. Hebei For. Res. 1986, 1, 29–31. [Google Scholar]
- Wang, H.; Li, C.; Bai, H.; Zhou, Z.; Wang, Y. Screening SSR Markers for Sex Identification in Populus davidiana Dode. J. Northeast. For. Univ. 2017, 45, 17–19+29. [Google Scholar] [CrossRef]
- Pakull, B.; Kersten, B.; Luneburg, J.; Fladung, M. A simple PCR-based marker to determine sex in aspen. Plant Biol. 2015, 17, 256–261. [Google Scholar] [CrossRef]
- Kim, G.; Leite Montalvão, A.P.; Kersten, B.; Fladung, M.; Müller, N.A. The genetic basis of sex determination in Populus provides molecular markers across the genus and indicates convergent evolution. Silvae Genet. 2021, 70, 145–155. [Google Scholar] [CrossRef]
- Geraldes, A.; Hefer, C.A.; Capron, A.; Kolosova, N.; Martinez-Nunez, F.; Soolanayakanahally, R.Y.; Stanton, B.; Guy, R.D.; Mansfield, S.D.; Douglas, C.J.; et al. Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol. Ecol. 2015, 24, 3243–3256. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wu, H.; Dai, X.; Li, W.; Qiu, Y.; Yang, Y.; Yin, T. Sex effect on growth performance and marker-aided sex discrimination of seedlings of Populus deltoides. J. For. Res. 2022, 34, 1639–1645. [Google Scholar] [CrossRef]
- Tong, C.; Yao, D.; Wu, H.; Chen, Y.; Yang, W.; Zhao, W. High-Quality SNP Linkage Maps Improved QTL Mapping and Genome Assembly in Populus. J. Hered. 2020, 111, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Yuan, G.; Lu, H.; Liu, Y.; Zhang, J.; Tuskan, G.A.; Muchero, W.; Chen, J.-G.; Yang, X. CRISPR/Cas9-based gene activation and base editing in Populus. Hortic. Res. 2023, 10, uhad085. [Google Scholar] [CrossRef] [PubMed]
- Kutsokon, N.K.; Jose, S.; Holzmueller, E. A Global Analysis of Temperature Effects on Populus Plantation Production Potential. Am. J. Plant Sci. 2015, 6, 23–33. [Google Scholar] [CrossRef]
- Xi, M.; Zhang, T.; Shang, D. A Male-Specific Genomic DNA Sequence of Populus simonii and Its Application. Patent No. CN111269974B, 4 December 2020. [Google Scholar]
- Qi, H.; Wu, L.; Shen, T.; Liu, S.; Cai, H.; Ran, N.; Wang, J.; Xu, M. Overexpression of the long non-coding RNA lncWOX5 negatively regulates the development of adventitious roots in Populus. Ind. Crops Prod. 2023, 192, 116054. [Google Scholar] [CrossRef]
- Wang, H.-L.; Li, L.; Tang, S.; Yuan, C.; Tian, Q.; Su, Y.; Li, H.-G.; Zhao, L.; Yin, W.; Zhao, R.; et al. Evaluation of Appropriate Reference Genes for Reverse Transcription-Quantitative PCR Studies in Different Tissues of a Desert Poplar via Comparision of Different Algorithms. Int. J. Mol. Sci. 2015, 16, 20468–20491. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Li, Z.; Bi, Y.; Wang, X.; Wang, Y.; Yang, S.; Zhang, Z.; Chen, J.; Lou, Q.; Schwarzacher, T. Chromosome identification in Cucumis anguria revealed by cross-species single-copy gene FISH. Genome 2018, 61, 397–404. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, T.; Wu, Y.; Zhang, W.; Zhang, P.; Xi, M.; Jiang, J. An extraordinarily stable karyotype of the woody Populus species revealed by chromosome painting. Plant J. 2019, 101, 253–264. [Google Scholar] [CrossRef]
- Muller, N.A.; Kersten, B.; Leite Montalvao, A.P.; Mahler, N.; Bernhardsson, C.; Brautigam, K.; Carracedo Lorenzo, Z.; Hoenicka, H.; Kumar, V.; Mader, M.; et al. A single gene underlies the dynamic evolution of poplar sex determination. Nat. Plants 2020, 6, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, M.; Jorge, V.; Paolucci, I.; Beritognolo, I.; Mugnozza, G.S.; Sabatti, M. Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet. Genomes 2007, 4, 25–36. [Google Scholar] [CrossRef]
- Zhou, R.; Macaya-Sanz, D.; Schmutz, J.; Jenkins, J.W.; Tuskan, G.A.; DiFazio, S.P. Sequencing and Analysis of the Sex Determination Region of Populus trichocarpa. Genes 2020, 11, 843. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, J.; Bachtrog, D.; An, N.; Huang, Q.; Jarvis, E.D.; Gilbert, M.T.P.; Zhang, G. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 2014, 346, 1246338. [Google Scholar] [CrossRef]
- Gao, W.-J.; Xie, L.; Lu, J.-W.; Deng, C.-L.; Lu, L.-D. Role of repetitive sequence and heterochromatize in recombination suppression of plant sex chromosomes. Hereditas 2010, 32, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yao, D.; Chen, Y.; Yang, W.; Zhao, W.; Gao, H.; Tong, C. De Novo Genome Assembly of Populus simonii Further Supports That Populus simonii and Populus trichocarpa Belong to Different Sections. G3 2020, 10, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, S.; Zhang, Y.; Tan, J.; Li, X.; Chu, X.; Xu, B.; Tian, Y.; Sun, Y.; Li, B.; et al. A telomere-to-telomere gap-free reference genome of watermelon and its mutation library provide important resources for gene discovery and breeding. Mol. Plant 2022, 15, 1268–1284. [Google Scholar] [CrossRef]
- Shi, X.; Cao, S.; Wang, X.; Huang, S.; Wang, Y.; Liu, Z.; Liu, W.; Leng, X.; Peng, Y.; Wang, N.; et al. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Hortic. Res. 2023, 10, uhad061. [Google Scholar] [CrossRef]
- Yue, J.; Chen, Q.; Wang, Y.; Zhang, L.; Ye, C.; Wang, X.; Cao, S.; Lin, Y.; Huang, W.; Xian, H.; et al. Telomere-to-telomere and gap-free reference genome assembly of the kiwifruit Actinidia chinensis. Hortic. Res. 2023, 10, uhac264. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, M.; Li, M.; Jiang, X.; Jiao, W.; Song, Q. A telomere-to-telomere gap-free assembly of soybean genome. Mol. Plant 2023, 16, 1711–1714. [Google Scholar] [CrossRef] [PubMed]
- You, C.; Wen, R.; Zhang, Z.; Cheng, G.; Zhang, Y.; Li, N.; Deng, C.; Li, S.; Gao, W. Development and applications of a collection of single copy gene-based cytogenetic DNA markers in garden asparagus. Front. Plant Sci. 2022, 13, 1010664. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Han, J.; Lin, Y.; Zhao, Y.; Lin, Q.; Ma, X.; Wang, J.; Zhang, M.; Zhang, L.; Yang, Q.; et al. Characterization of a Saccharum spontaneum with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theor. Appl. Genet. 2020, 133, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Yin, T.; Difazio, S.P.; Gunter, L.E.; Zhang, X.; Sewell, M.M.; Woolbright, S.A.; Allan, G.J.; Kelleher, C.T.; Douglas, C.J.; Wang, M.; et al. Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res. 2008, 18, 422–430. [Google Scholar] [CrossRef]
- Yang, W.; Wang, D.; Li, Y.; Zhang, Z.; Tong, S.; Li, M.; Zhang, X.; Zhang, L.; Ren, L.; Ma, X.; et al. A General Model to Explain Repeated Turnovers of Sex Determination in the Salicaceae. Mol. Biol. Evol. 2021, 38, 968–980. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Z.; Ma, D.; Zhai, J.; Han, X.; Jiang, Z.; Liu, S.; Xu, J.; Jiao, P.; Li, Z. Chromosome-scale assemblies of the male and female Populus euphratica genomes reveal the molecular basis of sex determination and sexual dimorphism. Commun. Biol. 2022, 5, 1186. [Google Scholar] [CrossRef]
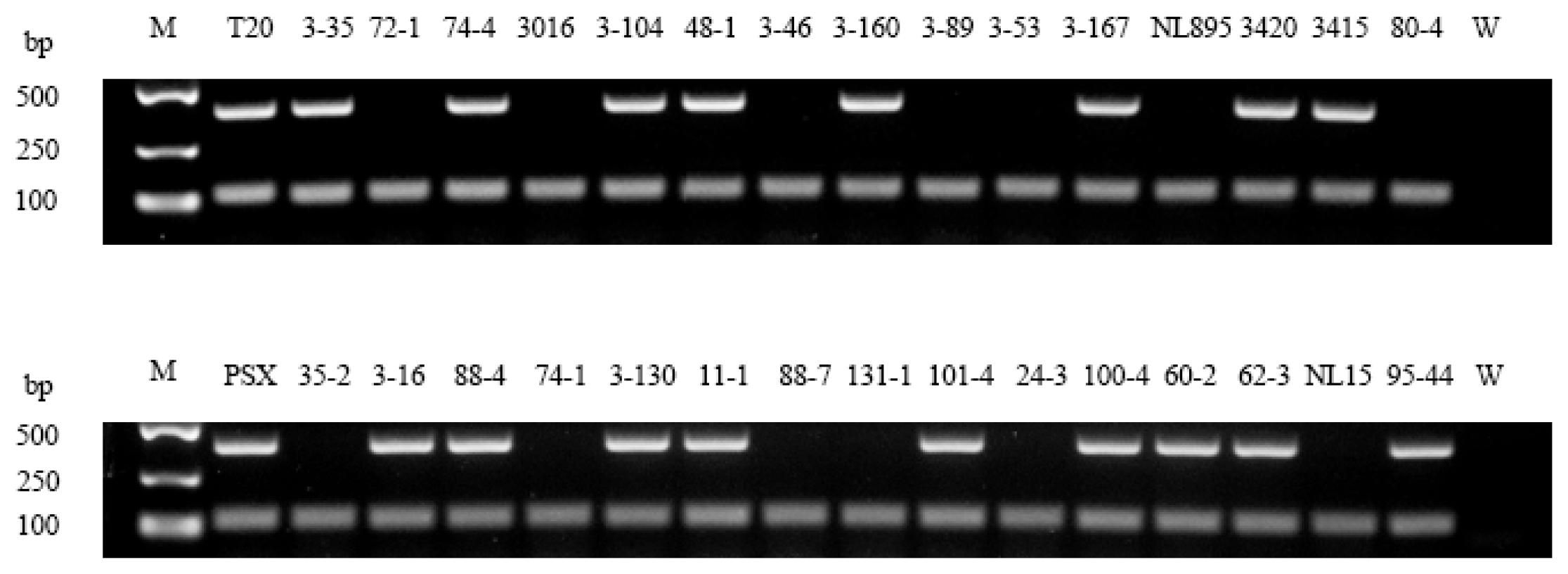
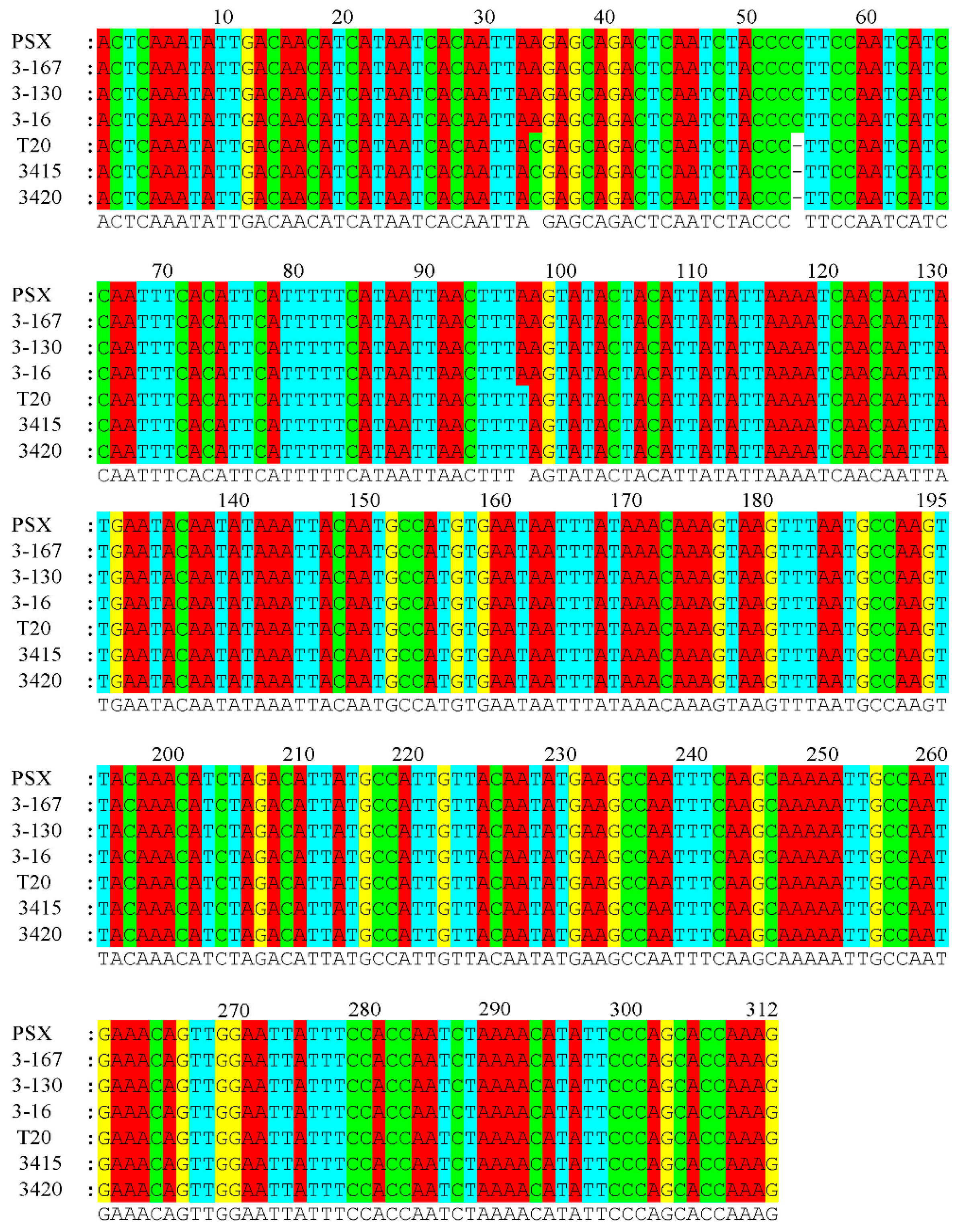
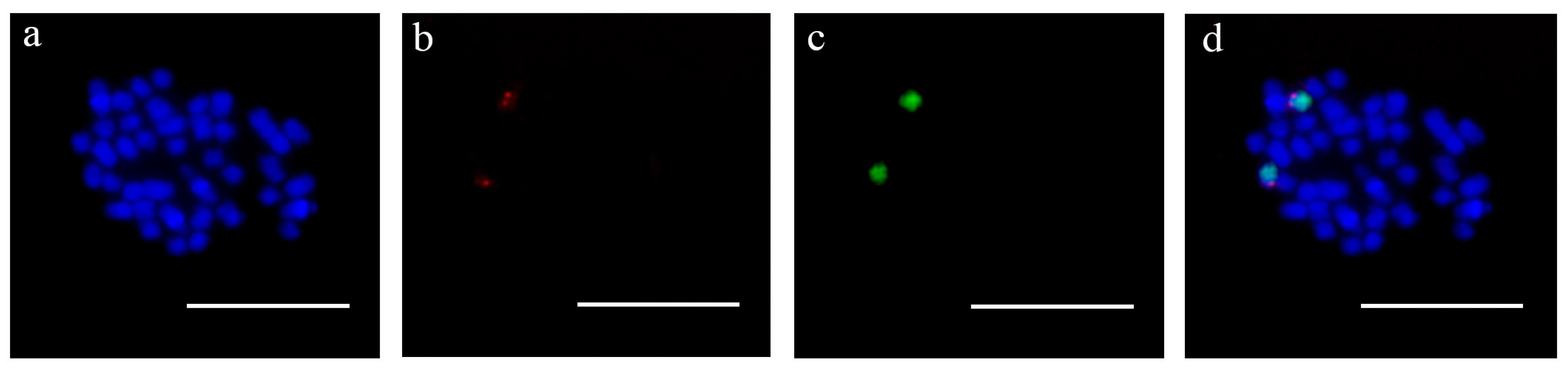
| Number | Species | Genotype | Sex |
|---|---|---|---|
| 1 | P. deltoides | T20 | male |
| 2 | P. deltoides ‘I-69′ × P. simonii F1 | 3-35 | male |
| 3 | P. deltoides | 72-1 | female |
| 4 | P. deltoides | 74-4 | male |
| 5 | P. deltoides | 3016 | female |
| 6 | P. deltoides ‘I-69′ × P. simonii F1 | 3-104 | male |
| 7 | P. deltoides | 48-1 | male |
| 8 | P. deltoides ‘I-69′ × P. simonii F1 | 3-46 | female |
| 9 | P. deltoides ‘I-69′ × P. simonii F1 | 3-160 | male |
| 10 | P. deltoides ‘I-69′ × P. simonii F1 | 3-89 | female |
| 11 | P. deltoides ‘I-69′ × P. simonii F1 | 3-53 | female |
| 12 | P. deltoides ‘I-69′ × P. simonii F1 | 3-167 | male |
| 13 | P. deltoides × P. euramericana | Nanlin895 (NL895) | female |
| 14 | P. deltoides | 3420 | male |
| 15 | P. deltoides | 3415 | male |
| 16 | P. deltoides | 80-4 | female |
| 17 | P. simonii | P. simonii | male |
| 18 | P. deltoides | 35-2 | female |
| 19 | P. deltoides ‘I-69′ × P. simonii F1 | 3-16 | male |
| 20 | P. deltoides | 88-4 | male |
| 21 | P. deltoides | 74-1 | female |
| 22 | P. deltoides ‘I-69′ × P. simonii F1 | 3-130 | male |
| 23 | P. deltoides | 11-1 | male |
| 24 | P. deltoides | 88-7 | female |
| 25 | P. deltoides | 131-1 | female |
| 26 | P. deltoides | 101-4 | male |
| 27 | P. deltoides | 24-3 | female |
| 28 | P. deltoides | 100-4 | male |
| 29 | P. deltoides | 60-2 | male |
| 30 | P. deltoides | 62-3 | male |
| 31 | P. deltoides | NanLin15 (NL15) | female |
| 32 | P. deltoides | 95-44 | male |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Lei, Y.; Liu, G.; Ning, Y.; Ni, R.; Zhang, T.; Xi, M. Male-Specific Sequence in Populus simonii Provides Insights into Gender Determination of Poplar. Forests 2023, 14, 2385. https://doi.org/10.3390/f14122385
Wang Z, Lei Y, Liu G, Ning Y, Ni R, Zhang T, Xi M. Male-Specific Sequence in Populus simonii Provides Insights into Gender Determination of Poplar. Forests. 2023; 14(12):2385. https://doi.org/10.3390/f14122385
Chicago/Turabian StyleWang, Ziyue, Yijing Lei, Guanqing Liu, Yihang Ning, Runxin Ni, Tao Zhang, and Mengli Xi. 2023. "Male-Specific Sequence in Populus simonii Provides Insights into Gender Determination of Poplar" Forests 14, no. 12: 2385. https://doi.org/10.3390/f14122385
APA StyleWang, Z., Lei, Y., Liu, G., Ning, Y., Ni, R., Zhang, T., & Xi, M. (2023). Male-Specific Sequence in Populus simonii Provides Insights into Gender Determination of Poplar. Forests, 14(12), 2385. https://doi.org/10.3390/f14122385






