Abstract
In this study, the PtWRKY33 gene sequence was cloned by PCR from cDNA, and systematic evolutionary analysis was performed. qRT–PCR was performed to measure the expression of the PtWRKY33 gene in the roots, stems and leaves of Populus tremuloides under drought and saline stresses, and overexpression vectors were constructed and genetically transformed into tobacco. The results showed that the WRKY transcription factors of Populus trichocarpa, Populus euphratica and Populus alba were in the same branch and were closely related to each other. Treating poplar seedlings with 20% PEG6000 solution to simulate drought stress showed that expression of the PtWRKY33 gene was induced and increased 1.89 times, 3.45 times and 11.6 times in leaves, stems and roots. The relative expression of the PtWRKY33 gene was slower to respond to 150 mM NaCl treatment but was nonetheless induced. At 48 h, the increase was 1–3 times in leaves, stems and roots, respectively. NaHCO3 (60 mM) was used to treat poplar seedlings with alkaline salt stress; the results indicated that the PtWRKY33 gene was sensitive to NaHCO3 stress treatment, and that its relative expression was significantly increased and increased 10.51 times (24 h), 6.56 times (6 h) and 5.16 times (24 h) in leaves, stems and roots, respectively. In this study, we found that NaCl and NaHCO3 stress treatments were able to induce an increase in the expression of the NtSOD1 and NtAPX2 genes using qRT–PCR, and the significant increase in expression under the treatments compared with WT may be caused by overexpression of the PtWRKY33 gene. Overexpression of the PtWRKY33 gene in tobacco enhanced the antioxidant stress capacity and improved the salinity tolerance of transgenic tobacco. These results indicate that the PtWRKY33 gene is a key gene for improved salinity-tolerant growth, which is important for future molecular breeding of tree resistance.
1. Introduction
WRKY proteins constitute one of the largest families of transcription factors in plants, with a zinc finger structure at the C-terminus and a WRKYGQK structural domain consisting of approximately 60 amino acid residues at the N-terminal end [1]. Transcription factors are essential for plant tolerance to drought and low temperatures and act in a variety of ways. First, plants can be influenced by signaling molecules such as abscisic acid (ABA) and H2O2 to respond to abiotic stresses such as low temperatures and drought [2]. Second, they are involved in regulating growth and development and biotic stress responses in a variety of plants [3,4,5]. For example, plants induce tolerance to drought by reducing lateral roots to change their root system structure [6].
Drought and salinity stress tolerance are polygenic traits influenced by genome–environment interactions [7]. To date, WRKY gene cloning and functional studies have been reported for a variety of plants, and 11 WRKY genes in Populus minor enhance plant heat tolerance through downregulated expression [8]. WRKY genes were found to be resistance candidates by analysis of WRKY genes in the model plant tobacco. However, the mechanisms of drought and salinity stress tolerance regulation are not well understood. For better cultivation of poplar in arid and saline areas of the northeastern Songnen Plain, application in urban greening, wind and sand control and planting in open saline lands in Heilongjiang Province, it is important to accelerate the improvement of poplar arid and saline germplasm and to mine arid and saline-related genes and their expression. Therefore, in this study, the PtWRKY33 gene (XM_002319046) was isolated and cloned from Populus trichocarpa, and the function of the PtWRKY33 gene involved in drought and salinity stress tolerance mechanisms was resolved by a transgenic tobacco strain with an overexpression vector (pGWB18). Exploring the physiological mechanism of drought and salinity tolerance in trees and its genetic factors is not only important in basic theory but also has wide application value to solving the practical problems of tree molecular breeding production.
2. Materials and Methods
2.1. Plant Materials
Poplar (Populus trichocarpa Torr. and Gray) plants, Arabidopsis thaliana Columbia-0 seeds and Nicotiana benthamiana seeds were obtained from the Key Laboratory of the Ministry of Education for Restoration and Reconstruction of Saline Vegetation in Northeast China (Northeast Forestry University). For cultivation and preservation, six-leaf seedlings and bottled seedlings were grown using tissue culture methods.
2.2. Methods
2.2.1. Cloning and Phylogenetic Analysis of the PtWRKY33 Gene
In this study, a one-step method was used to isolate total plant RNA with a kit from Coomei Century Biotechnology Ltd. [9,10]. Using total cDNA from the leaves and roots of Populus trichocarpa as a template, the specific primers PtF1: 5′-atggcatcttcagcttctgc-3′ and PtR2: 5′-ttagttaaggaagggatcaa-3′ (based on the mRNA sequence XM_002323601.3 from GenBank) were used, the target gene fragment was amplified by PCR with Blend-Taq DNA polymerase, and the DNA was recovered by agarose gel electrophoresis and ligated and inserted into PMD18-T vector, which was digested by the restriction endonucleases SalI and BamHI to confirm the successful construction of the PMD18-T-PtWRKY33 plasmid.
Phylogenetic analysis was performed by multiple sequence alignment and homology; similarity, comparison using DNAMAN 9 software and phylogenetic analysis was performed by the neighbor-joining method using MEGA 6.0 software (bootstrap value set to 1000).
2.2.2. Subcellular Localization of the PtWRKY33 Protein and Arabidopsis thaliana Protoplast Extraction and Recombinant Plasmid Transformation
The network bioinformatics software PSORT (http://psort.hgc.jp/form.html, accessed on 6 October 2023) and Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 6 October 2023) was used to predict subcellular localization, and an experiment was performed to verify the results. PCR amplification of the open reading frame (ORF) of PtWRKY33 was performed using the specific primers PtW33F1 and PtR6 (5′-gttaaggaagggatcaaaaa-3′). The fragment was fused upstream of green fluorescent protein (GFP) and under the control of the cauliflower mosaic virus (CaMV) 35S super promoter to create the pGWB5-PsWRKY33-GFP plasmid using the Gateway cloning system, as shown in Figure 1.

Figure 1.
Plant expression vector PGWB5-PSWRKY33-GFP containing a green fluorescent protein fusion. Fused upstream of green fluorescent protein (GFP) and under the control of the cauliflower mosaic virus (CaMV) 35S super promoter to create the pGWB5-PsWRKY33-GFP plasmid using the Gateway cloning system. Hyg R: Hygromycin B, GFP: Green fluorescent protein, Kan R: Kanamycin R.
Three treatments were applied for Arabidopsis protoplast extraction, following the methods of Fu-Hui Wu and Shu-Chen Shen et al. [11]. Arabidopsis protoplasts were transformed with PEG6000-mediated recombinant plasmids, as described by Yoo S D, Cho Y H, Sheen J. et al. [12]. Then, fluorescence microscopy was carried out to detect the fluorescent expression of the 35S-PtWRKY33-GFP fusion protein using the transient gene expression system of Arabidopsis thaliana leaf pulp protoplasts to verify the location where the subcellular expression of the PtWRKY33 protein plays a role.
2.2.3. Analysis of PtWRKY33 Expression by qRT–PCR
Two fluorescent quantitative primer pairs ((qPtW33F3: 5′-cgatttcgcttggaggat-3′; qPtW33R4: 5′-cctgctcccagcacctaaaa-3′) and (Actin2-F: 5′-ttctacaagtgctttgatggtgagttc-3′; Actin2-R: 5′-ctattcgatacatagaagatcagaatgttc-3′)) were synthesized using the Actin7 gene as an internal reference gene (XM_035067200) in Populus trichocarpa. The RNA of the corresponding treated poplar (Populus trichocarpa) roots, stems and leaves was extracted with Beijing Kangwei Biological Reagent Co., Ltd. plant RNA kits, and the total RNA was reverse-transcribed into cDNA according to the method of the TOYOBO reverse transcription kit. The relative expression of PtWRKY33 in was measured by qRT–PCR on an MxPro3000 using the SYBR Green method. Fluorescent quantitative PCR procedures were performed according to a previous method [13].
2.2.4. Expression Characteristics of PtWRKY33 under Simulated Drought Stress by PEG Treatment
Tissue culture seedlings of uniform size were selected, washed with static medium and cultured in sterilized Hoh nutrient solution with 8 h/16 h light/dark treatment at 25 ± 2 °C. After 15 days of nutrient solution culture, seedlings of uniform size with healthy growth were treated with 20% PEG6000 solution to simulate drought stress for 0 (using water as control), 6, 12, 24 and 48 h. Samples of roots, stems and leaves of seedlings were taken for freezing in liquid nitrogen.
2.2.5. Expression Characteristics of PtWRKY33 in Response to Salt (NaCl) Stress
Tissue culture seedlings of uniform size were selected, washed with static medium and cultured in sterilized Hoh nutrient solution with 8 h/16 h light/dark treatment at 25 ± 2 °C, and, after 15 days of nutrient solution culture, seedlings of uniform size with healthy growth were treated with a stress solution of 150 mM NaCl for 0 (using water as control), 6, 12, 24 and 48 h; samples of roots, stems and leaves of seedlings were taken for freezing in liquid nitrogen.
2.2.6. Expression Characteristics of PtWRKY33 in Response to Alkaline Salt (NaHCO3)
Tissue culture seedlings of uniform size were selected, washed with static medium and cultured in sterilized Hoh nutrient solution with 8 h/16 h light/dark treatment at 25 ± 2 °C, and, after 15 days of nutrient solution culture, seedlings of uniform size with healthy growth were treated with a stress solution of 60 mM NaHCO3 for 0 (using water as control), 6, 12, 24 and 48 h. Samples of roots, stems and leaves of seedlings were taken and frozen in liquid nitrogen.
2.2.7. Molecular Detection of Plant Expression Vector pGWB18-PtWRKY33 and PtWRKY33-Transformed Tobacco
PCR assays were performed to select clones with positive insertions, and the plant expression vector pGWB18-PtWRKY33, containing a CaM35S promoter and NOS terminator, was constructed by seamlessly cloning the DNA from the correct plasmid (confirmed by sequencing) into the pGWB18 vector via the Gateway system.
The DNA of plasmid pGWB18-PtWRKY33 was introduced into Agrobacterium tumefaciens EHA105 by electroshock, plated on YEP + 50 mg/L Hyg + 100 mg/L Rif plates for screening and further cultured in YEP medium with Hyg and Kana antibiotics. Then, AAM resuspension was used as the working solution, tobacco leaves were infiltrated with it and the infiltrated leaves were spread on AS medium for 72 h. The leaf pieces transferred to differentiation medium for screening, and the differentiated resistant shoots were rooted on 1/2 MS medium, washed, transplanted to grow in pots and harvested as T1-generation-resistant transformants [14]. Tobacco transgenic T1-generation strains with Hyg resistance were grown through 2 generations of thaumatin screening to obtain T3-generation-transformed tobacco. qRT–PCR was applied to measure the relative expression of the overexpressing T3-generation strains.
2.2.8. Resistance of Tobacco Overexpressing PtWRKY33 to Drought Stress Simulated by Mannitol
Full-bodied tobacco seeds of each strain were selected (T3-generation strain PtWRKY33:T3-#1, #2, #3 and WT seeds), sterilized with alcohol and sodium hypochlorite and sown in 1/2 MS medium for 15 days of germination; uniformly growing 2-leaf stage seedlings were selected and transplanted onto 1/2 MS + 0 (as control), 200, 300 and 400 mM mannitol (Mannitol) medium with three experimental replicates, then incubated in a tissue culture chamber at 25 ± 2 °C. Growth was observed and photographed after 14 days, and the fresh weight and root length were measured.
2.2.9. Tolerance Analysis of Seedling Growth to Salt (NaCl) Stress at the Seedling Stage
Salinity tolerance was analyzed in T3-generation tobacco strains overexpressing the trans-PtWRKY33 gene. The T3-generation strain (PtWRKY33: T3-#1, #2, #3) and 30 WT seeds were sterilized and sown in 7 cm × 7 cm pots and incubated in an artificial oxygen chamber at 25 ± 2 °C with a light level of 2600 LX and a light/dark time of 16/8 h. The salt (NaCl) root irrigation stress treatment was started at the 6-leaf stage by pouring 150 mM NaCl solution (50 mL) onto the roots. Phenotypes were investigated after 7 days of stress treatment, and the aboveground parts were examined for survival, fresh weight of individual plants, and malondialdehyde (MDA) and proline (Pro) contents.
2.2.10. Analysis of Seedling Growth Tolerance to Alkaline Salt (NaHCO3) Stress
Tobacco strains overexpressing the T3 generation of the PtWRKY33 gene were tested for alkaline salt tolerance. T3-generation strains (PtWRKY33: T3-#1, #2, #3) and WT seeds were sterilized and germinated on MS medium, and seedlings at the 3-leaf stage with consistent germination were washed and transplanted to 7 cm × 7 cm pots and cultured in a plant culture room at 25 ± 2 °C with a light level of 2600 LX and a light/dark time of 16/8 h. Alkaline salt treatment was started at the 7-leaf stage. Root irrigation stress treatments using 75 mM and 100 mM NaHCO3 were started at the 7-leaf stage, and clear water was used as the control (0 mM NaHCO3). The growth phenotype was observed for 14 days of stress treatment, and the fresh weight and malondialdehyde (MDA) and proline (Pro) contents of the individual plants were measured [15].
2.2.11. Characterization of NtSOD1 and NtAPX2 Gene Expression in Tobacco Leaves after Salinity Stress
The expression of adversity marker genes (NtCu/Zn-SOD1, NtAPX2) was measured in the leaves of the transgenic lines and WT plants after NaCl and NaHCO3 treatment. RNA was extracted from the top carpels, and reverse-transcribed cDNA was used as the template for the assay. qRT–PCR was performed to measure the expression of the genes, and the method was the same as that described in Section 2.2.3. The quantitative PCR primers are shown in the Supplementary Materials (Table S1).
3. Results
3.1. Cloning and Phylogenetic Analysis of the PtWRKY33 Gene in Populus trichocarpa
In this study, total RNA from the leaves and roots of Populus trichocarpa, extracted using a kit, was reverse-transcribed into cDNA and used as a template for PCR, and specific primers for RT–PCR amplification reaction products were electrophoresed to show that both had target gene bands of approximately 1800 bp (Figure 2a). An E. coli strain containing the recombinant plasmid pMD18-T-PtWRKY33 was created by transformation. The plasmid DNA was extracted using the plasmid mini-extraction kit from Conway Century and later underwent SalI/BamHI enzyme digestion and electrophoresis to confirm successful cloning (Figure 2b). Gel imaging revealed a DNA fragment of equal size to the expected target gene; the plasmid was named pMD18-T-PtWRKY33 and was sent for sequencing (#1, 2) to Coomassie Biologicals Ltd. for comparison with the XM_ 002323601.3 gene nucleotide sequence. It was found to be identical, and the PtWRKY33 gene was thus obtained.
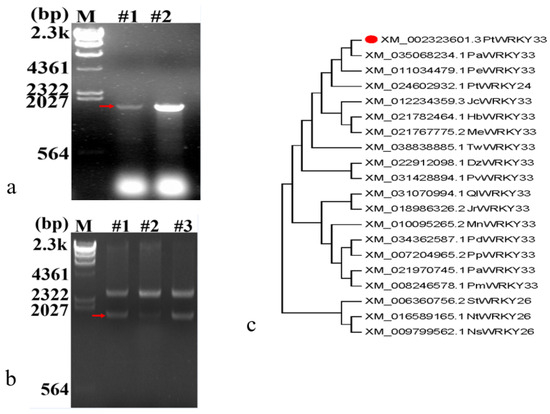
Figure 2.
Cloning and phylogenetic analysis of the PtWRKY33 gene in Populus trichocarpa. (a) Electrophoresis of PCR amplification products of full-length DNA of the PtWRKY33 gene; (b) Double digestion to identify the successful construction of pMD18-T-PtWRKY33; (c) Phylogenetic evolutionary tree of amino acid sequences of PtWRKY33 and WRKY33 transcription factors from other species. Note, Figure 2a: M is the HindIII cleavage of λDNA standard, and lanes #1 and #2 are PCR amplification products of the PtWRKY33 gene from the leaves and roots of Populus trichocarpa, respectively; Figure 2b: M is the HindIII cleavage of λDNA standard, and T-#1, T-#2 and T-#3 are the pMD18-T-PtWRKY33 gene identified by double cleavage of the plasmid DNA of Populus trichocarpa ligated into the pMD18-T vector; Figure 2c: Populus trichocarpa, PtWRKY33; Populus alba, PaWRKY33; Populus euphratica, PeWRKY33; Populus trichocarpa, PtWRKY24; Jatropha curcas, JcWRKY33; Hevea brasiliensis, HbWRKY33; Manihot esculenta, MeWRKY33; Tripterygium wilfordii, TwWRKY33; Durio zibethinus, DbWRKY33; Pistacia vera, PvWRKY33; Quercus fabri, QlWRKY33; Juglans regia, JrWRKY33; Morus notabilis, MnWRKY33; MnWRKY33; Prunus dulcis, PdWRKY33; Prunus persica, PpWRKY33; Prunus avium, PavWRKY33; Prunus mume, PmWRKY33; Solanum tuberosum, StWRKY26; Nicotiana tabacum, NtWRKY26; Nicotiana sylvestris, NsWRKY26.
An NCBI homology BlastP search of the PtWRKY33 gene, which encodes 579 amino acids, was conducted to obtain highly similar amino acid sequences from other species, and MEGA 6.0 software was used to perform a systematic analysis of these amino acid sequences. The results of the analysis were used to construct an evolutionary lineage tree, and the amino acid sequences of the WRKY proteins of poplar family species were highly homologous and in the same large branch. Poplar PtWRKY33 is in the same branch as those of poplar PaWRKY33 and poplar PeWRKY33, indicating that they are evolutionarily the most closely related (Figure 2c).
3.2. Subcellular Localization of PtWRKY33 Protein and Arabidopsis thaliana Protoplast Extraction and Recombinant Plasmid Transformation
In this study, the 579 amino acids of the protein encoded by the cloned PtWRKY33 gene were entered into the biological software PSORT, and the preliminary online plant protein subcellular localization predictions are shown. Localization in the nucleus had the highest probability, at 65.0%, followed by the microsomes at 10.0% probability, the mitochondrial matrix at 10% probability and the cell membrane at 0% probability. The Plant-mPLoc software similarly predicted that the PtWRKY33 protein would be localized to the nucleus.
The observation of pressed slices of transformed Arabidopsis protoplasts under a Caius fluorescence microscope (Olympus) revealed that the 35S-PtWRKY33-GFP fusion showed green fluorescent protein expression in the nucleus (Figure 3), demonstrating the nuclear localization of the PtWRKY33-GFP fusion protein, which is consistent with the above PSORT and Plant-mPLoc software that predicted subcellular localization to the nucleus. These results indicate that the PtWRKY33 transcriptional protein plays a transcriptional regulatory role in the nucleus.
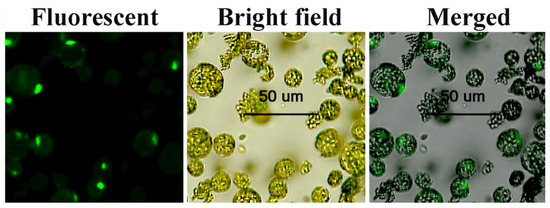
Figure 3.
PtWRKY33-GFP fusion protein localized to Arabidopsis thaliana Columbia-0 epidermal cells. The fusion proteins are shown under fluorescent, bright field and merged conditions, respectively.
3.3. Expression Characteristics of PtWRKY33 under Simulated Drought Stress by PEG Treatment
In this study, we treated poplar seedlings with 20% PEG6000 solution to simulate drought stress and analyzed the PtWRKY33 gene expression response using real-time quantitative PCR. Analysis of the qRT–PCR data showed that PEG drought stress induced the upregulated expression of the PtWRKY33 gene, and the expression in the roots, stems and leaves showed a general trend of elevation with increasing induction time (Figure 4a). PEG6000 stress induced a modest increase in the relative expression of this gene in the leaves, with the highest increase being 1.89-fold over that in untreated (0 h) leaves; in the stems, the relative expression increased 3.45-fold over that in untreated stems; and in the roots, the response was very large, reaching the highest value at 24 h, i.e., 11.6-fold over that in untreated roots. This result shows that the PtWRKY33 gene was induced by drought stress and that its expression was increased.
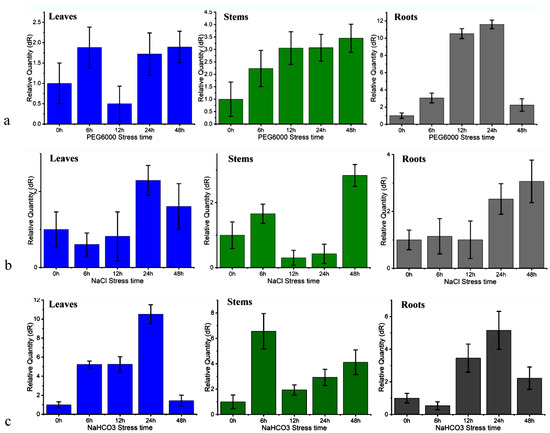
Figure 4.
Expression characteristics of PtWRKY33 under PEG, NaCl, NaHCO3 stress. (a) PtWRKY33 expression after treatment with 20% PEG6000 for 0, 6, 12, 24 and 48 h; (b) PtWRKY33 expression after treatment with 150 mM NaCl for 0, 6, 12, 24 and 48 h; (c) PtWRKY33 expression after treatment with 60 mM NaHCO3 for 0, 6, 12, 24 and 48 h, respectively.
3.4. Expression Characteristics of PtWRKY33 in Response to Salt (NaCl) Stress
In this study, the relative expression of the PtWRKY33 gene in 150 mM NaCl-stressed poplar seedlings was measured by qRT–PCR, with clear water-treated seedlings as the 0 h control, and the quantitative data obtained are shown in bar graphs in Figure 4b. As shown in the graphs, the PtWRKY33 gene responded slowly to NaCl treatment, with expression slightly decreasing in the leaves initially, and only after 24 h did salt (NaCl) induce expression. The expression of the PtWRKY33 gene in the stem was insensitive to salt treatment, with an initial decrease followed by an increase in expression to only slightly more than twice that of the control at 48 h of treatment. The expression in the root increased with the extension of salt stress time, and the expression was slightly decreased at 12 h of stress and started to respond to stress at 24 h, while PtWRKY33 expression started to be upregulated by 48 h and was still further induced significantly. The analysis of the histogram of PtWRKY33 expression induced by salinity stress in this study showed that salt could induce the expression of PtWRKY33 mRNA, but the PtWRKY33 gene was not very sensitive to salt stress.
3.5. Expression Characteristics of PtWRKY33 in Response to Alkaline Salt (NaHCO3)
In this study, 60 mM NaHCO3 was used to induce alkaline salt stress in poplar seedlings, as treatment with NaHCO3 includes both salt ions and high pH stress. The relative expression data from qRT–PCR showed that the PtWRKY33 gene was sensitive to NaHCO3 stress treatment, and its relative expression was significantly increased by NaHCO3. As shown in Figure 4c, expression of the PtWRKY33 gene increased rapidly in the leaves, with the highest expression at 24 h of treatment—10.51 times that of untreated leaves—and a decrease in expression occurring at 48 h of treatment, probably because the stress treatment was too injurious to affect the gene expression. The expression in the stems peaked at 6 h, 6.56 times that of untreated leaves, with more rapid decreases at 12 h and then 24 h. The overall trend of expression in the roots was consistent with that in the leaves, increasing and then decreasing with the prolongation of NaHCO3 stress treatment; the expression peaked at 24 h of stress, 5.16 times that in untreated roots. Although the expression of PtWRKY33 was downregulated after 48 h of treatment, it was still more than twice that in the untreated seedlings; alkaline salt stress induced the expression of this gene on the one hand, and affected its expression on the other hand, indicating that alkaline salt stress treatment may both induce and destroy the expression of PtWRKY33, and the mRNA synthesis and expression of the alkaline salt response gene are highly sensitive to it.
3.6. Molecular Detection of Plant Expression Vector pGWB18-PtWRKY33- and PtWRKY33-Transformed Tobacco
DNA fragments were obtained by using ORF cloning primers in PCR and cloned and inserted into the pQBV3 introductory vector; the sequenced plasmid DNA of the resulting plasmid, pQBV3-PtWRKY33, was mixed with pGWB18 plasmid DNA, followed by seamless fusion by adding LR reaction enzyme and incubating at 25 °C. The DNA was amplified by transforming into clone strain TOP10, and a kit was used to extract pGWB18-PtWRKY33 plasmid DNA. The constructed vector DNA was digested by XbaI and SacI restriction endonucleases, and agarose gel electrophoresis imaging showed a target gene band of approximately 1800 bp, indicating that the binary plant expression vector pGWB18-PtWRKY33 was successfully constructed. The DNA of plasmid pGWB18-PtWRKY33 was electrotransformed into Agrobacterium rhizogenes EHA105 and confirmed by PCR, and Agrobacterium strains containing pGWB18-PtWRKY33 were thus obtained. Identification of the binary plant expression vector pGWB18-PtWRKY33 by double restriction enzyme digestion and PCR detection of Agrobacterium tumefaciens EHA105containing pGWB18-PtWRKY33 are shown in the Supplementary Materials (Figures S1 and S2).
In this study, the leaves of transformed Nicotiana benthamiana were infiltrated with Agrobacterium rhizogenes EHA105, and thaumatin-resistant T1-generation-transformed strains (50 mg/L Hyg) were obtained on screening and differentiation culture during seedling cultivation; seeds were then germinated by screening on 50 mg/L Hyg and harvested in transplant culture to create T3-generation seeds. These T3-generation seeds (strains #1, #2 and #3) were sterilized and inactivated, sown on solid 1/2 MS medium + 35 mg/L Hyg and germinated and grown for 15 days, as shown in Figure 5a. Seedlings grown on 1/2 MS with consistent growth and development were used as a control. Wild-type tobacco seeds on Hyg selection medium were able to germinate, and two cotyledons unfolded, but the seedlings stopped growing and developing, whereas the transgenic T3 strains further grew and developed into plants that showed Hyg-resistant growth. Seedlings of WT and the transgenic T3-#1, 2 and 3 strains at the 6-leaf stage were grown in pots (WT was germinated on 1/2 MS); PCR confirmed the genomic integration of the PtWRKY33 gene in the leaves of the plants grown in cultivation for 20 days (Figure 5b). qRT–PCR revealed particularly high expression of the target gene relative to the WT in the transgenic T3-#1, 2 and 3 strains, with all three strains exceeding 12-fold higher expression (Figure 5c). These strains were used in the subsequent resistance test.
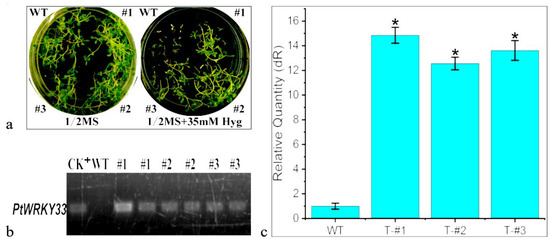
Figure 5.
Molecular detection of transgenic tobacco with the PtWRKY33 gene. (a) Germination of T3 transgenic tobacco lines with the PtWRKY33 gene and resistance to hygromycin; (b) PCR confirming genomic integration in T3 transgenic tobacco lines with the PtWRKY33 gene; (c) Gene expression of the PtWRKY33 transgenic line under the 35S promoter. * The difference was significant at the p < 0.05.
3.7. Drought Resistance of PtWRKY33-Transformed Tobacco
The resistance of the trans-PtWRKY33 gene was tested by simulating drought stress with mannitol. The germinated seedlings showed differences in resistance after 14 days of growth on a medium containing different concentrations of mannitol, and it was phenotypically evident that the transformed strains grew more robustly than the WT (Figure 6a); as indicated by the measured root length data, the transgenic strains differed significantly from the WT and grew longer roots than the WT (Figure 6b). By comparative analysis of the measured fresh weight of the plants, the fresh weight of the transgenic plants within the treatment groups was found to differ significantly from that of the WT (Figure 6c); the differences between the treatment groups were greater with increasing mannitol concentration, and the growth of the overexpressing plants outperformed that of the WT, indicating that the resistance to drought stress could be improved under PtWRKY33 overexpression driven by the 35S promoter.
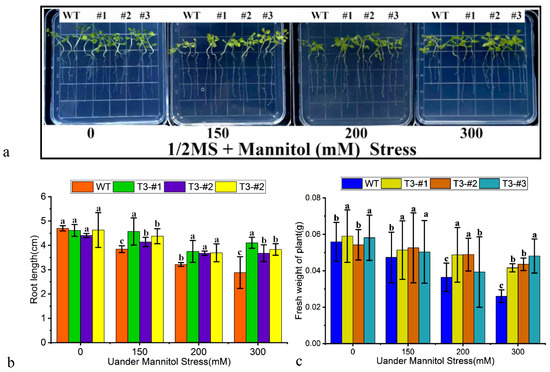
Figure 6.
Drought resistance of PtWRKY33-transformed tobacco. (a) Growth phenotypes of transgenic tobacco plants with the PTWRKY33 gene under drought stress simulated by mannitol. The phenotype under the addition of 0, 150, 200 and 300 mM mannitol to 1/2 MS was observed after 14 days; (b) Root growth of transgenic tobacco seedlings overexpressing the PTWRKY33 gene under drought stress simulated by mannitol. The main root length of the plants was measured after incubation on 1/2 MS containing 0, 150, 200 and 300 mM mannitol for 14 days (n = 3, p < 0.05); (c) Fresh weight of transgenic tobacco seedlings overexpressing the PTWRKY33 gene under drought stress simulated by mannitol. The fresh weight of the plants was measured after 14 days of growth under the stress of 0, 150, 200 and 300 mM mannitol in 1/2 MS (n = 3, p < 0.05). (a–c indicate p < 0.05).
3.8. Salt-Tolerant (NaCl) Growth of Tobacco Overexpressing PtWRKY33
Seedlings of wild-type and PtWRKY33-overexpressing tobacco were treated with a 150 mM NaCl solution by root irrigation to constitute an environment for growth under salt stress, thus enabling the analysis of the salt resistance of the overexpressing strains. The stress treatments showed differences in damage by salt stress after 7 days of growth, as shown in Figure 7a. The three transgenic strains had fewer dried, dead leaves than the wild type, and although all of the plants had died, the wild type had significantly more dried, dead leaves. The measured fresh weight data showed that each transgenic strain had a significantly higher fresh weight than the WT (Figure 7b). The reactive oxygen species-related malondialdehyde physiological index was measured in the leaves. Significance of difference analysis showed that the difference in MDA content between the positive control with the addition of water was not significant, the treatment group with the addition of NaCl showed a significant difference, and the transformed strain had less malondialdehyde content than the WT (Figure 7c), indicating an increase in oxidative damage produced by salt tolerance in the transgenic strain.
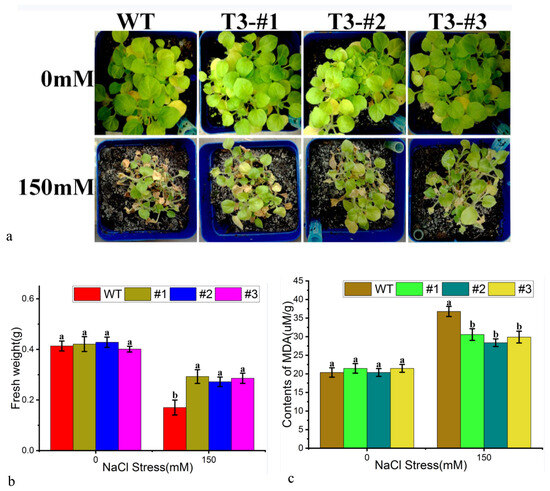
Figure 7.
Salt-tolerant (NaCl) growth of tobacco overexpressing PtWRKY33. (a) Plant growth of PtWRKY33-overexpressing and WT plants under 0 mM and 150 mM NaCl stress; (b) The fresh weight of plants overexpressing PtWRKY33 and WT plants under 0 mM or 150 mM NaCl stress; a and b indicate significant differences at the p < 0.05 level; (c) The malondialdehyde (MDA) contents in the leaves of plants overexpressing PtWRKY33 and WT plants under 0 mM or 150 mM NaCl stress; a and b indicate p < 0.05 level.
3.9. Alkaline Salt-Tolerant Growth of Tobacco Overexpressing PtWRKY33
Alkaline saline stress leads to both salt ion injury and high pH effects. In this study, wild-type and PtWRKY33-transformed tobacco plants were treated with an alkaline salt NaHCO3 solution by root irrigation to simulate an alkaline saline salt stress environment, and the alkaline saline tolerance was analyzed by testing the PtWRKY33 gene-expressing strains. The stress treatments showed growth differences after 14 days of growth, as shown in Figure 8a. The growth phenotypes of the three transgenic strains under increasing alkaline stress gradually showed increased injury, growth was inhibited and no buds appeared. The growth of each plant under the treatment also differed; single fresh weights were compared, revealing that the growth of the overexpressing strain was greater than that of the WT at 75 and 100 mM NaHCO3 (Figure 8b). The stress-affected physiological indicators—the malondialdehyde (MDA) and proline (Pro) contents—were examined, and the significance of differences by DPS analysis (Figure 8c,d) showed that there was no difference in the control and a significant difference between the treated transgenic strains and the WT. We speculate that overexpression of the PtWRKY33 gene may be related to reactive oxygen species homeostasis and is directly or indirectly involved in the ROS regulatory pathway.
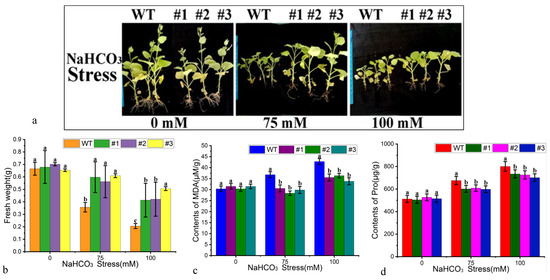
Figure 8.
Alkaline salt-tolerant growth of tobacco overexpressing PtWRKY33. (a) Plants overexpressing PtWRKY33 and WT plants were treated with 0 mM, 75 mM and 100 mM NaHCO3; (b) The fresh weight of plants overexpressing PtWRKY33 and WT plants treated with 0 mM, 75 mM and 100 mM NaHCO3 (a–c indicate p < 0.05); (c) Under the stress of 0 mM, 75 mM and 100 mM NaHCO3, the malondialdehyde (MDA) contents in the leaves of tobacco plants overexpressing PtWRKY33 and WT tobacco leaves were measured (a–c indicate p < 0.05); (d) Under the stress of 0 mM, 75 mM and 100 mM NaHCO3, the content of proline in the leaves of tobacco overexpressing PtWRKY33 and WT tobacco leaves was determined (a–c indicate p < 0.05).
3.10. Expression Characteristics of the NtSOD1 and NtAPX2 Genes in Tobacco Leaves after Salinity Stress
The most direct manifestation of the effects of salt (NaCl) and alkaline (NaHCO3) stress is the oxidative stress induced in cells. The genes encoding the key enzymes of the reactive oxygen species scavenging system are superoxide dismutase (SOD), ascorbate peroxidase (APX) and catalase. In this study, we determined using qRT–PCR that NaCl stress treatment was able to induce an increase in the expression of the NtSOD1 and NtAPX2 genes, and the expression of transgenic plants was significantly higher than that of WT plants in the treatment group by up to 4-fold. NaHCO3 treatment had a greater effect on tobacco, and the NtSOD1 and NtAPX2 genes responded strongly to the 75 mM stress. The expression of NtSOD1 in the transgenic strain was 4.12 times higher than that of the control WT under 75 mM stress and 6.18 times higher when the stress concentration was increased to 100 mM; the expression of NtAPX2 in the transgenic strain was 5.11 times higher than that of the control WT under 75 mM stress and 8.53 times higher when the stress concentration was increased to 100 mM; the significant difference between the expression of these two genes under the above two treatments and that of the WT may be caused by overexpression of the PtWRKY33 gene, which enhanced the antioxidant stress resistance of the tobacco strain and improved the salinity tolerance of the transgenic tobacco (Figure 9).
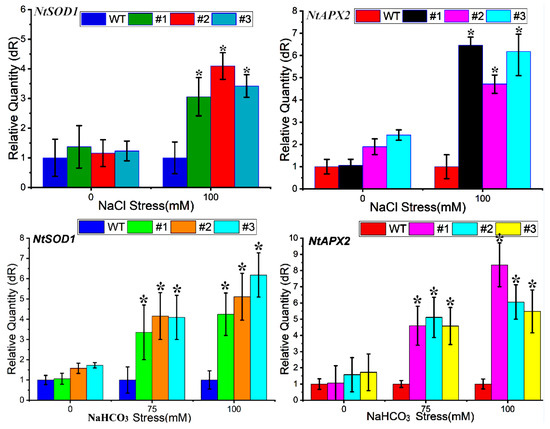
Figure 9.
Expression of the NtSOD1 and NtAPX2 genes in tobacco leaves under NaCl (0, 100 mM) and NaHCO3 (0, 75, 100 mM) stress. * p < 0.05.
4. Discussion
Transgenic engineering has become an important tool to improve plant stress tolerance. Water deficit (drought) and salinity are unfavorable factors affecting plant growth and development. Recent studies have shown that the regulation of TFs in response to adversity can alter stress tolerance phenotypes in different species. WRKY proteins are a class of transcriptional regulators unique to plants that are involved in development, senescence, and stress resistance and have received increasing attention for their role in the regulation of plant responses to various biotic and abiotic stresses [16,17,18,19,20]. In recent years, there have been an increasing number of reports of cloning WRKY genes to study their functions in model plants such as tobacco, but no papers have been published on the role of WRKY33 in drought and salinity tolerance in Populus trichocarpa. In this study, to investigate the role of the PtWRKY33 transcription factor in trees, the PtWRKY33 gene, with an open reading frame (ORF) of 1740 bp encoding 579 amino acids, was cloned from Populus trichocarpa. Homology comparison of the protein showed high similarity between Populus trichocarpa, Populus euphratica and Populus alba.
There are several immune systems, such as ETI and PTI, in plants, and the WRKY gene family plays an important role in these immune systems [21], while the activation of ETI and PTI resistance has similarities with systemic acquired resistance, both of which are signal transduction pathways that depend mainly on salicylic acid (SA) and jasmonic acid (JA) regulation. Most studies have shown that plants are regulated by WRKY transcription factors when they produce defense functions against the outside world [22]. The expression of the alfalfa MsWRKY33 protein in response to abiotic stress may have an important role in regulating the plant response to high salt stress [23]. In this study, the PtWRKY33 gene was determined to be induced and its mRNA expression relatively increased in PEG6000-, NaCl- and NaHCO3-treated poplar seedlings, but the PtWRKY33 gene was not very sensitive to salt stress; it was shown to be sensitive to NaHCO3 as a high-salt ion and high-pH stress in response to stress treatment and was relatively induced by NaHCO3. It is speculated that PtWRKY33 transcription factor expression in response to adverse stress conditions is involved in the stress tolerance regulation pathway.
Validation of the regulatory response function of WRKY transcription factors in plants has become an important research topic [24]. In the present study, tobacco lines overexpressing PtWRKY33 were obtained by genetic transformation, and PEG was used to assay their drought resistance characteristics at the germination stage; the transgenic lines had better drought resistance than the WT, which indicated that PtWRKY33 was able to improve drought stress tolerance and was closely related to drought tolerance regulation. Some studies have reported that overexpression of GhWRKY33, TaWRKY2 and TaWRKY19 in Arabidopsis enhances salt tolerance and drought tolerance in transgenic plants compared to controls due to their overexpression [24,25].
In this study, to analyze the salt (NaCl) and alkaline (NaHCO3) tolerance of plants overexpressing the PtWRKY33 gene, transformed plants and WT seedlings in pots were treated with NaCl and NaHCO3 solutions and found to be injured, with different phenotypes of growth inhibition and more growth inhibition with increasing treatment, but the overexpressing strain grew significantly more than WT, showing that overexpression produces plant salinity-tolerant growth characteristics. We speculate that PtWRKY33-mediated defense mechanisms may be involved in ROS system defense responses. qRT–PCR was used to determine that NaCl and NaHCO3 stress treatments were able to induce an increase in the expression of the NtSOD1 and NtAPX2 genes, and the significant increase in expression under treatment compared with WT may be caused by the overexpression of the PtWRKY33 gene. Overexpression of the PtWRKY33 gene in tobacco strains increased the antioxidant stress capacity and improved the salinity tolerance of transgenic tobacco. This result also suggests that overexpression of the PtWRKY33 gene enhances the role of the ROS system, and the significant difference in malondialdehyde content indicates the ability to enhance antioxidant stress injury and thus salinity tolerance under stress. The role of ROS in the defense response is mainly related to the SOD and APX enzymes, scavenging reactive oxygen species with the superoxide dismutation reaction and ascorbate peroxidase [26]. In this study, we found that NaCl and NaHCO3 stress treatments were able to induce an increase in the expression of the NtSOD1 and NtAPX2 genes, and the significant increase in expression under treatment compared to WT may be due to the overexpression of the PtWRKY33 gene, and overexpression of the PtWRKY33 gene in tobacco lines led to enhanced antioxidant stress capacity and improved salinity tolerance in transgenic tobacco.
In summary, we believe that the molecular mechanism in the defense response process mediated by PtWRKY33 is quite complex, and the PtWRKY33 transcription factor may be involved in multiple signaling pathways interacting with several defense genes or may be directly or indirectly involved in the reactive oxygen species regulatory system, which remains to be studied in depth.
5. Conclusions
We cloned the PtWRKY33 gene from the leaves and roots of Populus trichocarpa, and the amino acid homology comparison showed high similarity with Populus euphratica and Populus alba of the Populus family. Subcellular localization of PsWRKY33 protein expression in the nucleus was observed by fusion-linking the green fluorescent protein GFP to the PtWRKY33 gene with PEG6000-mediated infiltration of transformed protoplasts. We also examined the increase in PtWRKY33 gene expression induced by PEG6000, NaCl and NaHCO3 solution treatment of poplar seedlings. The response to salt stress was not very sensitive, and the response to NaHCO3 stress treatment was sensitive, and the relative expression was significantly increased by it. We detected that NaCl and NaHCO3 stress treatments induced an increase in the expression of NtSOD1 and NtAPX2 genes, which are regulatory genes associated with drought and salinity tolerance in plants, and overexpression of PtWRKY33 gene in tobacco lines enhanced antioxidant stress capacity and improved the salinity-tolerant growth of transgenic tobacco. This study provides a theoretical basis for exploring the physiological mechanisms of drought and salinity tolerance in trees and their genetic factors and can also be widely applied to solving practical problems in tree molecular breeding production. The future research may improve the breeding of poplars for resistance.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/f14102039/s1, Table S1: Table of quantitative PCR primers; Figure S1: Identification of the binary plant expression vector pGWB18-PtWRKY33 by double restriction enzyme digestion; Figure S2: PCR detection of Agrobacterium tumefaciens EHA105 containing pGWB18-PtWRKY33.
Author Contributions
C.Y. and Q.G. conceived and designed the experiments; M.J. and L.Z. performed the experiments, analyzed the data and wrote the paper. Q.G. and J.S. contributed reagents/materials/analysis tools. All authors have read and agreed to the published version of the manuscript.
Funding
We gratefully acknowledge financial support for this research from Key Laboratory of Saline-alkali Vegetation Ecology Restoration (Northeast Forestry University), Ministry of Education (0924210201); National Natural Science Foundation of China (32171989); and Science and Technology Fundamental Resources Investigation Special Project (2021FY100403).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Xie, Z.W.; Wang, L.J.; Chen, J.Y.; Wang, J.; Su, Y.J.; Yang, X.S. Studies on WRKY transcription factors and their biological functions in plants. J. Agric. Sci. Technol. Iran 2016, 18, 46–54. [Google Scholar] [CrossRef]
- Zhang, M.L.; Han, L.B. Research progress on plant response of ABA and H2O2 signals to cold-stress. Grassl. Turf. 2013, 33, 92–96. [Google Scholar] [CrossRef]
- Yu, D.Q.; Chen, L.G.; Zhang, L.P. Transcription factor WRKY superfamily: Origin, structure and function. Acta Bot. Yunnanica 2006, 28, 69–77. [Google Scholar] [CrossRef]
- Li, Y.Y.; Gao, Z.Q.; Cao, Q.H. Bioinformatics analysis of SPF1 transcription factors from sweet potato[Lpomoea batatas (L.) Lam]. Jiangsu J. Agric. Sci. 2017, 33, 760–767. [Google Scholar]
- Wang, Q.Y.; Xue, D.Q.; Chen, X.L.; Li, J.F.; Wang, A.X. Bioinformatics and functional analysis of WRKY transcription factors in interaction between tomato and Cladosporium fulvum. J. Henan Agric. Sci. 2017, 46, 74–79. [Google Scholar] [CrossRef]
- Kim, S.H.; Bahk, S.; Nguyen, N.T.; Pham, M.L.A.; Kadam, U.S.; Hong, J.C.; Chung, W.S. Phosphorylation of the auxin signaling transcriptional repressor IAA15 by MPKs is required for the suppression of root development under drought stress in Arabidopsis. Nucleic Acids Res. 2022, 50, 10544–10561. [Google Scholar] [CrossRef]
- Shelake, R.M.; Kadam, U.S.; Kumar, R.; Pramanik, D.; Singh, A.K.; Kim, J.-Y. Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives. Plant Commun. 2022, 3, 100417. [Google Scholar] [CrossRef]
- Chen, Q.Q. Isolation and Expression Analysis of Genes Induced under High Temperature and Chilling Stress in Populous simonii. Master’s Thesis, Beijing Forestry University, Beijing, China, 2013. [Google Scholar]
- Zheng, H. Functional Analysis of Rice Zinc Finger Protein Gene OsLOL2, OsC2HC-1 Responsed to Salinity Stress. Master’s Thesis, Northeast Forestry University, Harbin, China, 2013. [Google Scholar]
- Lu, Z.Q. The Studies on Function and Chaeacterization of APX and CAT Isoenzymes from Rice and Their Relationship with Salinity Stresses. Ph.D. Thesis, Northeast Forestry University, Harbin, China, 2005. [Google Scholar]
- Wu, F.H.; Shen, S.C.; Lee, L.Y.; Lee, S.H.; Chan, M.T.; Lin, C.S. Tape-Arabidopsis Sandwich—A Simpler Arabidopsis Protoplast Isolation Method. Plant Methods 2009, 5, 16. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Guan, Q.J.; Wang, L.F.; Bu, Q.Y.; Wang, Z.Y. The rice gene OsZFP6 functions in multiple stress tolerance responses in yeast and Arabidopsis. Plant Physiol. Biochem. 2014, 82, 1–8. [Google Scholar] [CrossRef]
- Niu, Y.T.; Li, X.T.; Xu, C.; Ajab, Z.; Liu, Q.; Majeed, Z.; Guan, Q.J. Analysis of drought and salt-alkali tolerance in tobacco by overexpressing WRKY39 gene from Populus trichocarpa. Plant Signal. Behav. 2021, 16, 1918885. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, N.; Qian, X.G.; Yang, W. Effects of different concentrations of heavy metals on tobacco proline content in four soil. Guizhou Agric. Sci. 2014, 42, 127–131. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Fu, Q.T.; Chen, L.G.; Huang, W.D.; Yu, D.Q. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.R.; Dong, Q.Y.; Yu, D.Q. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 2012, 185, 288–297. [Google Scholar] [CrossRef]
- Wang, C.; Deng, P.Y.; Chen, L.L.; Wang, X.T.; Ma, H.; Hu, W.; Yao, N.C.; Feng, Y.; Chai, R.H.; Yang, G.X.; et al. A wheat WRKY transcription factor TaWRKY10 confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS ONE 2013, 8, e65120. [Google Scholar] [CrossRef]
- Wang, Y.N.; Dang, F.F.; Liu, Z.Q.; Wang, X.; Eulgem, T.; Lai, Y.; Yu, L.; She, J.J.; Shi, Y.L.; Lin, J.H.; et al. CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol. Plant Pathol. 2013, 14, 131–144. [Google Scholar] [CrossRef]
- Eulgem, T.; Somssich, I.E. Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 2007, 10, 366–371. [Google Scholar] [CrossRef]
- Naoumkina, M.A.; He, X.Z.; Dixon, R.A. Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biol. 2008, 8, 1–14. [Google Scholar] [CrossRef]
- Zhang, J.J.; Wang, Y.P.; Wang, X.; Zhang, S.; Wang, X. Antibody preparation of MsWRKY33 protein and response to abiotic stress. Acta Agric. Boreali Sin. 2021, 36, 8. [Google Scholar] [CrossRef]
- Wei, X. Cloning and Functional Analysis of GhWRKY33 Related to Drought-and Senescence-Resistance in Gossypium hirsutum. Master’s Thesis, Hebei Agricultural University, Baoding, China, 2020. [Google Scholar]
- NIU, C.F.; Wei, W.; Zhou, Q.-Y.; Tian, A.-G.; Hao, Y.-J.; Zhang, W.-K.; Ma, B.; Lin, Q.; Zhang, Z.-B.; Zhang, J.-S.; et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012, 35, 1156–1170. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.; Dixon, R.A. The oxidative burst in plant disease resistance. Annu. Rev. Plant Biol. 1997, 48, 251–275. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).