Tree Species Classification over Cloudy Mountainous Regions by Spatiotemporal Fusion and Ensemble Classifier
Abstract
1. Introduction
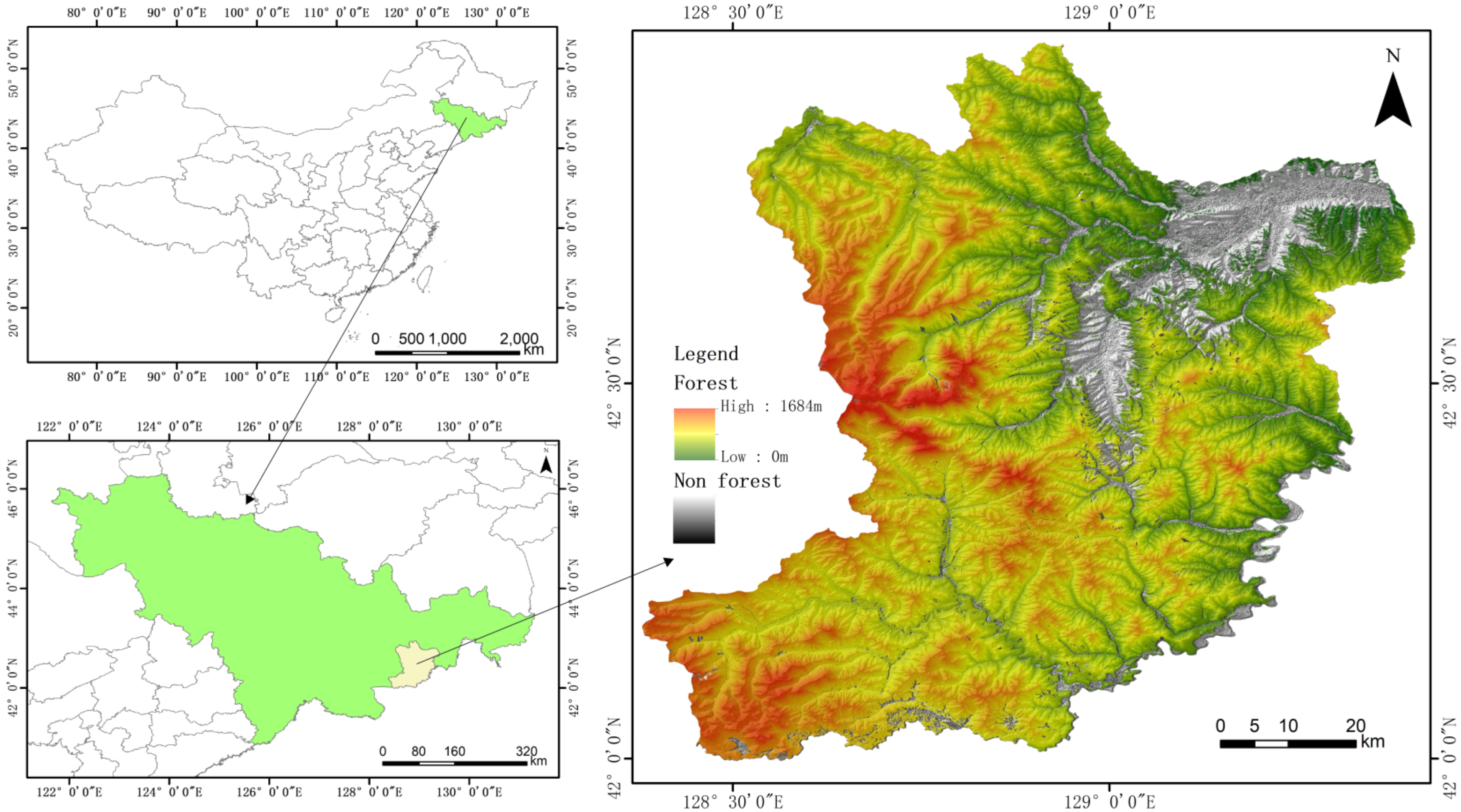
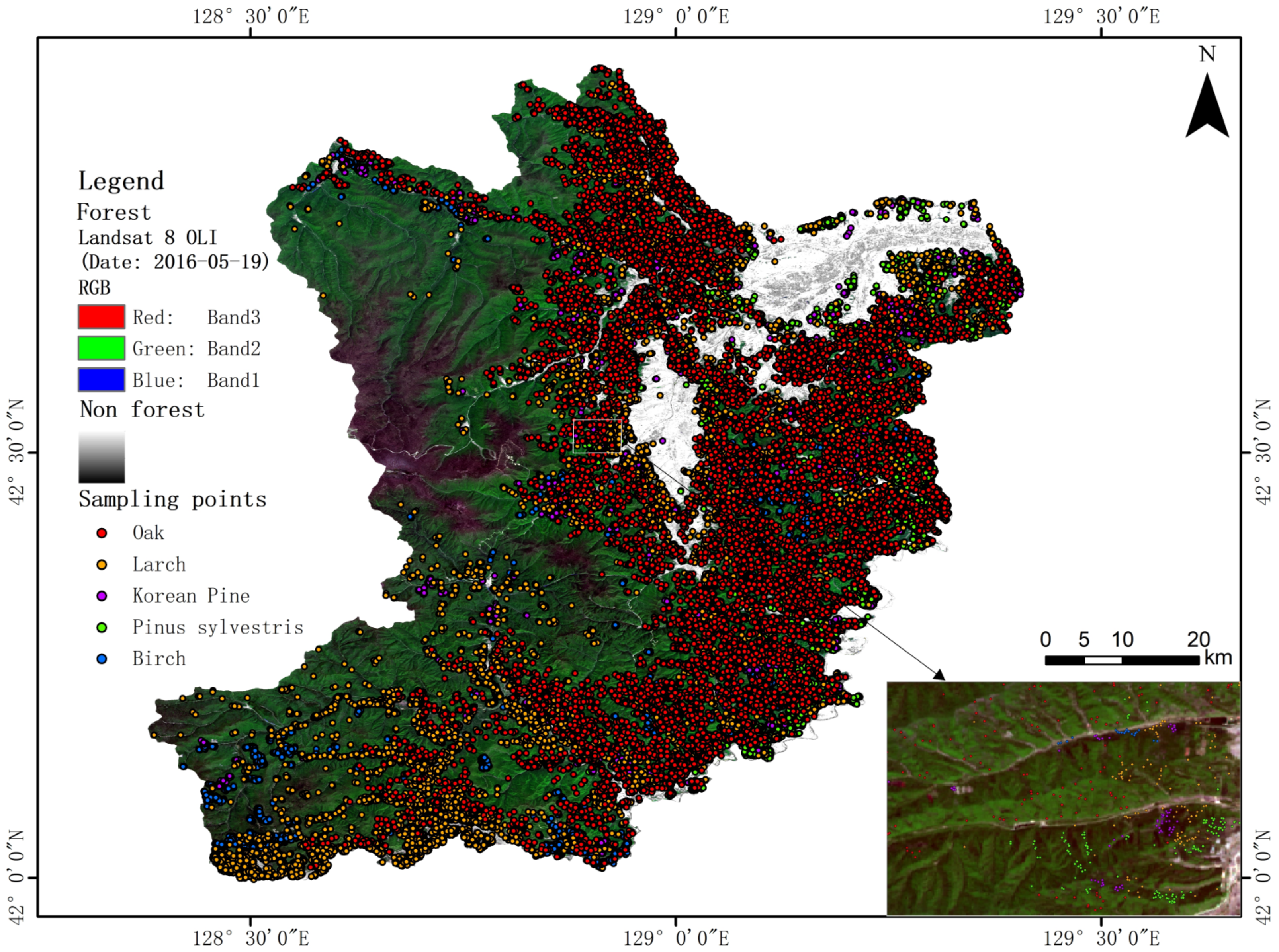
2. Study Area
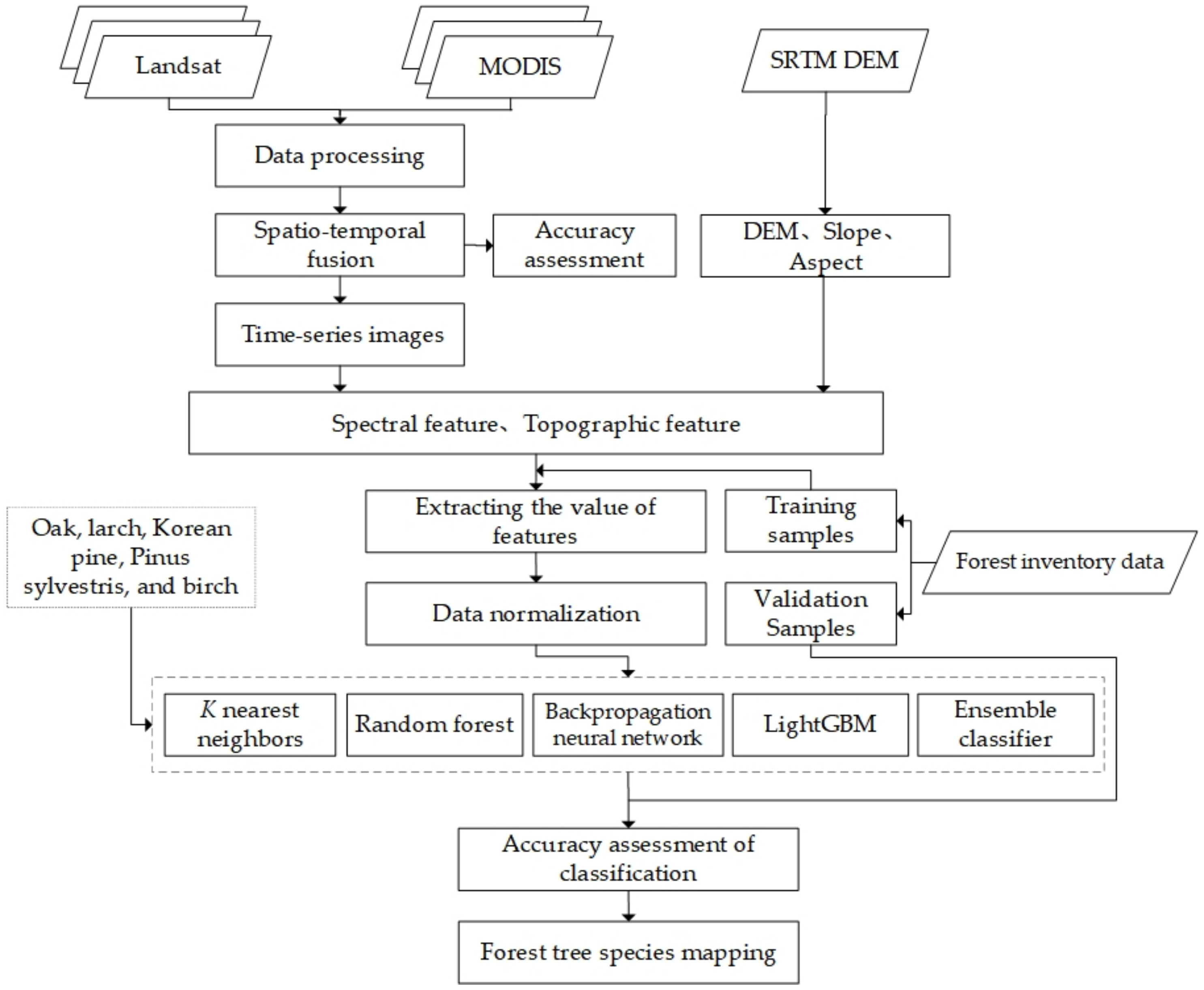
3. Data and Methods
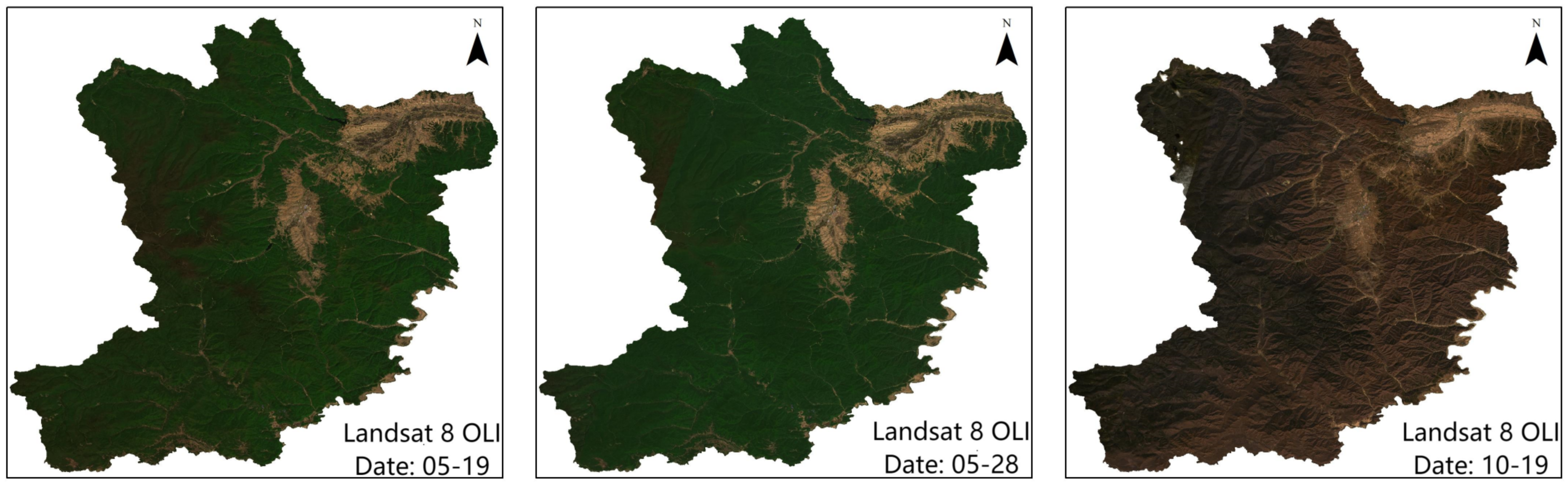
3.1. Remote Sensing Data and Preprocessing
3.2. Field Data
3.3. Spatiotemporal Fusion
3.3.1. STARFM
3.3.2. FSDAF
3.3.3. STNLFFM
3.4. Tree Species Classification
3.5. Accuracy Assessment
4. Results
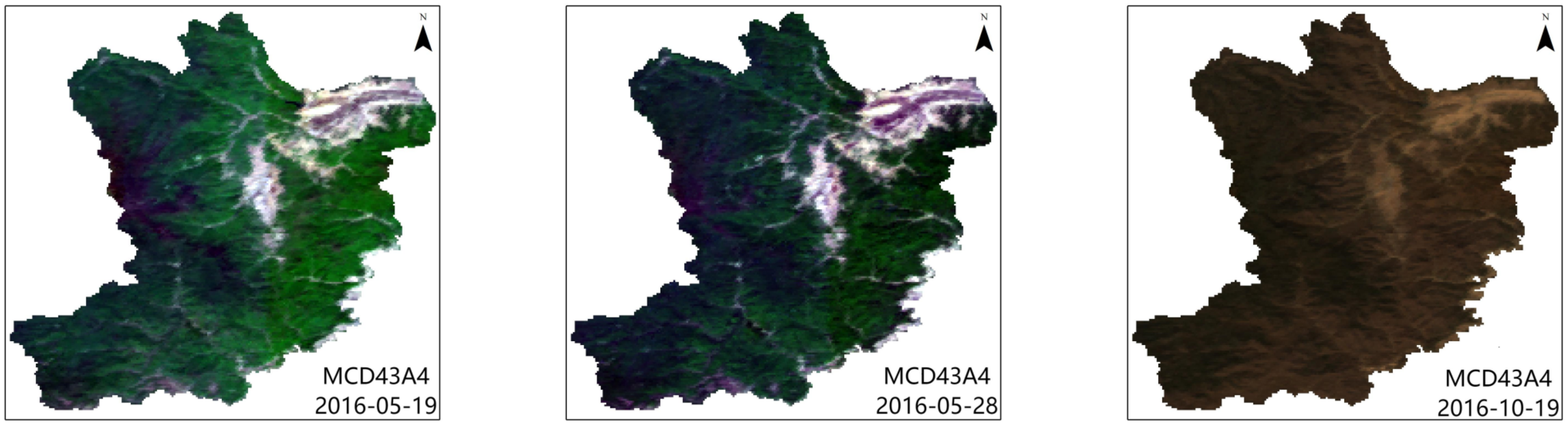
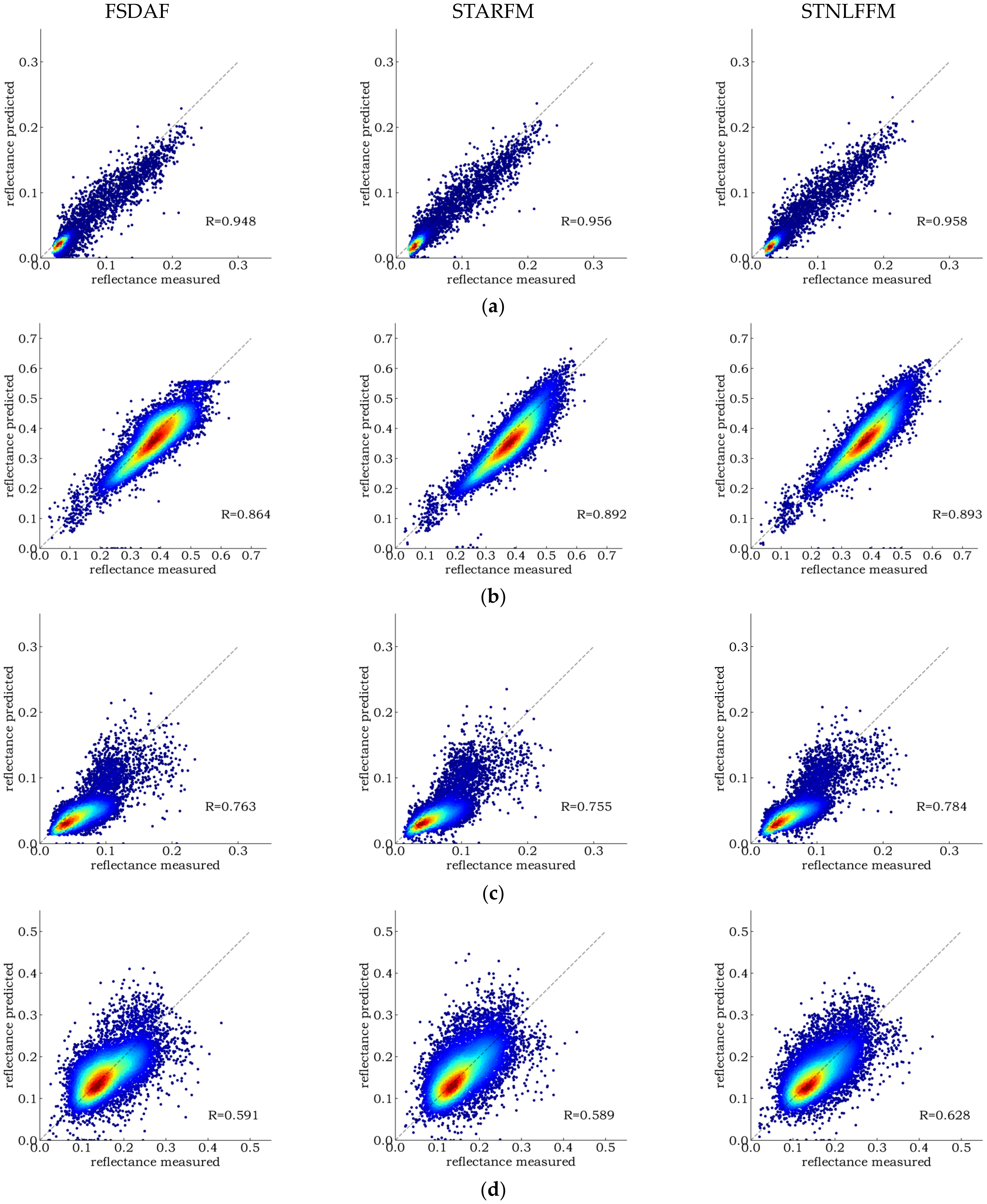
4.1. Image Fusion and Evaluation
4.2. Classification and Mapping of Tree Species
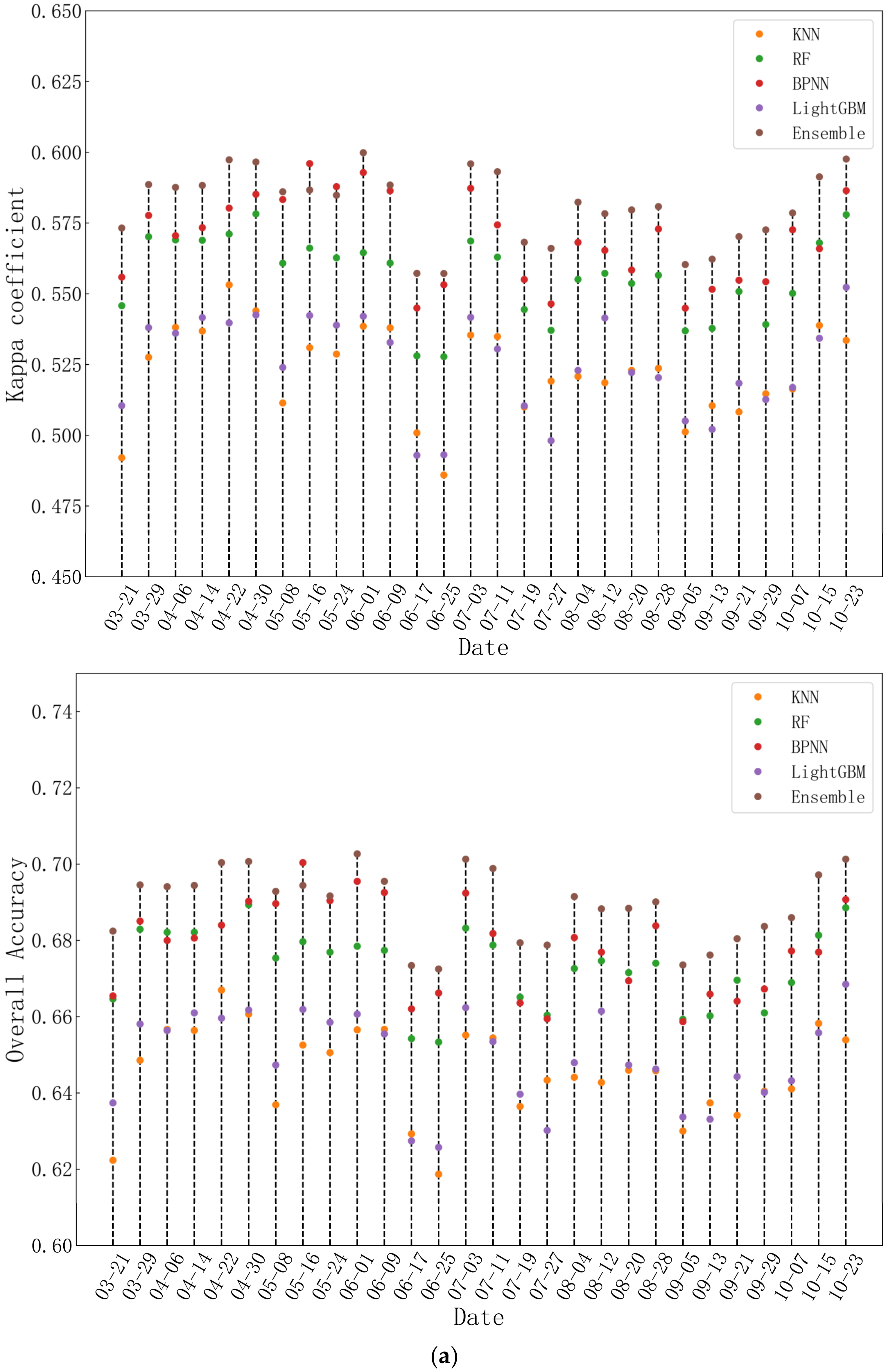
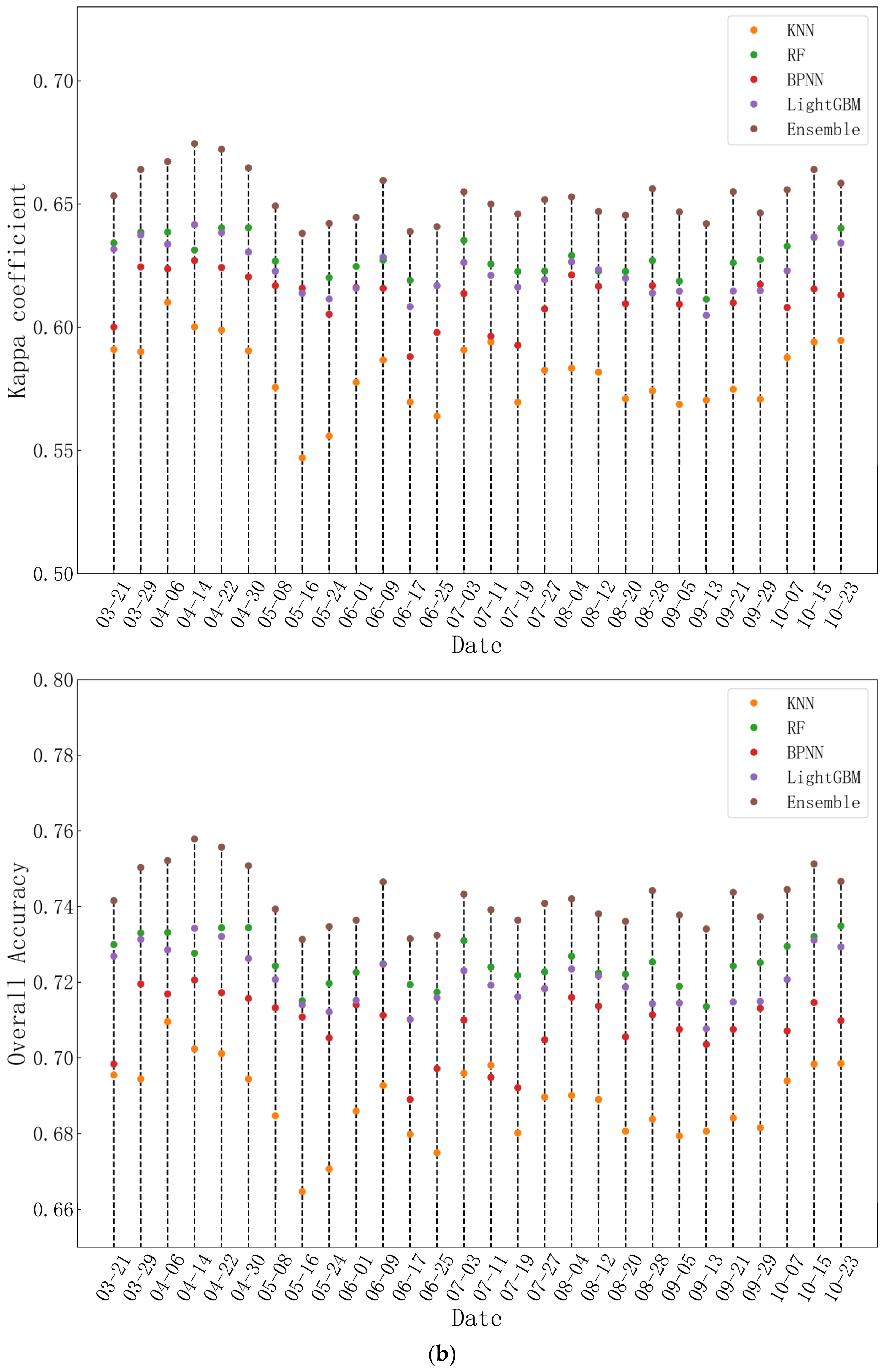
4.2.1. The Overall Accuracy of Five Tree Species
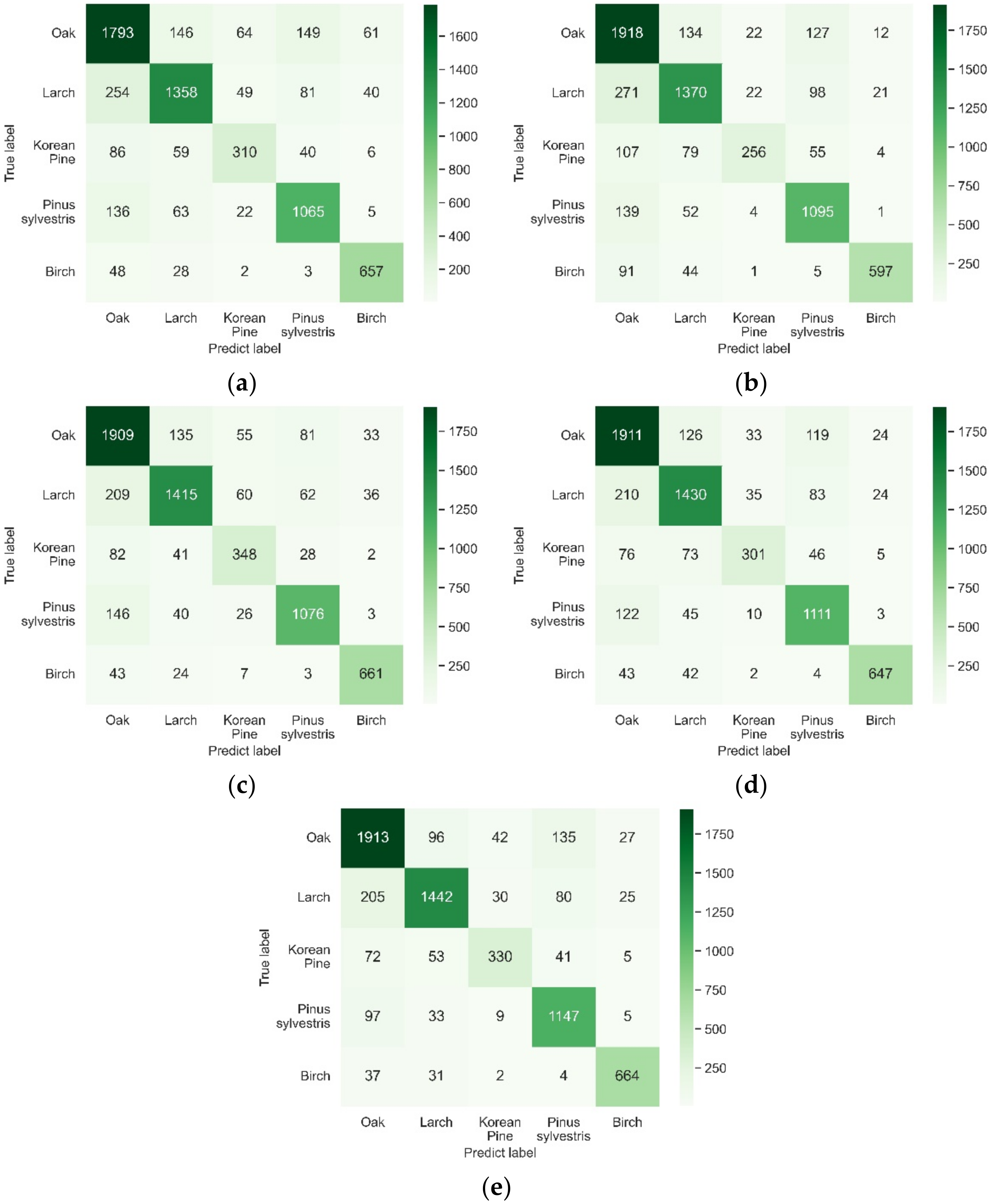
4.2.2. The Class-Wise Accuracies of Five Tree Species
4.2.3. Comparison with Other Ensemble Models
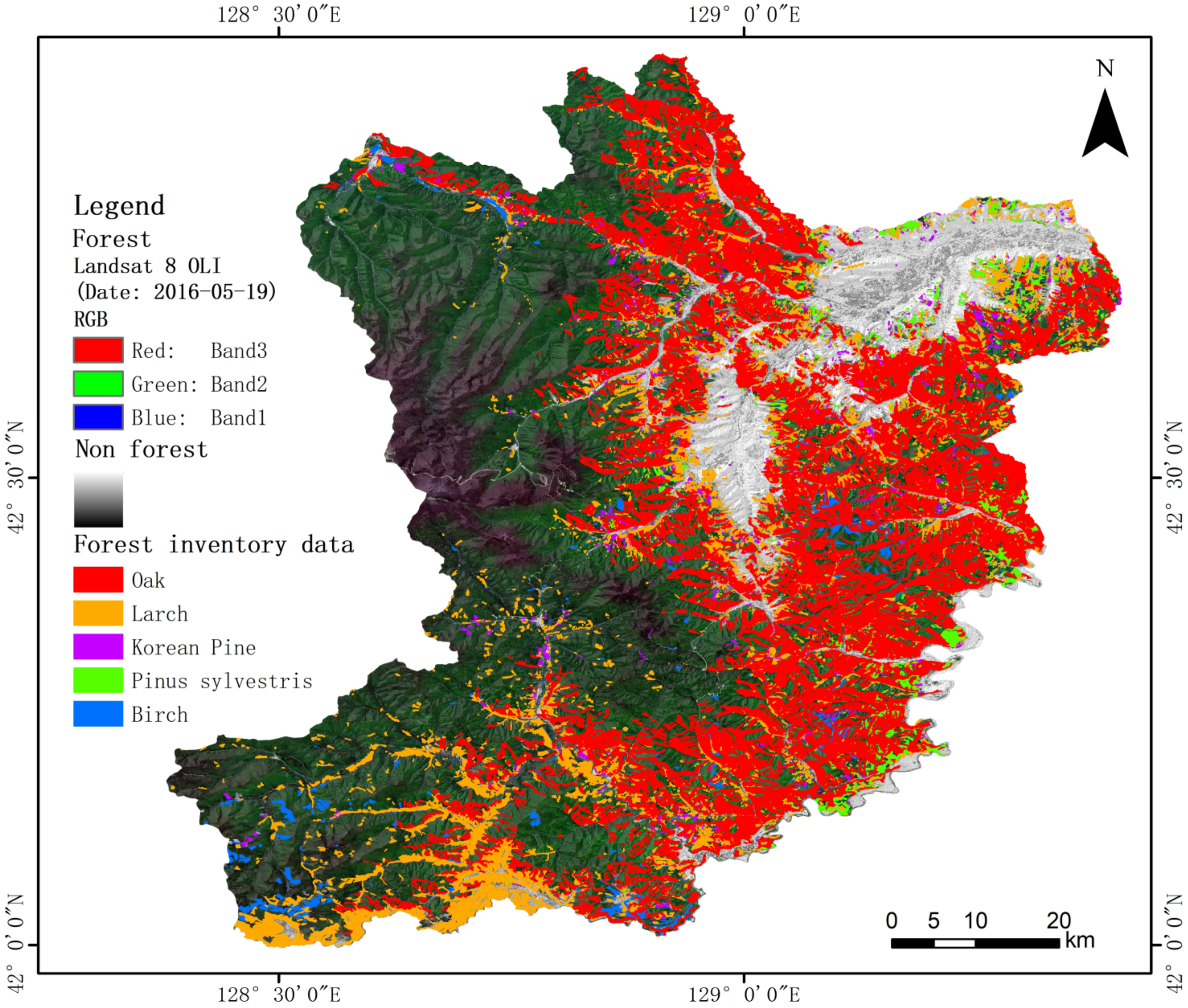
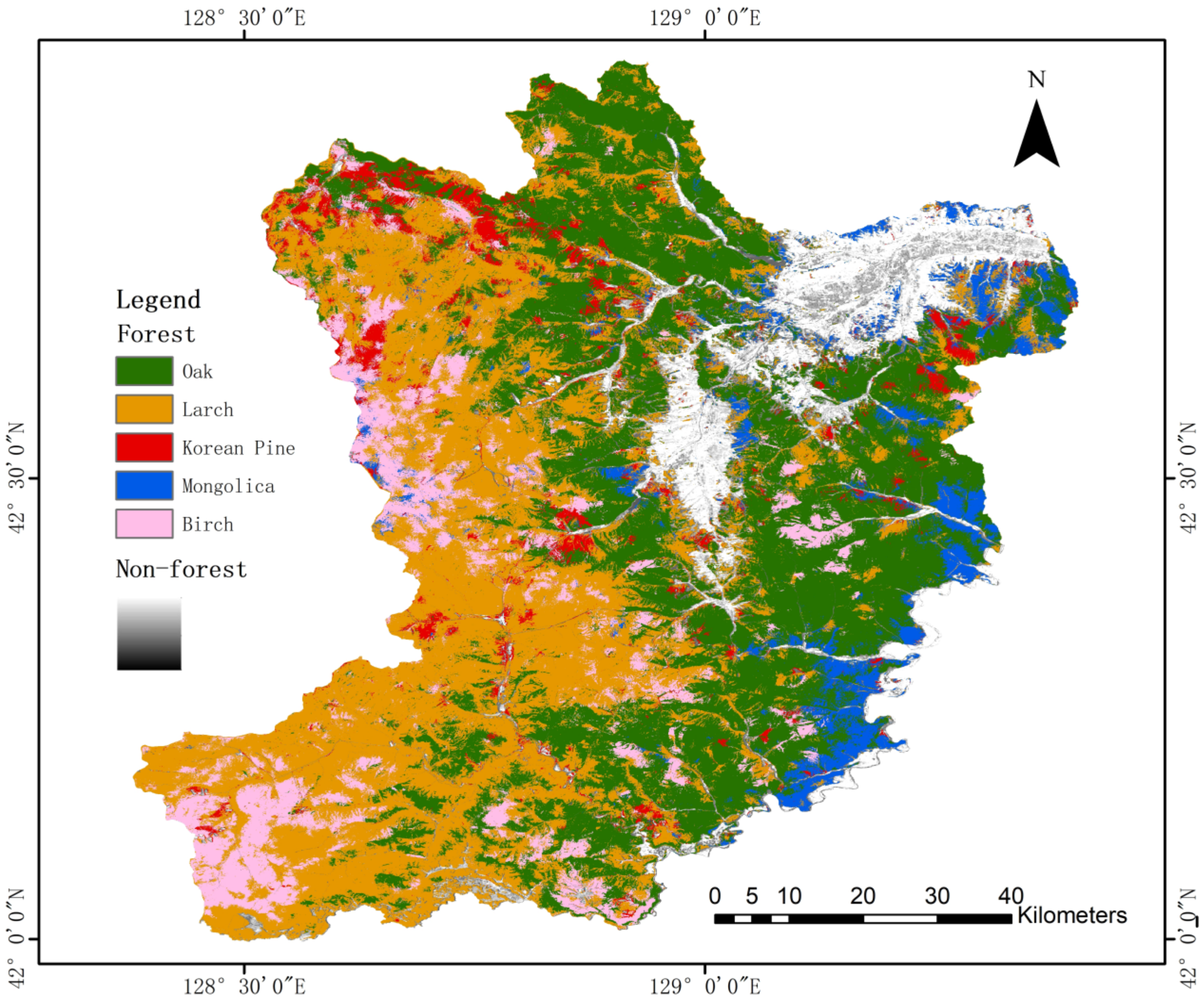
4.2.4. Tree Species Mapping
5. Discussion
5.1. Error Sources in Spatiotemporal Fusion Algorithms
5.2. The Influence of Different Features on Classification Accuracy
5.3. The Advantages and Limitations of Ensemble Classifiers
5.4. Comparison with Other Studies
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Portillo-Quintero, C.; Sanchez-Azofeifa, A.; Calvo-Alvarado, J.; Quesada, M.; do Espirito Santo, M.M. The role of tropical dry forests for biodiversity, carbon and water conservation in the neotropics: Lessons learned and opportunities for its sustainable management. Reg. Environ. Chang. 2015, 15, 1039–1049. [Google Scholar] [CrossRef]
- Watson, J.E.; Evans, T.; Venter, O.; Williams, B.; Tulloch, A.; Stewart, C.; Thompson, I.; Ray, J.C.; Murray, K.; Salazar, A. The exceptional value of intact forest ecosystems. Nat. Ecol. Evol. 2018, 2, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Führer, E. Forest functions, ecosystem stability and management. For. Ecol. Manag. 2000, 132, 29–38. [Google Scholar] [CrossRef]
- Chiarucci, A.; Piovesan, G. Need for a global map of forest naturalness for a sustainable future. Conserv. Biol. 2020, 34, 368–372. [Google Scholar] [CrossRef] [PubMed]
- McRoberts, R.E.; Tomppo, E.O. Remote sensing support for national forest inventories. Remote Sens. Environ. 2007, 110, 412–419. [Google Scholar] [CrossRef]
- Hemmerling, J.; Pflugmacher, D.; Hostert, P. Mapping temperate forest tree species using dense Sentinel-2 time series. Remote Sens. Environ. 2021, 267, 112743. [Google Scholar] [CrossRef]
- Lim, J.; Kim, K.-M.; Jin, R. Tree species classification using hyperion and sentinel-2 data with machine learning in South Korea and China. ISPRS Int. J. Geo-Inf. 2019, 8, 150. [Google Scholar] [CrossRef]
- Harikumar, A.; Paris, C.; Bovolo, F.; Bruzzone, L. A crown quantization-based approach to tree-species classification using high-density airborne laser scanning data. IEEE Trans. Geosci. Remote Sens. 2020, 59, 4444–4453. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Q.-Y.; Qiu, X.-C.; Peng, D.-L. Temporal stage and method selection of tree species classification based on GF-2 remote sensing image. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2019, 30, 4059–4070. [Google Scholar]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest stand species mapping using the Sentinel-2 time series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef]
- Bjerreskov, K.S.; Nord-Larsen, T.; Fensholt, R. Classification of nemoral forests with fusion of multi-temporal sentinel-1 and 2 data. Remote Sens. 2021, 13, 950. [Google Scholar] [CrossRef]
- Grybas, H.; Congalton, R.G. A comparison of multi-temporal RGB and multispectral UAS imagery for tree species classification in heterogeneous New Hampshire Forests. Remote Sens. 2021, 13, 2631. [Google Scholar] [CrossRef]
- Persson, M.; Lindberg, E.; Reese, H. Tree species classification with multi-temporal Sentinel-2 data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef]
- Liu, H. Classification of urban tree species using multi-features derived from four-season RedEdge-MX data. Comput. Electron. Agric. 2022, 194, 106794. [Google Scholar] [CrossRef]
- Sheeren, D.; Fauvel, M.; Josipović, V.; Lopes, M.; Planque, C.; Willm, J.; Dejoux, J.-F. Tree species classification in temperate forests using Formosat-2 satellite image time series. Remote Sens. 2016, 8, 734. [Google Scholar] [CrossRef]
- Wan, H.; Tang, Y.; Jing, L.; Li, H.; Qiu, F.; Wu, W. Tree species classification of forest stands using multisource remote sensing data. Remote Sens. 2021, 13, 144. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, Z.; Yu, W.; Zhao, P.; Yue, J.; Deng, Y.; Geng, J. How spatial resolution affects forest phenology and tree-species classification based on satellite and up-scaled time-series images. Remote Sens. 2021, 13, 2716. [Google Scholar] [CrossRef]
- Xu, K.; Tian, Q.; Zhang, Z.; Yue, J.; Chang, C.-T. Tree species (genera) identification with GF-1 time-series in a forested landscape, Northeast China. Remote Sens. 2020, 12, 1554. [Google Scholar] [CrossRef]
- Belgiu, M.; Stein, A. Spatiotemporal image fusion in remote sensing. Remote Sens. 2019, 11, 818. [Google Scholar] [CrossRef]
- Gao, F.; Hilker, T.; Zhu, X.; Anderson, M.; Masek, J.; Wang, P.; Yang, Y. Fusing Landsat and MODIS data for vegetation monitoring. IEEE Geosci. Remote Sens. Mag. 2015, 3, 47–60. [Google Scholar] [CrossRef]
- Chen, B.; Huang, B.; Xu, B. Comparison of spatiotemporal fusion models: A review. Remote Sens. 2015, 7, 1798–1835. [Google Scholar] [CrossRef]
- Zhu, X.; Cai, F.; Tian, J.; Williams, T.K.-A. Spatiotemporal fusion of multisource remote sensing data: Literature survey, taxonomy, principles, applications, and future directions. Remote Sens. 2018, 10, 527. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the Landsat and MODIS surface reflectance: Predicting daily Landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar]
- Cheng, Q.; Liu, H.; Shen, H.; Wu, P.; Zhang, L. A spatial and temporal nonlocal filter-based data fusion method. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4476–4488. [Google Scholar] [CrossRef]
- Zhu, X.; Helmer, E.H.; Gao, F.; Liu, D.; Chen, J.; Lefsky, M.A. A flexible spatiotemporal method for fusing satellite images with different resolutions. Remote Sens. Environ. 2016, 172, 165–177. [Google Scholar] [CrossRef]
- Gao, F.; Anderson, M.C.; Zhang, X.; Yang, Z.; Alfieri, J.G.; Kustas, W.P.; Mueller, R.; Johnson, D.M.; Prueger, J.H. Toward mapping crop progress at field scales through fusion of Landsat and MODIS imagery. Remote Sens. Environ. 2017, 188, 9–25. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, L.; Xie, D.; Yin, X.; Liu, C.; Liu, G. Application of Synthetic NDVI Time Series Blended from Landsat and MODIS Data for Grassland Biomass Estimation. Remote Sens. 2015, 8, 10. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Wei, X.; Yao, Y.; Su, Y.; Jiang, B.; Wang, X. Land Cover Classification of Landsat Data with Phenological Features Extracted from Time Series MODIS NDVI Data. Remote Sens. 2014, 6, 11518–11532. [Google Scholar] [CrossRef]
- Chaudhuri, P.; Ghosh, A.K.; Oja, H. Classification based on hybridization of parametric and nonparametric classifiers. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 31, 1153–1164. [Google Scholar] [CrossRef]
- Axelsson, A.; Lindberg, E.; Reese, H.; Olsson, H. Tree species classification using Sentinel-2 imagery and Bayesian inference. Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102318. [Google Scholar] [CrossRef]
- Zhang, M.-L.; Zhou, Z.-H. ML-KNN: A lazy learning approach to multi-label learning. Pattern Recognit. 2007, 40, 2038–2048. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Hepner, G.; Logan, T.; Ritter, N.; Bryant, N. Artificial neural network classification using a minimal training set- Comparison to conventional supervised classification. Photogramm. Eng. Remote Sens. 1990, 56, 469–473. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3147–3155. [Google Scholar]
- Li, D.; Yang, F.; Wang, X. Study on ensemble crop information extraction of remote sensing images based on SVM and BPNN. J. Indian Soc. Remote Sens. 2017, 45, 229–237. [Google Scholar] [CrossRef]
- Pham, H.; Olafsson, S. Bagged ensembles with tunable parameters. Comput. Intell. 2019, 35, 184–203. [Google Scholar] [CrossRef]
- Pham, H.; Olafsson, S. On Cesaro averages for weighted trees in the random forest. J. Classif. 2020, 37, 223–236. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Moon, H.; Ahn, H. A weight-adjusted voting algorithm for ensembles of classifiers. J. Korean Stat. Soc. 2011, 40, 437–449. [Google Scholar] [CrossRef]
- Du, P.; Xia, J.; Zhang, W.; Tan, K.; Liu, Y.; Liu, S. Multiple classifier system for remote sensing image classification: A review. Sensors 2012, 12, 4764–4792. [Google Scholar] [CrossRef]
- Ge, H.; Ma, F.; Li, Z.; Tan, Z.; Du, C. Improved Accuracy of Phenological Detection in Rice Breeding by Using Ensemble Models of Machine Learning Based on UAV-RGB Imagery. Remote Sens. 2021, 13, 2678. [Google Scholar] [CrossRef]
- Chen, H.; Liu, W.; Xiao, C.; Qin, R. Large-scale land cover mapping of satellite images using ensemble of random forests—IEEE Data Fusion Contest 2020 Track 1. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 7058–7061. [Google Scholar]
- Deepan, P.; Sudha, L. Scene Classification of Remotely Sensed Images using Ensembled Machine Learning Models. In Machine Learning, Deep Learning and Computational Intelligence for Wireless Communication; Springer: Berlin/Heidelberg, Germany, 2021; pp. 535–550. [Google Scholar]
- Qingchun, Z.; Guofeng, T.; Yong, L.; Liwei, G.; Huairong, C. River Detection in Remote Sensing Images Based on Multi-Feature Fusion and Soft Voting. Acta Opt. Sin. 2018, 38, 0628002. [Google Scholar] [CrossRef]
- Vermote, E.; Justice, C.; Claverie, M.; Franch, B. Preliminary analysis of the performance of the Landsat 8/OLI land surface reflectance product. Remote Sens. Environ. 2016, 185, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Huang, B.; Xu, B. Multi-source remotely sensed data fusion for improving land cover classification. ISPRS J. Photogramm. Remote Sens. 2017, 124, 27–39. [Google Scholar] [CrossRef]
- Farr, T.G.; Rosen, P.A.; Caro, E.; Crippen, R.; Duren, R.; Hensley, S.; Kobrick, M.; Paller, M.; Rodriguez, E.; Roth, L.; et al. The Shuttle Radar Topography Mission. Rev. Geophys. 2007, 45, RG2004. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Hauser, L.T.; An Binh, N.; Viet Hoa, P.; Hong Quan, N.; Timmermans, J. Gap-free monitoring of annual mangrove forest dynamics in ca mau province, vietnamese mekong delta, using the landsat-7-8 archives and post-classification temporal optimization. Remote Sens. 2020, 12, 3729. [Google Scholar] [CrossRef]
- Weiss, D.J.; Atkinson, P.M.; Bhatt, S.; Mappin, B.; Hay, S.I.; Gething, P.W. An effective approach for gap-filling continental scale remotely sensed time-series. ISPRS J. Photogramm. Remote Sens. 2014, 98, 106–118. [Google Scholar] [CrossRef]
- Hościło, A.; Lewandowska, A. Mapping forest type and tree species on a regional scale using multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 929. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Chen, X.; Zhu, X.; Qiu, Y.; Song, H.; Rao, Y.; Zhang, C.; Cao, X.; Cui, X. Sensitivity of six typical spatiotemporal fusion methods to different influential factors: A comparative study for a normalized difference vegetation index time series reconstruction. Remote Sens. Environ. 2021, 252, 112130. [Google Scholar] [CrossRef]
- Wang, Y.C.; Feng, C.C.; Vu Duc, H. Integrating Multi-Sensor Remote Sensing Data for Land Use/Cover Mapping in a Tropical Mountainous Area in Northern Thailand. Geogr. Res. 2012, 50, 320–331. [Google Scholar] [CrossRef]
- Zhou, Z.-H.; Feng, J. Deep forest. Natl. Sci. Rev. 2019, 6, 74–86. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Tetteh, G.O.; Gocht, A.; Conrad, C. Optimal parameters for delineating agricultural parcels from satellite images based on supervised Bayesian optimization. Comput. Electron. Agric. 2020, 178, 105696. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Khan, M.M.; Alparone, L.; Chanussot, J. Pansharpening quality assessment using the modulation transfer functions of instruments. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3880–3891. [Google Scholar] [CrossRef]
- Dennison, P.E.; Halligan, K.Q.; Roberts, D.A. A comparison of error metrics and constraints for multiple endmember spectral mixture analysis and spectral angle mapper. Remote Sens. Environ. 2004, 93, 359–367. [Google Scholar] [CrossRef]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good practices for estimating area and assessing accuracy of land change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Jia, D.; Cheng, C.; Song, C.; Shen, S.; Ning, L.; Zhang, T. A hybrid deep learning-based spatiotemporal fusion method for combining satellite images with different resolutions. Remote Sens. 2021, 13, 645. [Google Scholar] [CrossRef]
- Liao, C.; Wang, J.; Pritchard, I.; Liu, J.; Shang, J. A spatio-temporal data fusion model for generating NDVI time series in heterogeneous regions. Remote Sens. 2017, 9, 1125. [Google Scholar] [CrossRef]
- Ping, B.; Meng, Y.; Su, F. An enhanced linear spatio-temporal fusion method for blending Landsat and MODIS data to synthesize Landsat-like imagery. Remote Sens. 2018, 10, 881. [Google Scholar] [CrossRef]
- Wang, J.; Cai, X.; Chen, X.; Zhang, Z.; Tang, L. Classification of forest vegetation type using fused NDVI time series data based on STNLFFM. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 6047–6050. [Google Scholar]
- Xie, D.; Gao, F.; Sun, L.; Anderson, M. Improving spatial-temporal data fusion by choosing optimal input image pairs. Remote Sens. 2018, 10, 1142. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhou, J.; Chen, J.; Chen, X. Spatiotemporal fusion method to simultaneously generate full-length normalized difference vegetation index time series (SSFIT). Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102333. [Google Scholar] [CrossRef]
- Immitzer, M.; Neuwirth, M.; Böck, S.; Brenner, H.; Vuolo, F.; Atzberger, C. Optimal input features for tree species classification in Central Europe based on multi-temporal Sentinel-2 data. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef]
- Jia, K.; Liang, S.; Zhang, L.; Wei, X.; Yao, Y.; Xie, X. Forest cover classification using Landsat ETM+ data and time series MODIS NDVI data. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 32–38. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D. Accurate mapping of forest types using dense seasonal Landsat time-series. ISPRS J. Photogramm. Remote Sens. 2014, 96, 1–11. [Google Scholar] [CrossRef]
- Lim, J.; Kim, K.-M.; Kim, E.-H.; Jin, R. Machine learning for tree species classification using sentinel-2 spectral information, crown texture, and environmental variables. Remote Sens. 2020, 12, 2049. [Google Scholar] [CrossRef]
- Ma, X.; Shen, H.; Yang, J.; Zhang, L.; Li, P. Polarimetric-spatial classification of SAR images based on the fusion of multiple classifiers. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2013, 7, 961–971. [Google Scholar] [CrossRef]
- Li, W.; Wu, F.; Cao, D. Dual-Branch Remote Sensing Spatiotemporal Fusion Network Based on Selection Kernel Mechanism. Remote Sens. 2022, 14, 4282. [Google Scholar] [CrossRef]

| Landsat 8 | MODIS | ||
|---|---|---|---|
| Date (all in 2016) | 05-19, 05-28, 10-19 | 05-19, 05-28, 10-19 03-21, …,10-28 | |
| Wavelength (nm) | Blue Green Red NIR SWIR-1 SWIR-2 | 450–515 525–600 630–680 845–885 1560–1660 2100–2300 | 459–479 545–565 620–672 841–890 1628–1652 2105–2155 |
| Resolution | 30 m | 500 m | |
| Repeat cycle | 16 days | daily |
| Tree Species | Training Samples | Validation Samples |
|---|---|---|
| Oak | 8946 | 2236 |
| Larch | 6728 | 1682 |
| Korean pine | 2170 | 543 |
| Pinus sylvestris | 4944 | 1236 |
| Birch | 3145 | 786 |
| Experiment | Input Data | Number of Feature Variables |
|---|---|---|
| 1 | Single-temporal surface reflectance (28 groups) | 6 |
| 2 | Single-temporal surface reflectance, topographic features (28 groups) | 9 |
| 3 | 10 fused images in spring | 60 |
| 4 | 9 fused images in summer | 54 |
| 5 | 9 fused images in autumn | 54 |
| 6 | All fused images in the time series | 168 |
| 7 | All fused images in the time series, topographic features | 171 |
| 8 | 3 cloud-free Landsat images on May 19, May 28, and October 19 2016 | 18 |
| 9 | 3 seasonal composite images of spring, summer, and autumn 2016 | 18 |
| 10 | 9 seasonal composite images of spring, summer, and autumn 2015, 2016, and 2017 | 54 |
| Base Classifier | Parameter | Value |
|---|---|---|
| KNN | N_neighbors N_jobs | 3 −1 |
| RF | N_estimators criterion Max_depth Min_samples_split Min_samples_leaf Max_features N_jobs | 870 ‘gini’ None 2 1 ‘sqrt’ −1 |
| ANN | Hidden layer sizes Learning rate | (400, 200, 100, 50) 0.0005 |
| LightGBM | N_estimators Learning_rate Num_leaves Max_depth N_jobs | 1527 0.098 19 −1 −1 |
| Date | Correlation Coefficient | ||||||
|---|---|---|---|---|---|---|---|
| Blue | Green | Red | NIR | SWIR1 | SWIR2 | Average | |
| 28 May 2016 | 0.9689 | 0.9842 | 0.9717 | 0.9750 | 0.9862 | 0.9720 | 0.9763 |
| 19 October 2016 | 0.8123 | 0.9120 | 0.8051 | 0.8712 | 0.9121 | 0.8251 | 0.8563 |
| Date | Method | Metrics | ||||
|---|---|---|---|---|---|---|
| CC | RMSE | SSIM | SAM | ERGAS | ||
| 28 May 2016 | FSDAF | 0.9730 | 0.0165 | 0.9901 | 0.7942 | 2.3177 |
| STARFM | 0.9744 | 0.0158 | 0.9907 | 0.7915 | 2.2258 | |
| STNLFFM | 0.9746 | 0.0156 | 0.9904 | 0.7921 | 2.2794 | |
| 19 October 2016 | FSDAF | 0.9153 | 0.0258 | 0.9766 | 0.8344 | 3.2679 |
| STARFM | 0.9135 | 0.0260 | 0.9764 | 0.8427 | 3.2861 | |
| STNLFFM | 0.9226 | 0.0248 | 0.9781 | 0.8329 | 3.1740 | |
| Experiment | Kappa Coefficient | Overall Accuracy | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| KNN | RF | ANN | Light GBM | Ensemble | KNN | RF | ANN | Light GBM | Ensemble | |
| 1 | 0.5227 | 0.5561 | 0.5695 | 0.5252 | 0.5811 | 0.6456 | 0.6732 | 0.6783 | 0.6493 | 0.6895 |
| 2 | 0.5809 | 0.6276 | 0.6117 | 0.6229 | 0.6529 | 0.6884 | 0.7254 | 0.7086 | 0.7207 | 0.7420 |
| 3 | 0.6950 | 0.7141 | 0.7528 | 0.7367 | 0.7650 | 0.7710 | 0.7877 | 0.8144 | 0.8035 | 0.8241 |
| 4 | 0.6965 | 0.6826 | 0.7455 | 0.7111 | 0.7565 | 0.7726 | 0.7655 | 0.8083 | 0.7853 | 0.8178 |
| 5 | 0.6736 | 0.6879 | 0.7340 | 0.7066 | 0.7504 | 0.7556 | 0.7690 | 0.7991 | 0.7818 | 0.8092 |
| 6 | 0.7202 | 0.7303 | 0.7639 | 0.7601 | 0.7821 | 0.7897 | 0.7998 | 0.8215 | 0.8208 | 0.8368 |
| 7 | 0.7262 | 0.7338 | 0.7720 | 0.7693 | 0.7897 | 0.7943 | 0.8025 | 0.8290 | 0.8276 | 0.8432 |
| 8 | 0.5509 | 0.5915 | 0.5890 | 0.6121 | 0.6323 | 0.6702 | 0.7016 | 0.6915 | 0.7146 | 0.7292 |
| 9 | 0.5239 | 0.5581 | 0.5839 | 0.5727 | 0.6180 | 0.6497 | 0.6803 | 0.6879 | 0.6881 | 0.7184 |
| 10 | 0.6337 | 0.6317 | 0.6751 | 0.6736 | 0.7217 | 0.7275 | 0.7331 | 0.7596 | 0.7606 | 0.7945 |
| Class | Accuracy | KNN | RF | ANN | LightGBM | Ensemble |
|---|---|---|---|---|---|---|
| Oak | UA | 77.385 | 75.930 | 79.908 | 80.906 | 82.315 |
| PA | 81.021 | 86.670 | 86.263 | 86.353 | 86.444 | |
| F-value | 79.161 | 80.945 | 82.964 | 83.541 | 84.329 | |
| Larch | UA | 82.104 | 81.596 | 85.498 | 83.333 | 87.130 |
| PA | 76.207 | 76.880 | 79.405 | 80.247 | 80.920 | |
| F-value | 79.045 | 79.168 | 82.339 | 81.761 | 83.910 | |
| Korean Pine | UA | 69.351 | 83.934 | 70.161 | 79.003 | 79.903 |
| PA | 61.876 | 51.098 | 69.461 | 60.080 | 65.868 | |
| F-value | 65.401 | 63.524 | 69.809 | 68.254 | 72.210 | |
| Pinus sylvestris | UA | 79.596 | 79.348 | 86.080 | 81.511 | 81.521 |
| PA | 82.494 | 84.818 | 83.346 | 86.057 | 88.846 | |
| F-value | 81.019 | 81.992 | 84.691 | 83.723 | 85.026 | |
| Birch | UA | 85.436 | 94.016 | 89.932 | 92.034 | 91.460 |
| PA | 89.024 | 80.894 | 89.566 | 87.669 | 89.973 | |
| F-value | 87.193 | 86.963 | 89.749 | 89.799 | 90.710 |
| Class | Accuracy | KNN | RF | ANN | LightGBM | Ensemble |
|---|---|---|---|---|---|---|
| Oak | UA | 69.479 | 69.533 | 76.182 | 72.064 | 75.114 |
| PA | 77.03 | 83.57 | 76.713 | 81.652 | 82.612 | |
| F-value | 72.998 | 75.854 | 76.361 | 76.519 | 78.651 | |
| Larch | UA | 74.059 | 75.173 | 76.068 | 75.198 | 78.092 |
| PA | 69.193 | 71.237 | 74.251 | 73.31 | 74.994 | |
| F-value | 71.536 | 73.145 | 75.097 | 74.236 | 76.494 | |
| Korean Pine | UA | 58.703 | 74.728 | 60.113 | 68.342 | 71.236 |
| PA | 44.408 | 35.337 | 55.413 | 41.164 | 49.706 | |
| F-value | 50.241 | 47.48 | 57.227 | 51.062 | 58.237 | |
| Pinus sylvestris | UA | 74.778 | 75.212 | 75.968 | 76.23 | 77.876 |
| PA | 74.241 | 77.551 | 79.81 | 78.524 | 81.245 | |
| F-value | 74.463 | 76.274 | 77.759 | 77.323 | 79.493 | |
| Birch | UA | 76.916 | 85.99 | 81.762 | 83.96 | 85.898 |
| PA | 77.176 | 68.956 | 80.386 | 73.012 | 78.842 | |
| F-value | 77.018 | 76.452 | 81.002 | 78.032 | 82.189 |
| Method | Overall Accuracy | Kappa Coefficient |
|---|---|---|
| Deep Forest | 0.8256 | 0.7662 |
| Ensemble Classifier | 0.8423 | 0.7911 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Chen, S.; Mu, Y.; Xu, X.; Zhang, B.; Zhao, X. Tree Species Classification over Cloudy Mountainous Regions by Spatiotemporal Fusion and Ensemble Classifier. Forests 2023, 14, 107. https://doi.org/10.3390/f14010107
Cui L, Chen S, Mu Y, Xu X, Zhang B, Zhao X. Tree Species Classification over Cloudy Mountainous Regions by Spatiotemporal Fusion and Ensemble Classifier. Forests. 2023; 14(1):107. https://doi.org/10.3390/f14010107
Chicago/Turabian StyleCui, Liang, Shengbo Chen, Yongling Mu, Xitong Xu, Bin Zhang, and Xiuying Zhao. 2023. "Tree Species Classification over Cloudy Mountainous Regions by Spatiotemporal Fusion and Ensemble Classifier" Forests 14, no. 1: 107. https://doi.org/10.3390/f14010107
APA StyleCui, L., Chen, S., Mu, Y., Xu, X., Zhang, B., & Zhao, X. (2023). Tree Species Classification over Cloudy Mountainous Regions by Spatiotemporal Fusion and Ensemble Classifier. Forests, 14(1), 107. https://doi.org/10.3390/f14010107







