Abstract
A simple and efficient protocol for the genetic transformation of Japanese larch (Larix kaempferi (Lamb.) Carr.) was developed by altering the infection method and duration and the bacterial removal process. More than 600 hygromycin-resistant embryonal masses with the vector pCAMBIA1301 were obtained, with an average of 20 transgenic lines per gram of fresh weight. Nine hygromycin-resistant transformation events (designated P1–P9) were analyzed using PCR, quantitative real-time PCR, and histochemical β-glucuronidase (GUS) assays. The GUS transcript abundance in each transformed cell line ranged from 101 to 103 magnitudes, with a maximum abundance of 2.89 × 103. In addition, the pLaTCTP::GUS vector, which contains GUS under the control of the L. kaempferi LaTCTP promoter, led to constitutive expression of GUS in embryonal-suspensor mass and somatic embryos. The transcript abundance of the exogenous genes HPT and GUS, driven by the CaMV 35S or LaTCTP promoter, ranged from 101 to 104, which was equivalent to genes with moderate and low abundances in Japanese larch. The relatively low expression levels of exogenous genes in transformants might reflect the large genome of Japanese larch. Additional transgenic cell lines need to be screened to obtain transformants with higher expression levels of foreign genes for further functional research in Japanese larch.
1. Introduction
Larches (Larix spp.) are conifers indigenous to the Northern Hemisphere, comprising approximately 10 species distributed in North America, Europe, and Asia [1]. Among these species, Japanese larch (Larix kaempferi (Lamb.) Carr.) is characterized by rapid juvenile growth [2]. However, because of the long life history and complex genetic background of individual trees, the use of conventional breeding methods is difficult and propagation by seed inevitably leads to the segregation of desirable characteristics [3]. The application of modern breeding technologies to achieve rapid breeding of superior larch cultivars has become a top priority. Somatic embryogenesis may provide a novel procedure for the rapid multiplication of commercial hybrid larch genotypes.
Somatic embryogenesis is a valuable experimental system for the investigation of the regulatory mechanisms involved in plant development. However, somatic embryogenesis in conifers presents a number of challenges, such as difficulties with embryonal-suspensor mass (ESM) induction, a high frequency of abnormal embryo development during the maturation stage, and difficulties with in vitro synchronization [4,5,6]. Overcoming these problems requires knowledge of the molecular mechanisms of crucial genes that regulate the different developmental stages of somatic embryogenesis. MicroRNAs (miRNAs) of L. kaempferi have been identified using high-throughput sequencing [7,8] and might participate in the regulation of synchronism through responses to abscisic acid (ABA) during larch somatic embryogenesis [9]. Among these miRNAs, miR171, -159, -169, and -172 play important regulatory roles in the transformation of embryogenic tissue to non-embryogenic tissue [10]. Thus, an efficient transformation procedure is a prerequisite for functional studies of the aforementioned genes.
Somatic embryogenesis is the basis of the genetic transformation system used for the majority of gymnosperms. Particle bombardment and Agrobacterium-mediated transformation (AMT) are the transformation methods most frequently used in conifers [11]. The advantages of the latter over the former include the production of lower copy numbers, greater stability in transgene expression levels, and fewer instances of gene silencing [11,12,13,14,15]. The AMT approach has been successfully applied to a variety of tissues in numerous coniferous species, including Norway spruce (Picea abies (L.) Karst.), white spruce (Picea glauca (Moench) Voss), black spruce (Picea mariana (Mill.) Britt., Sterns and Poggenburg), loblolly pine (Pinus taeda L.) [14,16], maritime pine (Pinus pinaster Ait.) [17], and Korean fir (Abies koreana Wilson) [18], through the use of embryogenic masses. The first successful procedure for the stable transformation of hybrid larch (L. kaempferi × L. decidua Mill.) involved co-cultivation with an Agrobacterium tumefaciens strain C58/pMP90/pMRKE70Km [19]. However, the efficiency of stable transformations was low, averaging 1–2 transgenic lines from 50 co-cultivated embryogenic masses [19]. By modifying the initial protocol, such as the procedure by which the embryogenic masses were subcultured and co-cultivated on filter paper, and by pre-culturing the embryogenic masses with the bacterial suspension for 6 h, the transformation efficiency was significantly increased to 42 kanamycin-resistant embryogenic masses per gram of fresh weight (FW) [2]. However, the protocol involves multiple handling steps, such as multiplication procedures on filter paper and pre-culture for 6 h before co-cultivation, which are time consuming and increase the risk of culture contamination. Only three to five transformed lines per gram of FW of the co-cultivated embryogenic tissue were retrieved when a similar procedure was applied to other Pinaceae species, such as white pine (Pinus strobus) [20]. In our laboratory, only five LamiR166a overexpressed embryonic cell lines were recovered from the embryogenic tissue initially co-cultured with the A. tumefaciens strain GV3101 [21]. The embryogenic cells did not survive after co-cultivation and an excessive growth of bacterial cells occurred. Thus, the exploration of the specific AMT parameters required for the improved transformation of different larch species is necessary.
The present study aimed to establish a simple and efficient genetic transformation system for Larix kaempferi using the A. tumefaciens strain GV3101 in conjunction with the binary vector pCAMBIA1301 (Figure S1). This novel transformation system may accelerate the development of genetically modified L. kaempferi and promote further molecular functional studies.
2. Materials and Methods
2.1. Plant Materials and Culture Conditions
The highly embryogenic cell line S287 of Larix kaempferi was induced from immature embryos on the induction medium (S basal medium [22], supplemented with 450 mg/L glutamine, 500 mg/L casein hydrolysate, 1000 mg/L inositol, 2.2 mg/L 2,4-dichlorophenoxyacetic acid, and 0.8 mg/L 6-benzylaminopurine) [6,23] and subcultured at 3-week intervals on a solid proliferation medium (S + B basal medium [24], supplemented with 450 mg/L glutamine, 500 mg/L casein hydrolysate, 1000 mg/L inositol, 0.11 mg/L 2,4-dichlorophenoxyacetic acid, and 0.04 mg/L 6-benzylaminopurine). Approximately 2 g of embryogenic culture was dissociated into a suspension by transferal to a 250 mL flask containing 100 mL liquid proliferation medium and culture in a 10-day cycle at 25 °C in the dark on a gyratory shaker at 100–110 rpm [6,23]. Once grown to 200 mg/mL FW, the embryogenic suspensions were used for AMT.
2.2. Hygromycin Sensitivity of Embryogenic Line S287
The hygromycin sensitivity of line S287 was assessed by transferring the embryogenic cultures to the solid proliferation medium supplemented with hygromycin (0, 3, 4, 5, 7, or 9 mg/L). The FW of the ESM was recorded at the start of the culture period (day 0) and after 20 days. The percentage ESM proliferation was calculated to determine the hygromycin concentration that effectively prohibited cell proliferation using the following formula: ESM proliferation rate (%) = [(ESM FW after 20 days–ESM FW at day 0)/ESM FW at day 0] × 100. Three petri dishes, each containing seven pieces of callus as the inoculum, were used for each experiment, and the experiment was repeated twice. The means of three replicates were analyzed with Student’s t-test to assess the significance of the differences between hygromycin concentrations at α = 0.05 significance level. Data are presented as the mean ± standard deviation.
2.3. Agrobacterium Strains and Binary Vectors
The binary vector Pcambia1301, 35S::GUS, and recombinant Pcambia-LaTCTP promoter-GUS binary vector (pLaTCTP::GUS) were used. The first two vectors harbor the hygromycin phosphotransferase (HPT) gene as a selection marker and the β-glucuronidase (GUS) reporter gene driven by the Cauliflower mosaic virus (CaMV) 35S promoter. The latter vector harbors the GUS gene driven by the endogenous LaTCTP promoter (pLaTCTP). The Agrobacterium tumefaciens strain GV3101, harboring one of the three binary vectors, was cultured at 28 °C for 36 h on a solid LB medium supplemented with 50 mg/L kanamycin, 50 mg/L rifampicin, and 50 mg/L gentamicin. A single Agrobacterium clone was selected and cultured in 2 mL liquid LB medium supplemented with the same antibiotics at 28 °C for 24 h on a rotary shaker (200 rpm). The culture was transferred to 50 mL LB medium and grown under the same conditions for 5–6 h to OD600 = 0.6–0.8. The bacterial cells were collected by centrifugation at 4000 rpm for 10 min. The pellet was resuspended to OD600 = 0.05–0.1 in 100 mL sterile water, and 200 µM acetosyringone was added for transformation.
2.4. Agrobacterium-Mediated Transformation
Approximately 10 g (~50 mL) of the embryogenic suspension was added to an equal volume of the agrobacterial suspension. Based on the characteristics of the embryogenic cell suspension cultures, we improved the vacuum filter (Figure S2), which comprised a Buchner funnel with 400-mesh stainless steel screens instead of filter paper and a microvacuum pump, for the removal of excess liquid from the embryogenic cell suspensions. The filtered cultures were spread on a solid proliferation medium supplemented with 100 µM acetosyringone and co-cultivated for ~46–120 h in the dark at 25 °C. Agrobacterium cells were then removed from the cultures with the vacuum filter. The co-cultivated embryogenic suspensions were washed three times with 100 mL sterile distilled water, and then three times with 100 mL cefotaxime solution (500 mg/L). The suspensions were transferred to a selective medium (a solid proliferation medium supplemented with 5 mg/L hygromycin and 500 mg/L cefotaxime) for the selection of transgenic materials. After ~4–8 weeks, hygromycin-resistant embryonal masses 3 mm in diameter were transferred to a fresh medium for further selection. The surviving putative transformants were maintained on the selective medium during all of the following steps until the completion of somatic embryogenesis. Nine independent putative hygromycin-resistant lines representing transformation events with the binary vector pCAMBIA1301, designated P1–P9, were selected for PCR detection, qRT-PCR, and histochemical GUS analysis.
2.5. PCR Detection
The selected hygromycin-resistant lines were subjected to molecular analysis using a PCR approach. Genomic DNA was isolated from 100 mg fresh culture of the hygromycin-resistant lines and the non-transgenic ESM using PlantZol (TransGen, Beijing, China). For the PCR analysis, three combinations of primers were used (Table 1). The primers HPT-F and HPT-R amplified a 480 bp fragment of the HPT gene. The primers virG-F and virG-R amplified a 485 bp fragment of the virG sequence, which is a sequence of the Ti plasmid located outside of the T-DNA. The primers GUS-F and GUS-R amplified a 450 bp fragment of the GUS gene. The amplifications were conducted using the following PCR program: one cycle at 94 °C for 5 min, then 30 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, followed by one cycle at 72 °C for 5 min. The PCR products were analyzed by electrophoresis in 1% agarose gel. Control DNA from the non-transformed ESM and Agrobacterium strain GV3101 harboring pCAMBIAI301 were included in the analysis to ensure that the reagents were not contaminated.

Table 1.
Oligonucleotide primers used in the study.
2.6. Histochemical GUS Assay
Histochemical assays for GUS staining were performed using the β-Galactosidase Reporter Gene Staining Kit (HUAYUEYANG, Beijing, China). The putatively transformed cell lines and somatic embryos at different developmental stages were stained by incubation for 12 h at 37 °C. Chlorophyll was removed by soaking in 70% ethanol for 2–3 times until the negative control material was white.
Based on the results of qRT-PCR analyses and GUS staining of the transformed cell lines, the lines P2, P5, and pLaTCTP::GUS-4 were selected for somatic embryogenesis. The ESM of the lines P2 and P5 were cultured on the proliferation medium for 16 days and were then transferred to a differentiation medium (S + B basal medium, supplemented with 450 mg/L glutamine, 500 mg/L casein hydrolysate, 100 mg/L inositol, 7.5% (w/v) PEG4000, and 20 mg/L ABA) and cultured in the dark at 25 ± 2 °C [23]. Based on morphological traits, somatic embryos at different developmental stages were sampled for the GUS assay and qRT-PCR.
2.7. Gene Expression Analysis by Quantitative Real-Time PCR
Total RNA was extracted from various tissues with the RNAiso Plus and RNAiso-mate for Plant Tissue kits (TaKaRa, Dalian, China). Then, 1 µg RNA was reverse transcribed using the iScript™ gDNA Clear cDNA Synthesis Kit (Bio-Rad, Cat. No. 172-5035) following the manufacturer’s protocol. The GUS expression level was determined by conducting quantitative real-time PCR (qRT-PCR) analyses with the SYBR Premix EX Taq Kit (TaKaRa) and the CFX96™ Real-Time System. Four fragments, 1638 bp of Elongation factor (EF; GenBank accession No. JX157845), 782 bp of Translation initiation factor (eIF; GenBank accession No. JR153705), 805 bp of Adaptor-related protein complex subunit mu (APm; GenBank accession No. JR140253), and 504 bp of Translationally controlled tumor protein (TCTP; GenBank accession No. JR140253), were amplified and independently cloned into the pEASY-Blunt Zero vector (TransGen, Beijing, China), which were used as plasmid standards for the above endogenous genes in absolute quantitative analysis, respectively. The EF, eIF, APm, HPT, GUS, and TCTP gene-specific primers used are listed in Table 1. The specificity of the primers designed for qRT-PCR was confirmed by separating the PCR products on agarose gels and sequencing clones. Each reaction was performed with 2 μL diluted cDNA in a total reaction volume of 25 μL. Using the aforementioned four plasmid DNAs and pCAMBIAI301 as standard materials, standard curves for EF, APm, eIF, TCTP, HPT, and GUS were generated using a gradient serial dilution of the plasmid as the template. Relative quantifications were normalized against the expression of EF [7]. Expression levels were estimated using absolute and relative quantification methods. Data are presented as the mean ± standard deviation of three replicates.
3. Results
3.1. Optimization of the Transformation Procedure
Liquid medium-based suspension cultures have been established and used for the AMT of many gymnosperm species, such as Norway spruce, loblolly pine, and white pine [16,20]. The approach has the advantage of providing a large amount of high-quality materials with vigorous growth within a short period. In the present study, a suspension culture for larch embryogenic cells in a liquid medium was established. The embryogenic suspensions cultured to 200 mg/mL FW were mixed with a lower concentration of bacteria (OD600 = 0.05–0.1) to achieve infection without the pre-culturing of mixtures [2,25,26]. The co-cultivation time was determined according to whether the turbidity of the embryogenic masses was slightly turbid. When the turbidity of the culture became turbid or too turbid, the Agrobacterium cells had grown excessively and the embryonic cells were damaged, which affected the screening of the transformed cells in the next step. In most experiments performed, the co-cultivation period of 46–120 h was conducive to cell recovery and acquiring hygromycin-resistant cell lines. To eliminate the Agrobacterium cells, the vacuum filter was replaced to remove excess liquid containing the bacteria (Figure S2). The improved filter enabled the large-scale removal of bacteria from the suspension cultures in less than 15 min. It was found that 500 mg/L cefotaxime was conducive to the recovery of embryogenic cells co-cultured with Agrobacterium, whereas 300 mg/L timentin was detrimental to the recovery (Figure 1). Therefore, 500 mg/L cefotaxime was used to inhibit bacterial growth. The adjustment of these factors did not increase the transformation efficiency, but enabled the development of a simplified system that achieved a reasonable transformation efficiency (Figure S3).

Figure 1.
The recovery of embryogenic cell co-cultured with Agrobacterium with different combinations of Timentin and cefotaxime. (a) 300 mg/L Timentin, (b) 300 mg/L Timentin and 200 mg/L cefotaxime, (c) 500 mg/L cefotaxime. It was conducive to the recovery of embryonic cells on medium supplemented with 500 mg/L cefotaxime.
3.2. Selection of Transformed Embryogenic Lines
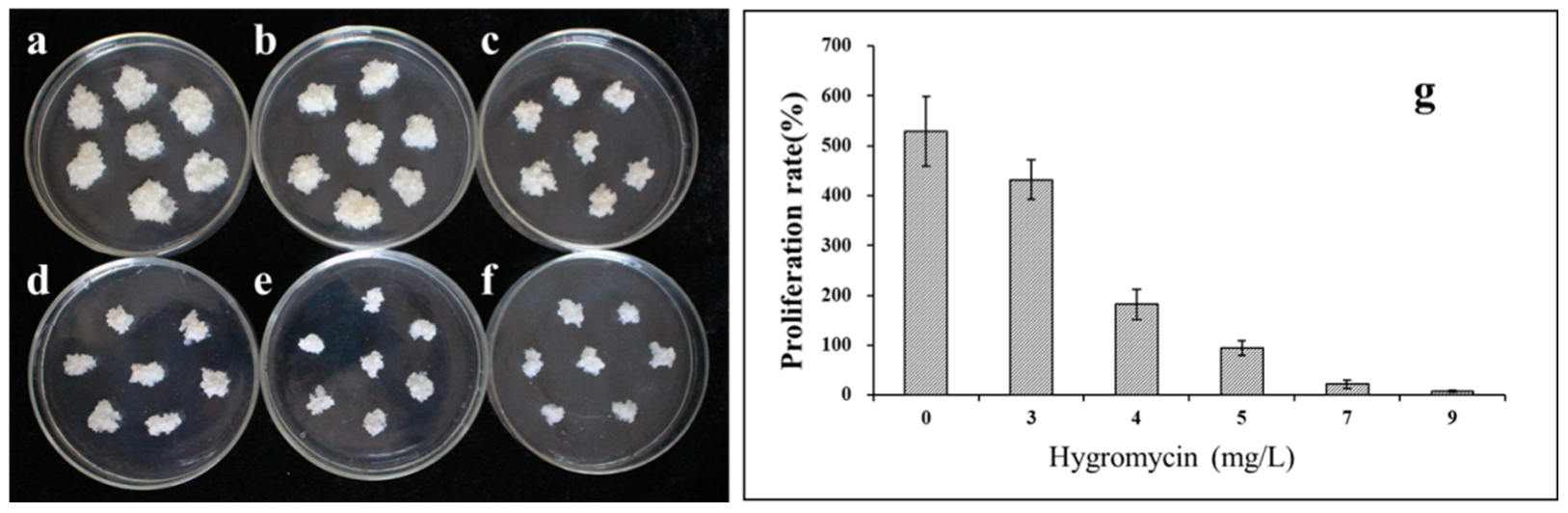
To determine the optimal concentration of the antibiotic for the selection of transformed lines, the sensitivity of the S287 line was evaluated by culturing callus pieces on a proliferation medium supplemented with hygromycin at concentrations ranging from 0 to 10 mg/L. The proliferation of the untransformed embryogenic masses was slightly inhibited by a hygromycin concentration as low as 3 mg/L (Figure 2a–g). An increase in the antibiotic concentration to 5 mg/L was sufficient to prevent cell proliferation. Thus, the latter concentration was chosen for the selection of putative transformed embryogenic cell lines in the transformation procedure.
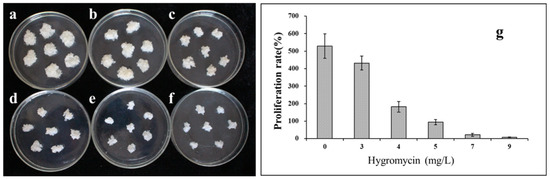
Figure 2.
Proliferation frequency of non-transformed embryogenic tissue from hybrid larch line S287 on medium supplemented with various hygromycin concentrations. (a) 0, (b) 3, (c) 4, (d) 5, (e) 7, and (f) 9 mg/L of hygromycin. When the antibiotic concentration was raised to 5 mg/L, it was sufficient to prevent cell proliferation. (g) The proliferation rate of embryogenic cultures after 20 days of culturing on medium containing various hygromycin concentrations.
We performed three independent transformation experiments using the pCAMBIA1301 vector. After the co-cultivation of the S287 line with the A. tumefaciens strain GV3101 harboring the binary vector pCAMBIA1301, the embryogenic cells were washed with sterile distilled water and cefotaxime solution. The embryogenic cells were then cultured on the selective medium. The number of transformation events per gram FW was recorded after 60 days (Figure S4). In three separate experiments, ~600 hygromycin-resistant embryonal masses were obtained, with an average of 20 transgenic lines per gram FW (Table 2).

Table 2.
Transformation efficiency in three separate experiments with pCAMBIA1301. Experiments with the pCAMBIA1301 vectors were carried out with 9.2, 10.3, and 11.7 g fresh weight of embryogenic tissue, respectively. The transformation efficiencies are expressed as the numbers of hygromycin-resistant cell lines per gram fresh weight of co-cultivated embryogenic tissue. Oligonucleotide primers used in the study.
3.3. Molecular Analysis of the Transformed Lines
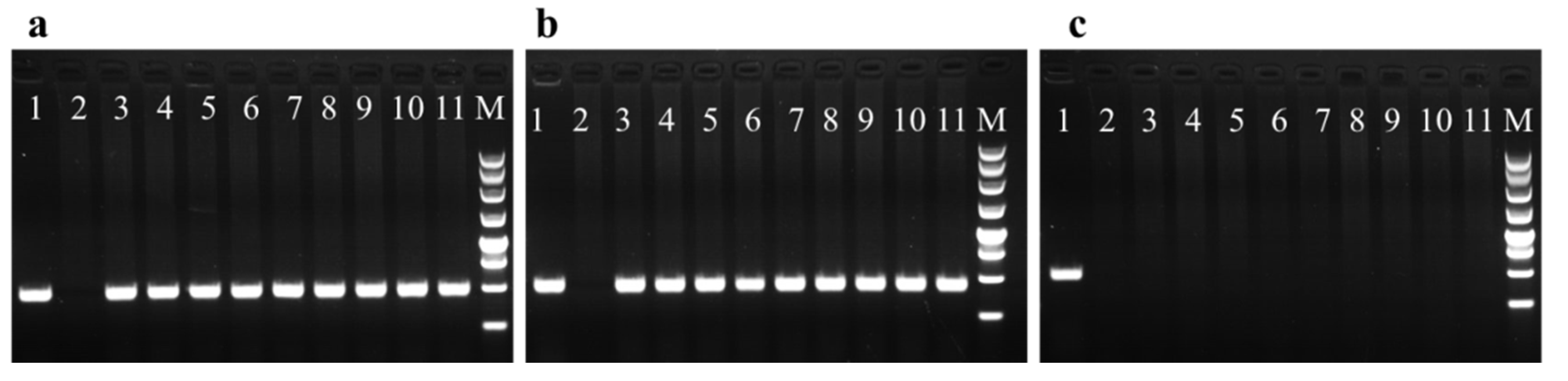
After more than three rounds of selection, more than 80% of the hygromycin-resistant embryonal masses were lost. In addition, only 5% of the putative transformants were stably grown on the selective medium, which may reflect the HPT expression level. Nine hygromycin-resistant cell lines harboring the pCAMBIA1301 vector were chosen for molecular characterization (Figure 3). The HPT- and GUS-specific primers amplified a 480 bp fragment for HPT and a 450 bp fragment for GUS (Figure 3a,b). No fragment was amplified in the non-transformed controls. In addition, the amplification of virG was not detected in any of the hygromycin-resistant lines (Figure 3c). Thus, the hygromycin-resistant lines were not contaminated with Agrobacterium.
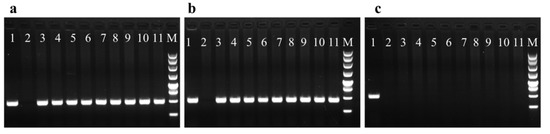
Figure 3.
PCR analysis of hygromycin-resistant lines P1 to P9 transformed by Agrobacterium tumefaciens GV3101. (a) PCR amplification with primers for HPT (480 bp). (b) PCR amplification with primers for GUS (450 bp). (c) PCR amplification with primers for virG (485 bp). Lane 1 positive control (GV3101 containing the plasmid pCAMBIA1301), Lane 2 negative control (Total DNA from non-transformed embryogenic tissue), Lanes 3–11 Total DNA from the nine different hygromycin-resistant lines P1 to P9, M 250-bp marker.
3.4. GUS Assay and Gus Transcript Abundance in Stably Transformed Lines
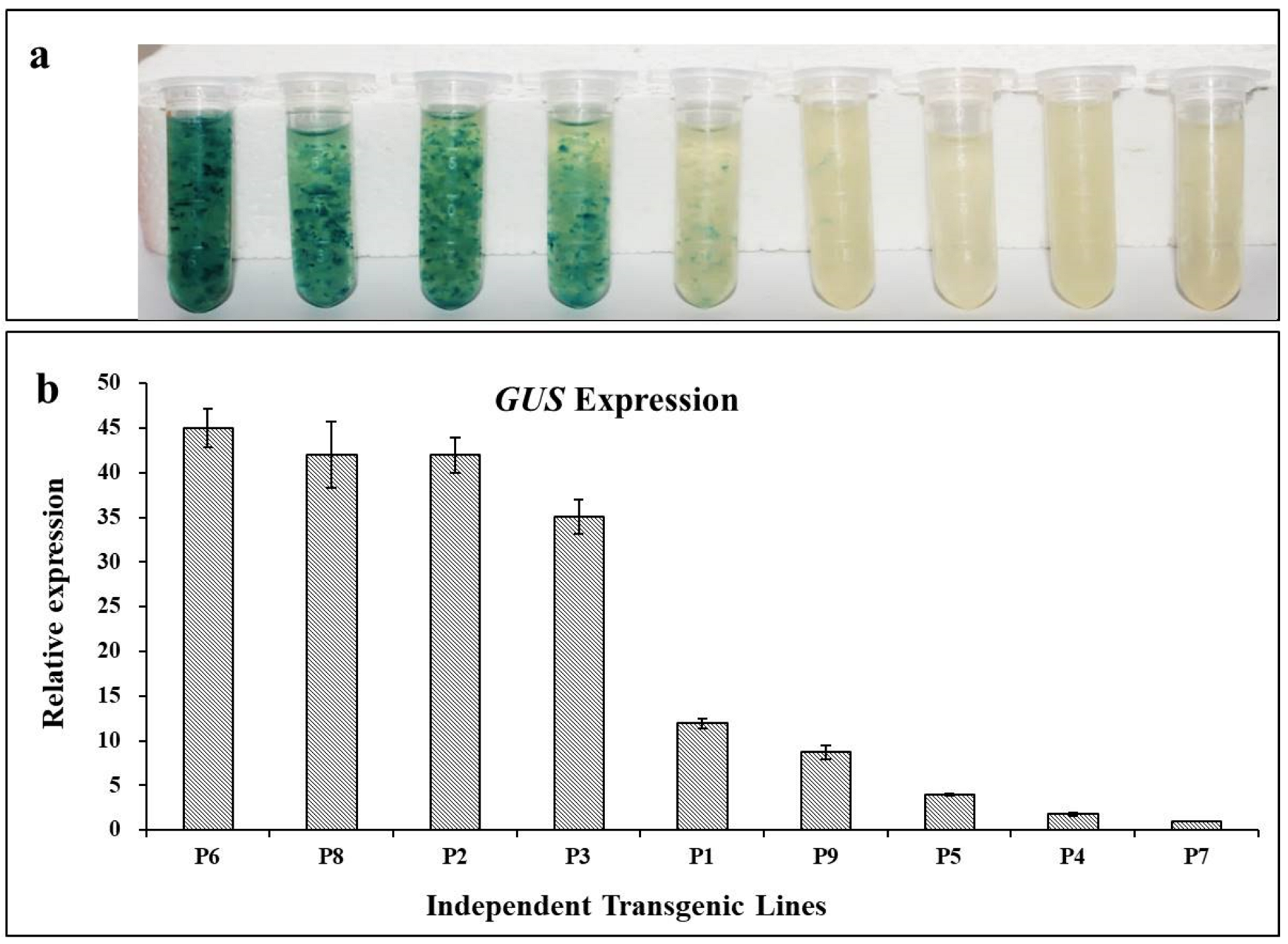
The nine hygromycin-resistant cell lines were used to visualize GUS staining. Four cell lines (P6, P8, P2, and P3) showed intense GUS staining and two cell lines (P1 and P9) exhibited lighter staining. No GUS staining was detected in the remaining three cell lines (P5, P4, and P7) (Figure 4a). Based on the staining intensity (Figure 4), the GUS activity differed in each cell line. In addition, the GUS transcript abundance was determined in comparison with the endogenous control EF using qRT-PCR (Figure 4b). The GUS transcript level in each putative transformed cell line was consistent with the GUS staining intensity (Figure 4).
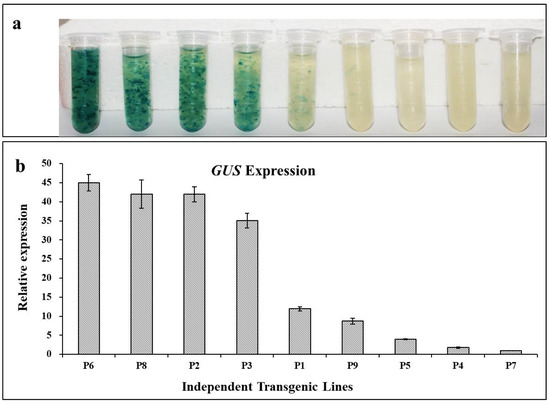
Figure 4.
Histological GUS assay and GUS expression in different transformed lines. (a) Histochemical staining for GUS staining in transgenic lines P1 to P9. (b) Expression of GUS in transgenic lines P1 to P9 as assessed by qRT-PCR.
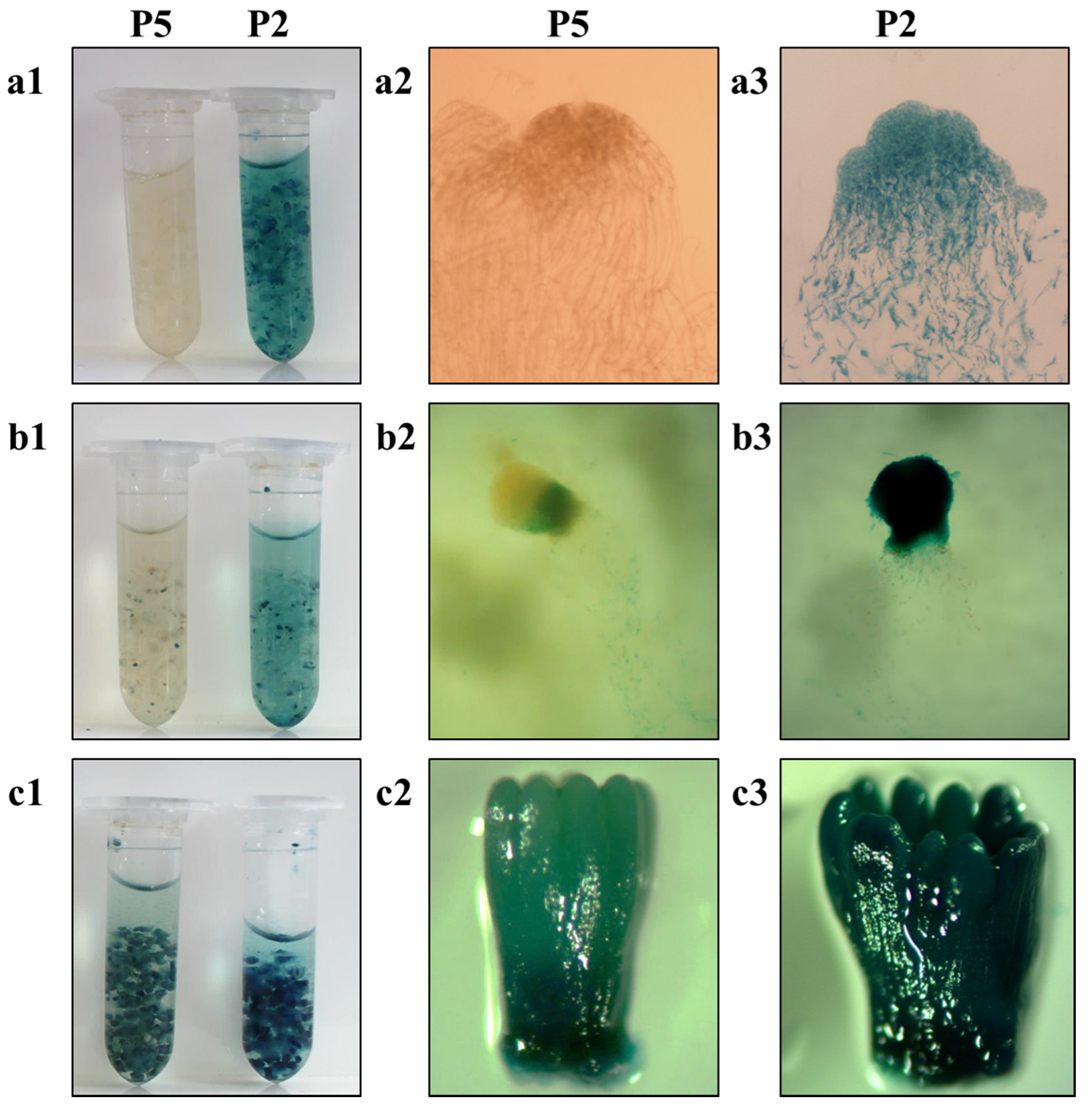
Based on the qRT-PCR results, the transformed cell lines that did not show GUS staining (P5, P4, and P7) still exhibited a low level of GUS expression (Figure 4b). Thus, the cell lines P2 (intense GUS staining) and P5 (no GUS staining) were selected for somatic embryogenesis on a differentiation medium supplemented with ABA, and GUS staining was performed at different developmental stages (Figure 5a–c). When the cell line P5 was cultured for 15 days, early single embryos had formed and GUS staining was visible at the junction of the embryo and suspensor, which might be related to its vigorous physical activity. The GUS staining of P5 was also detected in mature somatic embryos and was significantly weaker than that in P2. The GUS transcript abundance in P2 and P5 did not change significantly at different stages of somatic embryogenesis as assessed by qRT-PCR (Figure S5).
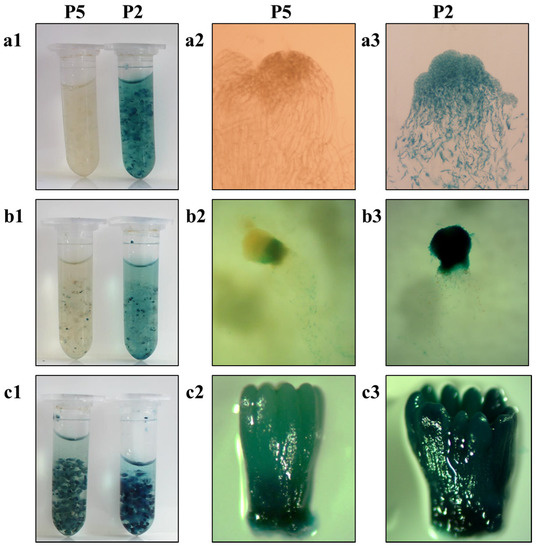
Figure 5.
Histochemical detection of the GUS staining in somatic embryos at different developmental stages from transgenic cell lines P2 and P5. (a) Embryogenic tissues, (b) early embryos cultured for 15 days, (c) mature somatic embryos cultured for 42 days on differentiation medium supplemented with abscisic acid. (a1,b1,c1) Panoramic images of GUS staining of the above three tissues of P2 and P5, (a2,b2,c2) microscopic images of GUS staining of the above three tissues of P5, (a3,b3,c3) microscopic images of GUS staining of the above three tissues of P2.
The expression level of the CaMV 35S-driven GUS gene in the stable transformation lines was further estimated using an absolute quantification method. A standard curve for GUS expression was generated using a gradient serial dilution of the plasmid as the qRT-PCR template. Using the measured Ct values, the GUS transcript abundance in each transformed cell line was calculated as shown in Table 3. The GUS transcript abundance in each transformed cell line ranged from 101 to 103 magnitudes, with the highest transcript abundance of 2.89 × 103, which was consistent with the histochemical assessment of GUS staining (Table 3).

Table 3.
Transcript abundance of GUS, HPT, EF, eIF, and APm by qRT-PCR in stably transformed lines.
In addition, an intronless GUS gene from 35S::GUS was transferred and integrated into the genome of larch embryogenic cells (Figure S1), resulting in resistant cell lines (Figure S6). Two hygromycin-resistant cell lines harboring the vector 35S::GUS were selected for the detection of GUS staining (Figure S7). The two cell lines were stained and the GUS staining intensity was generally consistent with the GUS transcript abundance in each cell line (Table 3).
3.5. HPT Transcript Abundance in Stable Transformation Lines
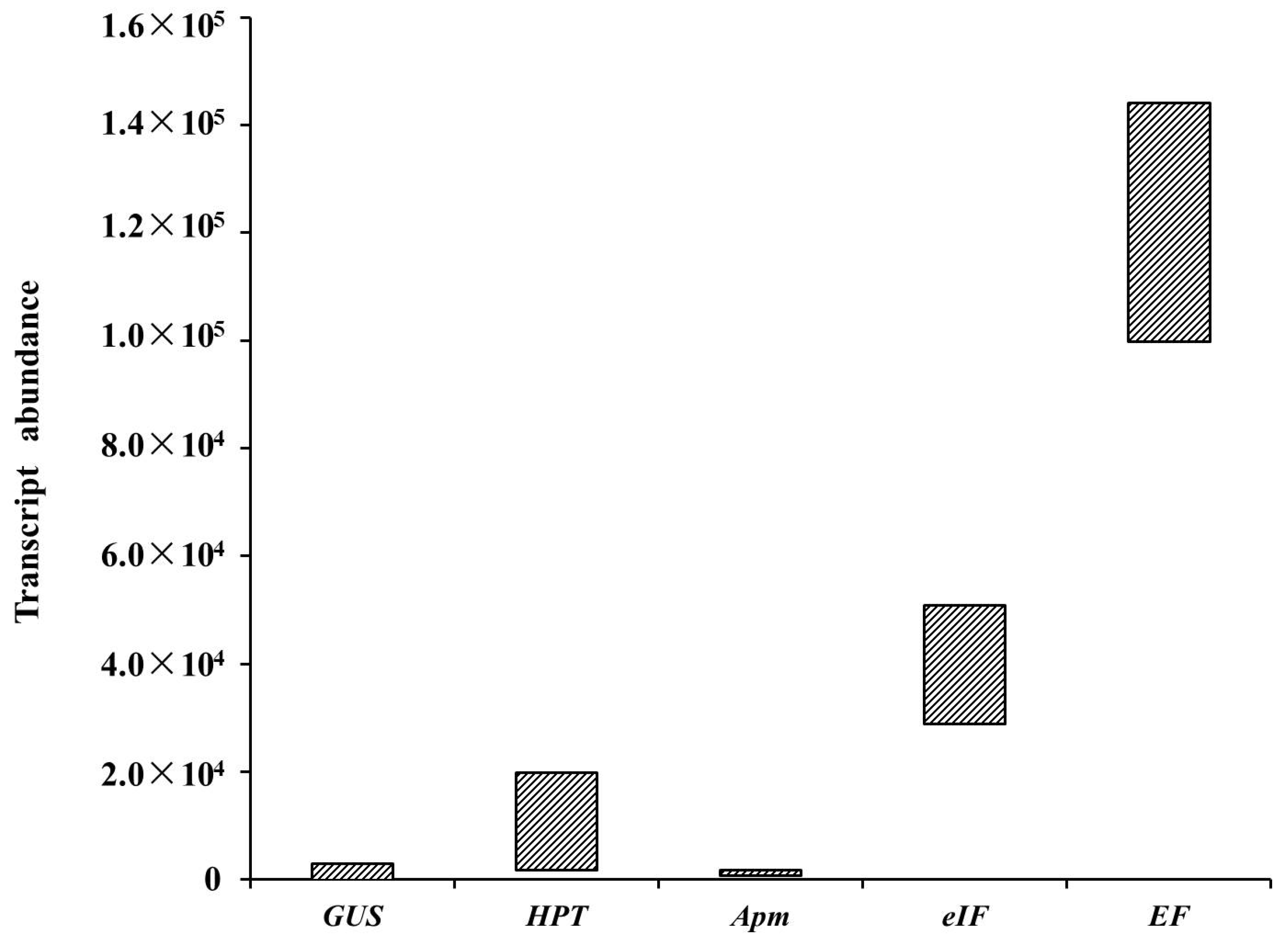
The HPT transcript abundance in different samples was calculated using the measured Ct values (Table 3). The HPT transcript abundance in each transformed cell line ranged from 103 to 104 magnitudes, with the highest and lowest transcript abundances of 1.98 × 104 and 1.80 × 103, respectively. Three candidate reference genes (EF, eIF, and APm) differed in abundance, as assessed by qRT-PCR, which was used to quantify their mRNA levels (Table 3). EF had the highest transcript abundance, eIF was moderately abundant, and the APm transcripts were detected at a low level (Figure 6). The transcript levels of HPT and GUS driven by the CaMV 35S promoter were equivalent to those of reference genes with moderate and low transcript abundances, respectively (Figure 6).
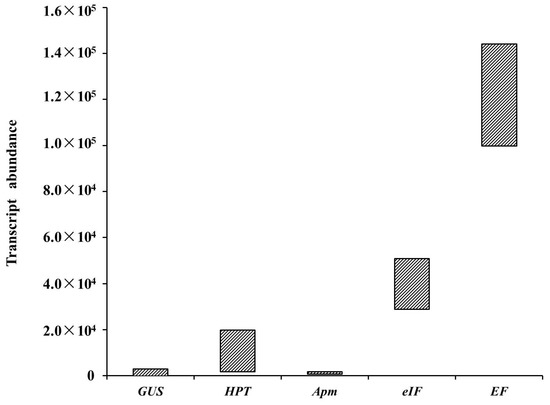
Figure 6.
Schematic diagram of transcript abundance distributions of GUS, HPT, EF, eIF, and APm as assessed by qRT-PCR in stably transformed lines.
3.6. GUS Transcript Abundance under the Control of the LaTCTP Promoter during Somatic Embryogenesis
To further validate the accuracy of the aforementioned results, GUS was expressed under the control of the L. kaempferi pLaTCTP promoter in the ESM and somatic embryos at different developmental stages (Figure S1). The detailed flowchart of transformation is shown in Supplementary Figure S3. TCTPs are ubiquitously expressed in many eukaryotic organisms and are multifunctional proteins that perform vital functions, particularly in a variety of complex life processes [27]. Expression analysis has previously demonstrated that LaTCTP is constitutively and highly expressed during somatic embryogenesis [5]. The TCTP promoter can be used as a constitutive promoter to drive gusA expression in monocotyledons and dicotyledons [28], and thus is useful for the production of recombinant proteins that require constitutive expression in a plant system.
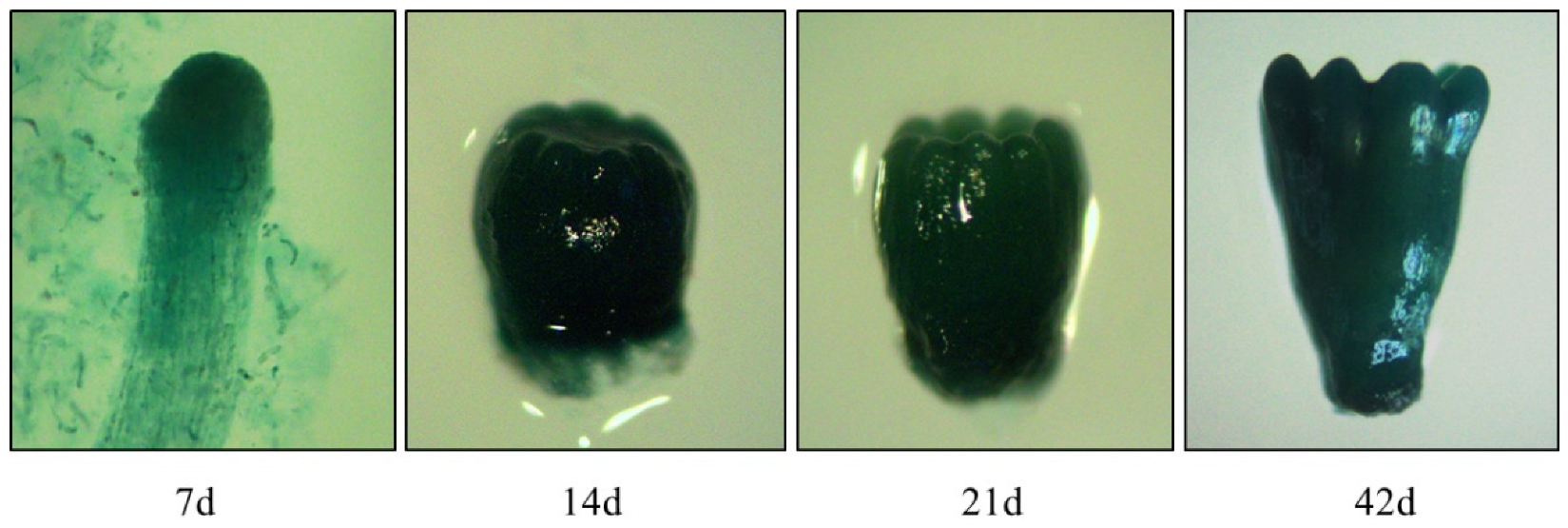
In the present study, five hygromycin-resistant cell lines harboring the pLaTCTP::GUS vector were chosen for further analysis. The transgenic ESM and somatic embryos at different developmental stages were stained intensely with X-Gluc, indicating that GUS expression was driven by the LaTCTP promoter (Figure 7). Based on the staining intensity, the GUS activity differed in each cell line and showed lighter staining (Figure S8). Except for pLaTCTP::GUS-5, the early embryos of the other four cell lines cultured for 15 days showed dark GUS staining, which was similar to that of the P5 cell line. Dark GUS staining was detected at different stages of somatic embryogenesis in pLaTCTP::GUS-4 (Figure 7).
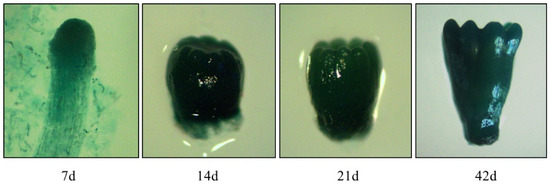
Figure 7.
Histochemical detection of the GUS staining in somatic embryos from transgenic cell lines pLaTCTP:GUS at different developmental stages detected in 7 days (7 days, 14 days, 21 days, and 42 days).
The transcript abundances of HPT, LaTCTP, and GUS driven by the LaTCTP promoter in the five hygromycin-resistant cell lines (pLaTCTP::GUS) were estimated (Table 4). The LaTCTP transcript abundance in each transformed cell line was at 105 magnitudes, whereas the GUS transcript abundance driven by the LaTCTP promoter was of 102 magnitudes, which was similar to the transcript abundance of GUS in cell lines transformed with the pCAMBIA1301 vector.

Table 4.
Transcript abundance of GUS, TCTP by qRT-PCR in stably transformed lines.
4. Discussion
In this study, several factors affecting the Agrobacterium-mediated genetic transformation of Japanese larch were adjusted, including the infection method and duration, and the bacterial removal process (Figure S3). The co-cultivation time is an important factor that affects transformation frequency [29,30]. A two-day co-cultivation period was previously considered to be the maximum because excessive bacterial growth causes culture death [16]. In the present study, under infection with a reduced concentration of bacteria, it can prevent excessive bacterial growth causing culture death in co-cultivation, and the co-cultivation time can be appropriately extended. Because embryogenic cell suspension cultures are sensitive to bacteria, a higher concentration of bacteria or prolonged infection time will increase the likelihood of excessive bacterial growth causing culture death in co-cultivation and will increase the difficulty of maintaining uncontaminated cultures. Thus, infection with a reduced concentration of bacteria and elimination of the pre-culture step can appropriately prolong the co-cultivation time and achieve reasonable transformation efficiency in Larix kaempferi, which can be attained with an average of 20 transgenic lines per gram FW (Table 2).
The antibiotics kanamycin and hygromycin are efficient selective agents for the AMT of conifer embryonic cultures [2,16]. In spruce and loblolly pine, 2.5 mg/L hygromycin has been used for the selection of transformed embryonic lines [16]. However, in hybrid larch (L. kaempferi × L. decidua), 5 mg/L hygromycin is sufficient to inhibit the growth of non-transformed embryonal masses [2,19], which was subsequently chosen for further selection (Figure 2). This is consistent with the optimal hygromycin concentration for the screening of L. kaempferi in the present study, and slightly higher than the optimal concentration (4 mg/L hygromycin) for L. olgensis and hybrid larch (L. kaempferi 3 × L. gmelinii 9) [25,26]. The cell lines that were able to grow on the selective medium were transformation positive, which indicated that hygromycin is an efficient selective agent for larch transformation [2,19,25,26].
In the present experiments, the percentage detection of GUS staining was 66.67% among the nine hygromycin-resistant cell lines harboring the pCAMBIA1301 vector, indicating that the GUS expression level differed in each cell line. The GUS staining intensity in each transformed cell line was consistent with the GUS transcript abundance (Figure 4). The transcript abundance of GUS in the transformed lines P2 and P5 did not change significantly at different stages of somatic embryogenesis, as assessed by qRT-PCR (Figure S5), which demonstrated that GUS expression was not dependent on the developmental stage of the somatic embryo.
Previous studies have shown that the GUS intron-containing reporter gene [31] has an extremely low expression level, probably because the intron is poorly spliced in larch and thus cannot be used to monitor early transformation events following co-cultivation, such as the appearance of transformed cells and their multiplication [19]. In the current study, the GUS gene, with or without introns, was normally expressed in larch. However, most of the hygromycin-resistant embryonal masses were lost during further selection processes, which may be associated with the HPT expression level. The transformed lines were used to detect the transcript abundance of foreign genes. It was observed that the expression level of HPT in the surviving transformation lines must be greater than 103, and the GUS transcript level in each transformed cell line ranged from 101 to 103 (Table 3). Thus, the GUS expression level, as represented by the GUS staining intensity, observed with the naked eye must be greater than 103 in the ESM (Figure 4). Based on the HPT and GUS expression levels, it was observed that the expression levels of exogenous genes driven by the CaMV 35S promoter ranged from 101 to 104 (Figure 6). Compared with the expression of a housekeeping gene, the expression levels of HPT and GUS driven by the CaMV 35S promoter were equivalent to those of genes with moderate and low abundances, respectively, in Japanese larch (Figure 6). The expression levels of exogenous genes driven by the CaMV 35S promoter in the Japanese larch ESM were relatively low, which might be related to the insertion site and GUS copy number in the genome, or to the large genome of Japanese larch.
The GUS gene under the control of the pLaTCTP promoter was integrated into the genome of L. kaempferi embryogenic cells by Agrobacterium-mediated genetic transformation. The LaTCTP promoter could effectively drive the ectopic expression of GUS in larch, and constitutive expressions of GUS was observed in the ESM and somatic embryos at different developmental stages (Figure 7). The GUS transcript abundance driven by the LaTCTP promoter was of 102 magnitudes (Table 4), which was similar to the abundance in nine hygromycin-resistant cell lines harboring the pCAMBIA1301 vector, whereas the LaTCTP transcript abundance in each transformed cell line was of 105 magnitudes. The expression levels of GUS or HPT driven by the CaMV 35S or pLaTCTP promoters in Japanese larch were relatively low, which may be due to the relatively small number of transgenic samples. Therefore, additional transgenic cell lines need to be screened to obtain transformants with higher expression levels of foreign genes in Japanese larch for further functional research. This may be the reason why there are few reports of gene overexpression in gymnosperms.
5. Conclusions
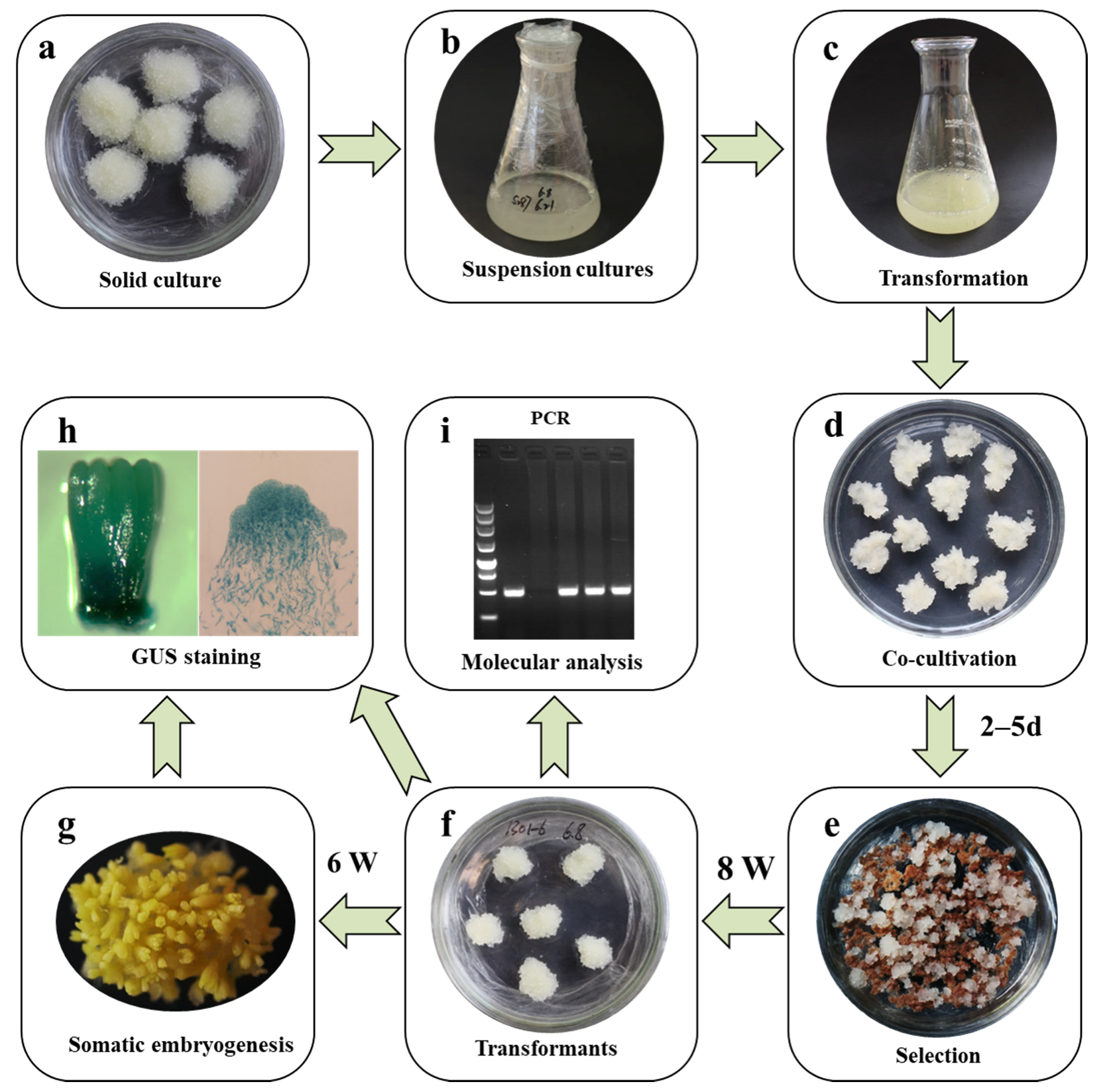
In the present study, a simple and efficient protocol for the laboratory-scale genetic transformation of Larix kaempferi was established using the A. tumefaciens strain GV3101 in conjunction with the binary vector pCAMBIA1301(Figure 8a–i). Several factors affecting the Agrobacterium-mediated genetic transformation of Japanese larch were adjusted, including the infection method and duration, and the bacterial elimination process. A relatively stable transformation efficiency was achieved by simplifying the infection method and improving the bacterial removal procedure. In the current study, the exogenous genes HPT and GUS, driven by the CaMV 35S, were normally expressed in Japanese larch. Additionally, its transcript abundances ranged from 101 to 104, which was equivalent to genes with moderate and low abundances, respectively. In addition, the endogenous pLaTCTP drove the constitutive expression of GUS in the ESM and somatic embryos at different developmental stages. The transcript abundance of GUS driven by the pLaTCTP promoter in Japanese larch verified the aforementioned results, which might be related to the insertion site and GUS copy number in the genome, or to a large genome of Japanese larch. A higher level of GUS expression was not detected, which may be due to the relatively small number of transgenic samples. Therefore, it is necessary to develop a new vector construction strategy or efficiently expressed promoter that can drive the ectopic expression of crucial genes to facilitate studies of gene function and quality modifications in Japanese larch.
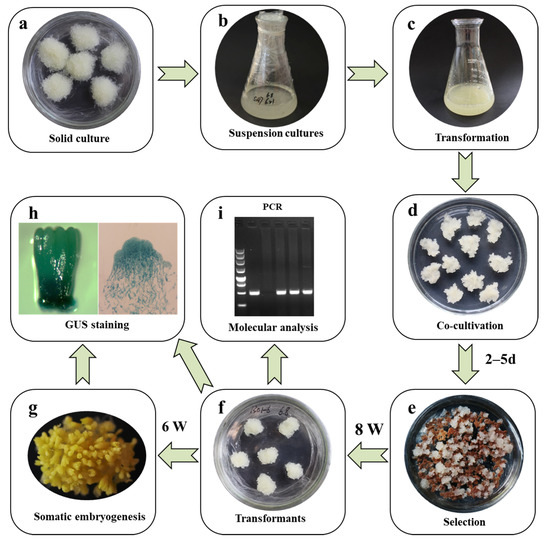
Figure 8.
Overview of the simplified protocol for the genetic transformation of Larix kaempferi. (a) Embryogenic masses, (b) suspension cells, (c) suspension cells mixed with Agrobacterium (OD600 = 0.05–0.1), (d) embryogenic masses co-cultivated with Agrobacterium for 46–120 h, (e) the selection of transformed embryogenic masses with cefotaxime (500 mg/L) and hygromycin (5 mg/L), (f) transgenic embryogenic masses, (g) somatic embryo maturation, (h) histochemical detection of GUS staining in somatic embryos, (i) molecular analysis of transgenic embryogenic lines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13091436/s1, Figure S1: Physical map of T-DNA region of the binary plasmids pCAMBIA1301, 35S::GUS, and pLaTCTP::GUS used for genetic transformation of Larix kaempferi; Figure S2: Schematic diagram of the improved vacuum filter; Figure S3: Principle of the simplified protocol for genetic transformation of Larix kaempferi; Figure S4: Development of hygromycin-resistant embryonal masses derived from line S287 co-cultivated with A. tumefaciens GV3101 harboring the binary vector pCAMBIA1301 on selective medium (supplemented with hygromycin 5 mg/L) after 60 day of culture; Figure S5: Accumulation of GUS transcripts at different developmental stages of somatic embryos from transgenic cell lines P2 (a) and P5 (b); Figure S6: Development of hygromycin-resistant embryonal masses derived from line S287 co-cultivated with A. tumefaciens GV3101 harboring the binary vector 35S::GUS on selective medium (supplemented with hygromycin 5 mg/L) after 60 days of culture; Figure S7: Histochemical staining for GUS activity in transgenic lines 35S::GUS-1 and 35S::GUS-2; Figure S8: GUS staining in the transgenic lines pLaTCTP:GUS.
Author Contributions
S.D. and L.Z. designed the experiments and drafted the manuscript; L.Z. carried out all the experiments and data analysis; S.H. and L.Q. provided suggestions on the experimental design and analyses. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (CAFYBB2019SY011), the National Natural Science Foundation of China (32171811 and 31600544), and the National Transgenic Major Program of China (2018ZX08020-003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pâques, L.E.; Foffová, E.; Heinze, B.; Lelu-Walter, M.A.; Liesebach, M.; Philippe, G. Larches (Larix sp.). In Forest Tree Breeding in Europe: Current State-of-the-Art and Perspectives, Managing Forest Ecosystems; Pâques, L.E., Ed.; Springer: Dordrecht, The Netherlands, 2013; Volume 25, pp. 13–122. [Google Scholar]
- Marie-Anne, L.; Pilate, G. Transgenic in Larix. In Molecular Biology of Woody Plants; Jain, S.M., Minocha, S.C., Eds.; Springer: Dordrecht, The Netherlands, 2000; Volume 2, pp. 119–134. [Google Scholar]
- Zhang, L.F.; Li, W.F.; Xu, H.Y.; Qi, L.W.; Han, S.Y. Cloning and characterization of four differentially expressed cDNAs encoding NFYA homologs involved in responses to ABA during somatic embryogenesis in Japanese larch (Larix leptolepis). Plant Cell Tissue Organ Cult. PCTOC 2014, 117, 293–304. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Han, S.; Li, X.; Qi, L. Transcriptome profiling and in silico analysis of somatic embryos in Japanese larch (Larix leptolepis). Plant Cell Rep. 2012, 31, 1637–1657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Li, W.F.; Han, S.Y.; Yang, W.H.; Qi, L.W. cDNA cloning, genomic organization and expression analysis during somatic embryogenesis of the translationally controlled tumor protein (TCTP) gene from Japanese larch (Larix leptolepis). Gene 2013, 529, 150–158. [Google Scholar] [CrossRef]
- Zhang, L.F.; Lan, Q.; Han, S.Y.; Qi, L.W. A GH3-like gene, LaGH3, isolated from hybrid larch (Larix leptolepis × Larix olgensis) is regulated by auxin and abscisic acid during somatic embryogenesis. Trees 2019, 33, 1723–1732. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Han, S.; Wu, T.; Li, X.; Li, W.; Qi, L. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 2012, 236, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Han, S.; Li, X.; Tong, Z.; Qi, L. Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in larches (Larix leptolepis). PLoS ONE 2013, 8, e81452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Zhang, S.G.; Li, S.G.; Han, S.Y.; Li, W.F.; Li, X.M.; Qi, L.W. Regulation of synchronism by abscisic-acid-responsive small noncoding RNAs during somatic embryogenesis in larch (Larix leptolepis). Plant Cell Tissue Organ Cult. PCTOC 2014, 116, 361–370. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Han, S.; Yang, W.; Li, W.; Wei, H.; Li, X.; Qi, L. Four abiotic stress-induced miRNA families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem. Biophys. Res. Commun. 2010, 398, 355–360. [Google Scholar] [CrossRef]
- Sarmast, M.K. Genetic transformation and somaclonal variation in conifers. Plant Biotechnol. Rep. 2016, 10, 309–325. [Google Scholar] [CrossRef]
- Tang, W.; Newton, R. Genetic transformation of conifers and its application in forest biotechnology. Plant Cell Rep. 2003, 22, 1–15. [Google Scholar] [CrossRef]
- Klimaszewska, K.; Devantier, Y.; Lachance, D.; Lelu, M.A.; Charest, P.J. Larix laricina (tamarack): Somatic embryogenesis and genetic transformation. Can. J. For. Res. 1997, 27, 538–550. [Google Scholar] [CrossRef]
- Klimaszewska, K.; Lachance, D.; Pelletier, G.; Lelu, M.A.; Séguin, A. Regeneration of transgenic Picea glauca, P. Mariana, and P. abies after cocultivation of embryogenic tissue with Agrobacterium tumefaciens. Vitr. Cell. Dev. Biol. Plant 2001, 37, 748–755. [Google Scholar] [CrossRef]
- Newton, R.J.; Bloom, J.C.; Bivans, D.H.; Mohan Jain, S. Stable genetic transformation of conifers. Phytomorphology 2001, 51, 421–434. [Google Scholar]
- Wenck, A.R.; Quinn, M.; Whetten, R.W.; Pullman, G.; Sederoff, R. High-efficiency Agrobacterium-mediated transformation of Norway spruce (Picea abies) and loblolly pine (Pinus taeda). Plant Mol. Biol. 1999, 39, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Ordás, R.J. Stable Agrobacterium-mediated transformation of maritime pine based on kanamycin selection. Sci. World J. 2013, 2013, 681792. [Google Scholar] [CrossRef]
- Lee, H.; Moon, H.K.; Park, S.Y. Agrobacterium-mediated Transformation via Somatic Embryogenesis System in Korean fir (Abies koreana Wil.), A Korean Native Conifer. Korean J. Plant Res. 2014, 27, 242–248. [Google Scholar] [CrossRef][Green Version]
- Levée, V.; Lelu, M.A.; Jouanin, L.; Cornu, D.; Pilate, G. Agrobacterium tumefaciens-mediated transformation of hybrid larch (Larix kaempferi × L. decidua) and transgenic plant regenerationn. Plant Cell Rep. 1997, 16, 680–685. [Google Scholar] [CrossRef]
- Levee, V.; Garin, E.; Klimaszewska, K.; Seguin, A. Stable genetic transformation of white pine (Pinus strobus L.) after cocultivation of embryogenic tissues with Agrobacterium tumefaciens. Mol. Breed. 1999, 5, 429–440. [Google Scholar] [CrossRef]
- Li, Z.X.; Li, S.G.; Zhang, L.F.; Han, S.Y.; Li, W.F.; Xu, H.Y.; Yang, W.H.; Liu, Y.L.; Fan, Y.R.; Qi, L.W. Over-expression of miR166a inhibits cotyledon formation in somatic embryos and promotes lateral root development in seedlings of Larix leptolepis. Plant Cell Tissue Organ Cult. PCTOC 2016, 127, 461–473. [Google Scholar] [CrossRef]
- Ewald, D.; Weckwerth, W.; Naujoks, G.; Zocher, R. Formation of Embryo-like Structures in Tissue Cultures of Different Yew Species. J. Plant Physiol. 1995, 147, 139–143. [Google Scholar] [CrossRef]
- Li, S.G.; Li, W.f.; Han, S.Y.; Yang, W.H.; Qi, L.W. Stage-specific regulation of four HD-ZIP III transcription factors during polar pattern formation in Larix leptolepis somatic embryos. Gene 2013, 522, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Ewald, D.; Kretzschmar, U.; Chen, Y. Continuous micropropagation of juvenile larch from different species via adventitious bud formation. Biol. Plant. 1997, 39, 321–329. [Google Scholar] [CrossRef]
- Song, Y.; Bai, X.; Dong, S.; Yang, Y.; Dong, H.; Wang, N.; Zhang, H.; Li, S. Stable and Efficient Agrobacterium-Mediated Genetic Transformation of Larch Using Embryogenic Callus. Front. Plant Sci. 2020, 11, 584492. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Yan, S.; An, P.; Cao, Q.; Wang, C.; Wang, J.; Zhang, H.; Zheng, L. Embryogenic callus induction from immature zygotic embryos and genetic transformation of Larix kaempferi 3x Larix gmelinii 9. PLoS ONE 2021, 16, e0258654. [Google Scholar] [CrossRef]
- Bommer, U.A.; Thiele, B.J. The translationally controlled tumour protein (TCTP). Int. J. Biochem. Cell Biol. 2004, 36, 379–385. [Google Scholar] [CrossRef]
- Masura, S.S.; Parveez, G.K.; Ti, L.L. Isolation and characterization of an oil palm constitutive promoter derived from a translationally control tumor protein (TCTP) gene. Plant Physiol. Biochem. 2011, 49, 701–708. [Google Scholar] [CrossRef]
- Ghimire, B.K.; Seong, E.S.; Lim, J.D.; Heo, K.; Kim, M.J.; Chung, I.M.; Juvik, J.A.; Yu, C.Y. Agrobacterium-mediated transformation of Codonopsis lanceolata using the γ-TMT gene. Plant Cell Tissue Organ Cult. PCTOC 2008, 95, 265–274. [Google Scholar] [CrossRef]
- Mishra, S.; Sangwan, R.S.; Bansal, S.; Sangwan, N.S. Efficient genetic transformation of Withania coagulans (Stocks) Dunal mediated by Agrobacterium tumefaciens from leaf explants of in vitro multiple shoot culture. Protoplasma 2013, 250, 451–458. [Google Scholar] [CrossRef]
- Vancanneyt, G.; Schmidt, R.; O’Connor-Sanchez, A.; Willmitzer, L.; Rocha-Sosa, M. Construction of an intron-containing marker gene: Splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol. Gen. Genet. MGG 1990, 220, 245–250. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).