A Key Study on Pollen-Specific SFB Genotype and Identification of Novel SFB Alleles from 48 Accessions in Japanese Apricot (Prunus mume Sieb. et Zucc.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. DNA and RNA Extraction
2.2.2. PCR Amplification of SFB Alleles
2.2.3. RT-PCR Amplification
2.2.4. PCR Products Sequencing
2.2.5. Sequence and Phylogenetic Analysis
3. Results
3.1. SFB Alleles Identification from P. mume Accessions
3.2. Specific Expression Analysis of SFB Alleles
3.3. Comparative Analysis of SFB Alleles Identified in P. mume and Other Prunus Species
3.4. Conserved Motifs Analysis
3.5. Phylogenetic Analysis of Pollen S genes (SLFL, SFB) in Prunus Species
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PmSFB | SFB allele of Prunus mume |
| bp | Base pairs |
| SC | Self-compatibility |
| SI | Self-incompatibility |
| PCR | Polymerase chain reaction |
| AS-PCR | Allele-specific polymerase chain reaction |
| RT-PCR | Reverse-transcription polymerase chain reaction |
| CTAB | Cetyltrimethylammonium bromide |
References
- Muñoz-Sanz, J.V.; Zuriaga, E.; Cruz-García, F.; McClure, B.; Romero, C. Self-(in) compatibility systems: Target traits for crop-production, plant breeding, and biotechnology. Front. Plant Sci. 2020, 11, 195. [Google Scholar] [CrossRef]
- Nasrallah, J.B. Self-incompatibility in the Brassicaceae: Regulation and mechanism of self-recognition. Curr. Top. Dev. Biol. 2019, 131, 435–452. [Google Scholar] [PubMed]
- Raduski, A.R.; Haney, E.B.; Igić, B. The expression of self-incompatibility in angiosperms is bimodal. Evol. Int. J. Org. Evol. 2012, 66, 1275–1283. [Google Scholar] [CrossRef]
- Bedinger, P.A.; Broz, A.K.; Tovar-Mendez, A.; McClure, B. Pollen-pistil interactions and their role in mate selection. Plant Physiol. 2017, 173, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Hiscock, S.J.; Tabah, D.A. The different mechanisms of sporophytic self–incompatibility. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2003, 358, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.B.; Meagher, T.R.; Gibbs, P.E. Do s genes or deleterious recessives control late-acting self-incompatibility in Handroanthus heptaphyllus (Bignoniaceae)? A diallel study with four full-sib progeny arrays. Ann. Bot. 2021, 127, 723–736. [Google Scholar] [CrossRef]
- Aguiar, B.; Vieira, J.; Cunha, A.E.; Fonseca, N.A.; Iezzoni, A.; van Nocker, S.; Vieira, C.P. Convergent evolution at the gametophytic self-incompatibility system in Malus and Prunus. PLoS ONE 2015, 10, e0126138. [Google Scholar] [CrossRef]
- Sheick, R.; Serra, S.; Tillman, J.; Luby, J.; Evans, K.; Musacchi, S. Characterization of a novel S-RNase allele and genotyping of new apple cultivars. Sci. Hortic. 2020, 273, 109630. [Google Scholar] [CrossRef]
- Morimoto, T. Insights into the Evolution and Establishment of the Prunus-Specific Self-Incompatibility Recognition Mechanism. Ph.D. Thesis, Kyoto University, Kyoto, Japan, 2017. [Google Scholar]
- Yamane, H.; Tao, R. Molecular and Developmental Biology: Self-incompatibility. In The Prunus Mume Genome; Springer: Cham, Switzerland, 2019; pp. 119–135. [Google Scholar]
- Newbigin, E.D.; Joshua, T.P.; Kohn, R. RNase-Based Self-Incompatibility: Puzzled by Pollen S. Plant Cell 2008, 20, 2286–2292. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, C.; Zhang, B.; Tang, F.; Li, F.; Liao, Q.; Tang, D.; Peng, Z.; Jia, Y.; Gao, M. A nonS-locus F-box gene breaks self-incompatibility in diploid potatoes. Nat. Commun. 2021, 12, 4142. [Google Scholar] [CrossRef]
- Claessen, H.; Keulemans, W.; Van de Poel, B.; De Storme, N. Finding a compatible partner: Self-incompatibility in European pear (Pyrus communis); molecular control, genetic determination, and impact on fertilization and fruit set. Front. Plant Sci. 2019, 10, 407. [Google Scholar] [CrossRef]
- Morimoto, T.; Akagi, T.; Tao, R. Evolutionary analysis of genes for S-RNase-based self-incompatibility reveals S locus duplications in the ancestral Rosaceae. Hortic. J. 2015, 84, 233–242. [Google Scholar] [CrossRef]
- Meng, X.; Sun, P.; Kao, T. S-RNase-based self-incompatibility in Petunia inflata. Ann. Bot. 2011, 108, 637–646. [Google Scholar] [CrossRef]
- Orlando Marchesano, B.M.; Chiozzotto, R.; Baccichet, I.; Bassi, D.; Cirilli, M. Development of an HRMA-Based Marker Assisted Selection (MAS) Approach for Cost-Effective Genotyping of S and M Loci Controlling Self-Compatibility in Apricot (Prunus armeniaca L.). Genes 2022, 13, 548. [Google Scholar] [CrossRef]
- McClure, B.A.; Haring, V.; Ebert, P.R.; Anderson, M.A.; Simpson, R.J.; Sakiyama, F.; Clarke, A.E. Style self-incompatibility gene products of Nicotlana alata are ribonucleases. Nature 1989, 342, 955–957. [Google Scholar] [CrossRef]
- Ushijima, K.; Sassa, H.; Dandekar, A.M.; Gradziel, T.M.; Tao, R.; Hirano, H. Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. Plant Cell 2003, 15, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, S.N.; Croxford, A.E.; March, T.J.; Wirthensohn, M.G.; Hrmova, M.; Mather, D.E. Variation among S-locus haplotypes and among stylar RNases in almond. Sci. Rep. 2020, 10, 583. [Google Scholar] [CrossRef] [PubMed]
- Yamane, H.; Ikeda, K.; Ushijima, K.; Sassa, H.; Tao, R. A pollen-expressed gene for a novel protein with an F-box motif that is very tightly linked to a gene for S-RNase in two species of cherry, Prunus cerasus and P. avium. Plant Cell Physiol. 2003, 44, 764–769. [Google Scholar] [CrossRef]
- Ikeda, K.; Igic, B.; Ushijima, K.; Yamane, H.; Hauck, N.R.; Nakano, R.; Sassa, H.; Iezzoni, A.F.; Kohn, J.R.; Tao, R. Primary structural features of the S haplotype-specific F-box protein, SFB, in Prunus. Sex. Plant Reprod. 2004, 16, 235–243. [Google Scholar] [CrossRef]
- Sonneveld, T.; Tobutt, K.R.; Vaughan, S.P.; Robbins, T.P. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype–specific F-Box gene. Plant Cell 2005, 17, 37–51. [Google Scholar] [CrossRef]
- Vaughan, S.; Russell, K.; Sargent, D.; Tobutt, K. Isolation of S-locus F-box alleles in Prunus avium and their application in a novel method to determine self-incompatibility genotype. Theor. Appl. Genet. 2006, 112, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Duan, X.; Wu, C.; Yu, J.; Liu, C.; Wang, J.; Zhang, X.; Yan, G.; Jiang, F.; Li, T. Ubiquitination of S4-RNase by S-LOCUS F-BOX LIKE2 contributes to self-compatibility of Sweet Cherry ‘Lapins’. Plant Physiol. 2020, 184, 1702–1716. [Google Scholar] [CrossRef]
- Romero, C.; Vilanova, S.; Burgos, L.; Martinez-Calvo, J.; Vicente, M.; Llácer, G.; Badenes, M.L. Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Mol. Biol. 2004, 56, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, S.; Badenes, M.L.; Burgos, L.; Martínez-Calvo, J.; Llácer, G.; Romero, C. Self-compatibility of two apricot selections is associated with two pollen-part mutations of different nature. Plant Physiol. 2006, 142, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Gu, C.; Zhang, S.; Zhang, S.; Wu, H.; Heng, W. Identification of S-haplotype-specific S-RNase and SFB alleles in native Chinese apricot (Prunus armeniaca L.). J. Hortic. Sci. Biotechnol. 2009, 84, 645–652. [Google Scholar] [CrossRef]
- Entani, T.; Iwano, M.; Shiba, H.; Che, F.S.; Isogai, A.; Takayama, S. Comparative analysis of the self-incompatibility (S-) locus region of Prunus mume: Identification of a pollen-expressed F-box gene with allelic diversity. Genes Cells 2003, 8, 203–213. [Google Scholar] [CrossRef]
- Hu, G.; Ni, Z.; Couliblay, D.; Gao, Z. Analysis of S Genotypes of 11 Plum Cultivars and Identification of New S Genes. J. Plant Genet. Resour. 2021, 22, 860–872. [Google Scholar]
- Lai, Z.; Ma, W.; Han, B.; Liang, L.; Zhang, Y.; Hong, G.; Xue, Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol. Biol. 2002, 50, 29–41. [Google Scholar] [CrossRef]
- Yamane, H.; Ushijima, K.; Sassa, H.; Tao, R. The use of the S haplotype-specific F-box protein gene, SFB, as a molecular marker for S-haplotypes and self-compatibility in Japanese apricot (Prunus mume). Theor. Appl. Genet. 2003, 107, 1357–1361. [Google Scholar] [CrossRef]
- Makovics-Zsohár, N.; Halász, J. Self-incompatibility system in polyploid fruit tree species—A review. Int. J. Plant Reprod. Biol. 2016, 8, 1–10. [Google Scholar]
- Ikeda, K.; Ushijima, K.; Yamane, H.; Tao, R.; Hauck, N.R.; Sebolt, A.M.; Iezzoni, A.F. Linkage and physical distances between the S-haplotype S-RNase and SFB genes in sweet cherry. Sex. Plant Reprod. 2005, 17, 289–296. [Google Scholar] [CrossRef]
- Lomax, J. Investigating Pollen Compatibility of Commercial Sweet Cherry Cultivars by DNA Analysis. Bachelor’s Thesis, University of Tasmania, Tasmania, Australia, 2021. [Google Scholar]
- Zhang, S.-L.; Huang, S.-X.; Kitashiba, H.; Nishio, T. Identification of S-haplotype-specific F-box gene in Japanese plum (Prunus salicina Lindl.). Sex. Plant Reprod. 2007, 20, 1–8. [Google Scholar] [CrossRef]
- Abdallah, D.; Baraket, G.; Ben Mustapha, S.; Angeles Moreno, M.A.; Salhi Hannachi, A. Molecular and Evolutionary Characterization of Pollen S Determinant (SFB Alleles) in Four Diploid and Hexaploid Plum Species (Prunus spp.). Biochem. Genet. 2021, 59, 42–61. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Wu, J.; Zhang, S.-J.; Yang, Y.-N.; Wu, H.-Q.; Khan, M.A.; Zhang, S.-L.; Liu, Q.-Z. Molecular analysis of eight SFB alleles and a new SFB-like gene in Prunus pseudocerasus and Prunus speciosa. Tree Genet. Genomes 2011, 7, 891–902. [Google Scholar] [CrossRef]
- Chin, S.-W.; Shaw, J.; Haberle, R.; Wen, J.; Potter, D. Diversification of almonds, peaches, plums and cherries—Molecular systematics and biogeographic history of Prunus (Rosaceae). Mol. Phylogenet. Evol. 2014, 76, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Pan, Z.; Hayat, F.; Bai, Y.; Coulibaly, D.; Ali, S.; Ni, X.; Shi, T.; Gao, Z. Comprehensive transcriptome profiling to identify genes involved in pistil abortion of Japanese apricot. Physiol. Mol. Biol. Plants 2021, 27, 1191–1204. [Google Scholar] [CrossRef]
- Yaegaki, H.; Shimada, T.; Moriguchi, T.; Hayama, H.; Haji, T.; Yamaguchi, M. Molecular characterization of S-RNase genes and S-genotypes in the Japanese apricot (Prunus mume Sieb. et Zucc.). Sex. Plant Reprod. 2001, 13, 251–257. [Google Scholar] [CrossRef]
- Habu, T.; Matsumoto, D.; Fukuta, K.; Esumi, T.; Tao, R.; Yaegaki, H.; Yamaguchi, M.; Matsuda, M.; Konishi, T.; Kitajima, A. Cloning and characterization of twelve S-RNase alleles in Japanese apricot (Prunus mume Sieb. et Zucc.). J. Jpn. Soc. Hortic. Sci. 2008, 77, 374–381. [Google Scholar] [CrossRef][Green Version]
- Xu, J.; Gao, Z.; Zhang, Z. Identification of S-genotypes and novel S-RNase alleles in Japanese apricot cultivars native to China. Sci. Hortic. 2010, 123, 459–463. [Google Scholar] [CrossRef]
- Wang, P.; Gao, Z.; Ni, Z.; Zhuang, W.; Zhang, Z. Isolation and identification of new pollen-specific SFB genes in Japanese apricot (Prunus mume). Genet. Mol. Res. 2013, 12, 3286–3295. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Tong, Z.; Zhang, Z.; Chen, Q.; Jiang, L.; Zhao, J. Improved method of DNA extraction from young leaves of wild peach. Jiangsu Agric. Sci. 2006, 5, 66–69. [Google Scholar]
- Tao, R.; Yamane, H.; Sugiura, A.; Murayama, H.; Sassa, H.; Mori, H. Molecular typing of S-alleles through identification, characterization and cDNA cloning for S-RNases in sweet cherry. J. Am. Soc. Hortic. Sci. 1999, 124, 224–233. [Google Scholar] [CrossRef]
- Xu, J.; Gao, Z.; Hou, J.; Wang, S.; Zhang, Z. Optimization of Guomei AS-PCR reaction system. Jiangsu Agric. Sci. 2008, 2, 69–71. [Google Scholar]
- Junxia, X. S Genotype Identification and New S Gene Cloning of Plum Varieties Native to China. Thesis of Mater, Nanjing Agricultural University, Nanjing, China, 2011. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Chen, Q.; Meng, D.; Gu, Z.; Li, W.; Yuan, H.; Duan, X.; Yang, Q.; Li, Y.; Li, T. SLFL genes participate in the ubiquitination and degradation reaction of S-RNase in self-compatible peach. Front. Plant Sci. 2018, 9, 227. [Google Scholar] [CrossRef]
- Abdallah, D.; Baraket, G.; Perez, V.; Ben Mustapha, S.; Salhi-Hannachi, A.; Hormaza, J.I. Analysis of self-incompatibility and genetic diversity in diploid and hexaploid plum genotypes. Front. Plant Sci. 2019, 10, 896. [Google Scholar] [CrossRef]
- Zhang, J. Study and Use Advance about Plum and Apricot Resource, 5th ed.; China Forestry Publishing House: Beijing, China, 2008. [Google Scholar]
- Ushijima, K.; Yamane, H.; Watari, A.; Kakehi, E.; Ikeda, K.; Hauck, N.R.; Iezzoni, A.F.; Tao, R. The S haplotype-specific F-box protein gene, SFB, is defective in self-compatible haplotypes of Prunus avium and P. mume. Plant J. 2004, 39, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, D.; Baraket, G.; Perez, V.; Hannachi, A.S.; Hormaza, J.I. Self-compatibility in peach [Prunus persica (L.) Batsch]: Patterns of diversity surrounding the S-locus and analysis of SFB alleles. Hortic. Res. 2020, 7, 170. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.H.R. Transcriptome Analysis of Self- and Cross-pollinated Pistils of Japanese Apricot (Prunus mume Sieb. et Zucc.). J. Jpn. Soc. Hort. Sci. 2014, 82, 95–107. [Google Scholar]
- Yamane, H.; Tao, R. Molecular basis of self-(in) compatibility and current status of S-genotyping in Rosaceous fruit trees. J. Jpn. Soc. Hortic. Sci. 2009, 78, 137–157. [Google Scholar] [CrossRef]
- Tao, R.; Watari, A.; Hanada, T.; Habu, T.; Yaegaki, H.; Yamaguchi, M.; Yamane, H. Self-compatible peach (Prunus persica) has mutant versions of the S haplotypes found in self-incompatible Prunus species. Plant Mol. Biol. 2007, 63, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Sanz, J.V.; Zuriaga, E.; López, I.; Badenes, M.L.; Romero, C. Self-(in) compatibility in apricot germplasm is controlled by two major loci, S and M. BMC Plant Biol. 2017, 17, 82. [Google Scholar] [CrossRef] [PubMed]
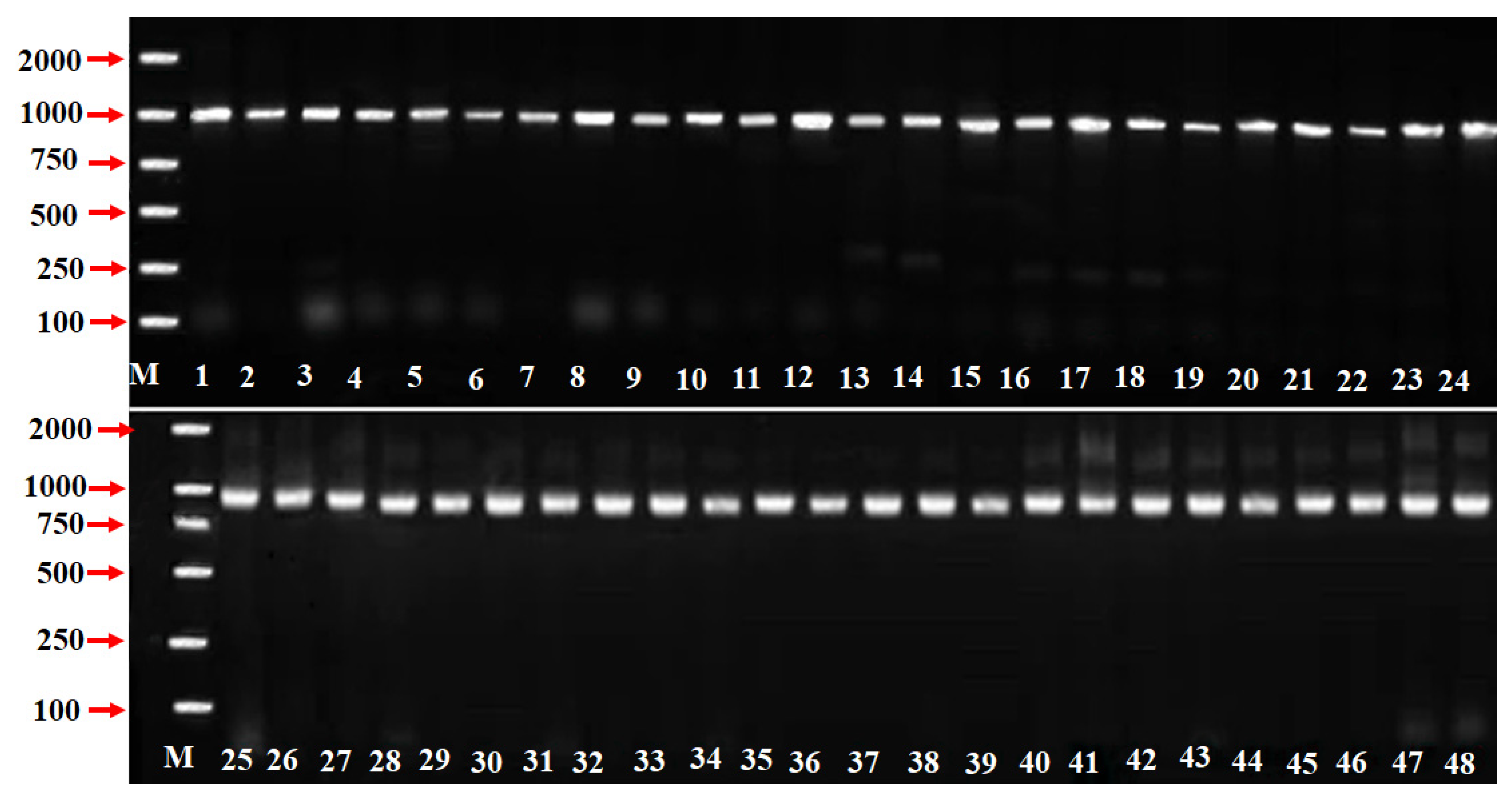


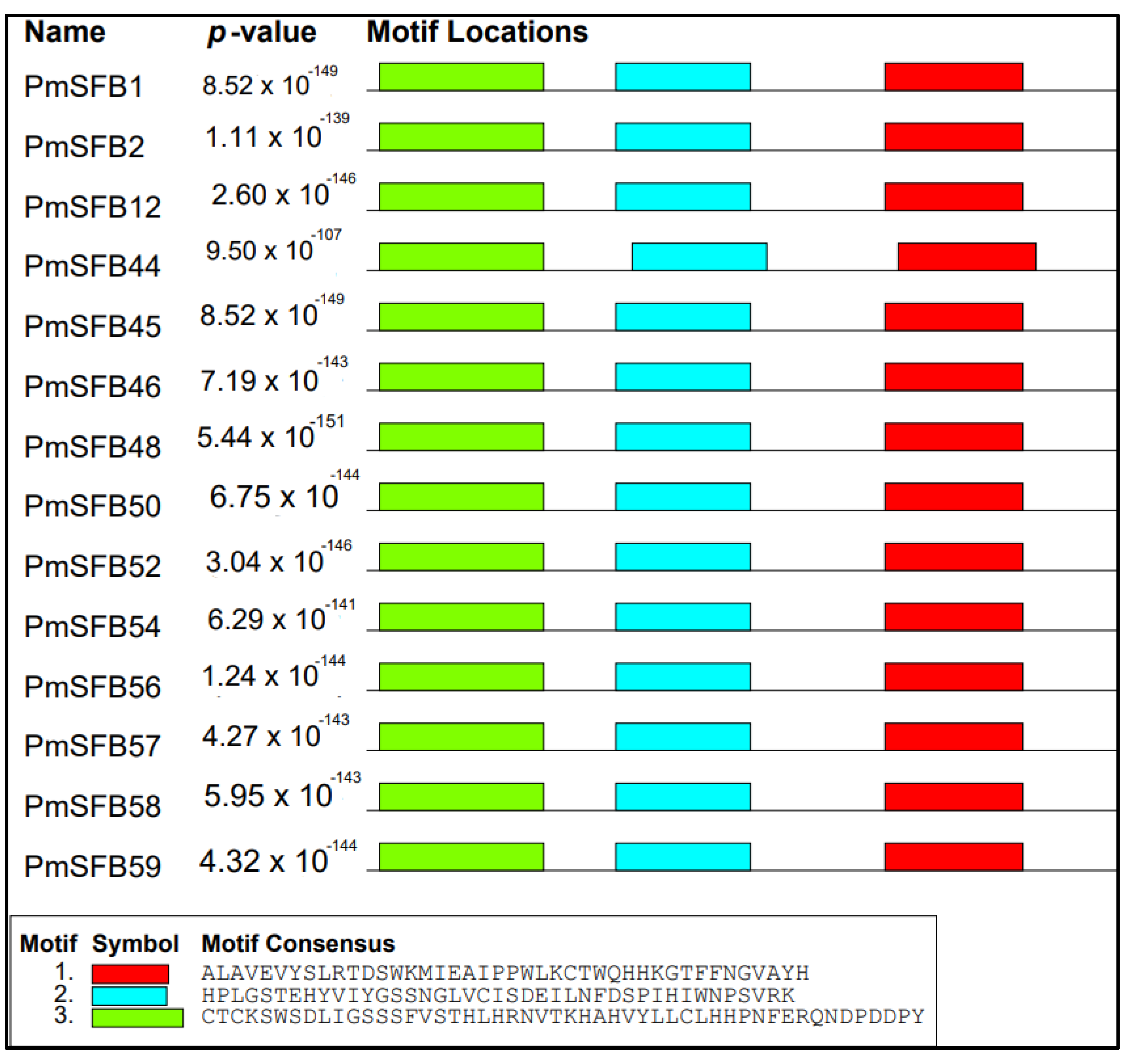
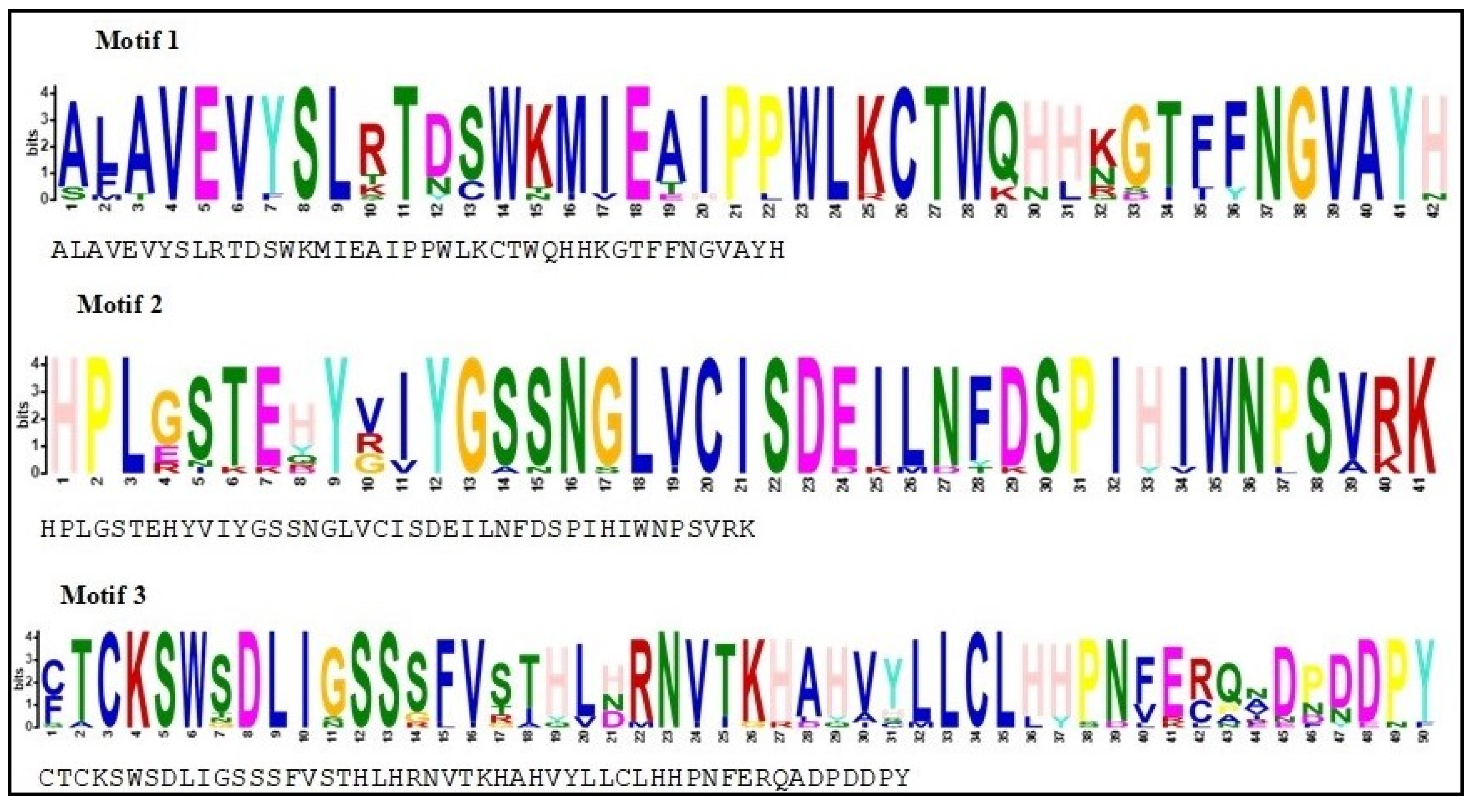
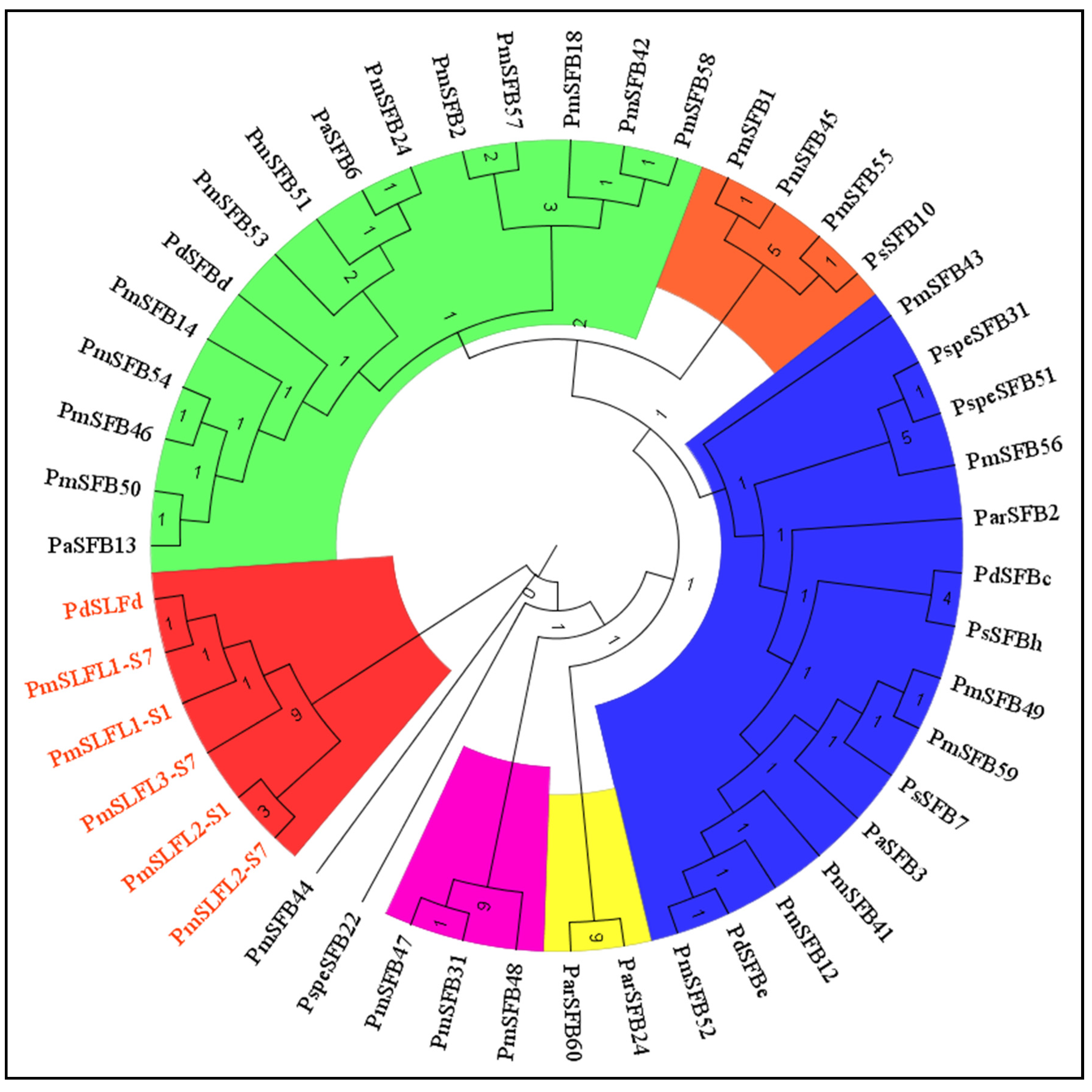
| Cultivars | SFB Genes | Novel Genes | Accession Numbers | Origin |
|---|---|---|---|---|
| Meilinhuang | SFB2/SFB42 | Zhejiang | ||
| Changnong No. 17 | SFB14/SFB42 | Zhejiang | ||
| Changxing No. 1 | SFB18/SFB42 | Zhejiang | ||
| Changxing No. 2 | SFB14/SFB41 | Zhejiang | ||
| Changxing No. 3 | SFB7/SFB54 | SFB54 | MW186470 | Zhejiang |
| Changxing No. 4 | SFB7/SFB42 | Zhejiang | ||
| Changxing No. 5 | SFB2/SFB42 | Zhejiang | ||
| Changxing No. 6 | SFB12/SFB41 | Zhejiang | ||
| Xianjulvmei | SFB2/SFB41 | Zhejiang | ||
| Longyoubaimei | SFB2/SFB14 | Zhejiang | ||
| Lizimei | SFB2/SFB41 | Zhejiang | ||
| Hongding | SFB18/SFB42 | Zhejiang | ||
| Xinbaimei | SFB24/SFB31 | Guangdong | ||
| Puningqingzhumei | SFB24/SFB41 | Guangdong | ||
| Guangdonghuangpi | SFB43/SFB56 | SFB56 | MW186472 | Guangdong |
| Daheqing | SFB24/SFB43 | Guangdong | ||
| Huanghoumei | SFB24/SFB31 | Guangdong | ||
| Dalizhong | SFB12/SFB55 | Guangdong | ||
| Henghe | SFB2/SFB42 | Guangdong | ||
| Yuanjiangroumei | SFB2/SFB14 | Hunan | ||
| Yuanjiangdaqing | SFB2/SFB47 | Hunan | ||
| Siyuemei | SFB14/SFB18 | Hunan | ||
| Yunnanyanmei | SFB14/SFB31 | Yunnan | ||
| Yunnanzhaoshuimei | SFB14/SFB50 | SFB50 | MW186466 | Yunnan |
| Zhaoanshuimei | SFB2/SFB14 | Fujian | ||
| Fujianqingmei | SFB14/SFB44 | SFB44 | MW186460 | Fujian |
| Sichuanbaimei | SFB31/SFB52 | SFB52 | MW186468 | Sichuan |
| Sichuanhuangmei | SFB12/SFB24 | Sichuan | ||
| Xinxiangxingmei | SFB14/SFB24 | Henan | ||
| Kaidi | SFB2/SFB45 | SFB45 | MW186461 | Jiangsu |
| Xiaolve | SFB1/SFB14 | Jiangsu | ||
| Yadanmei | SFB2/SFB42 | Jiangsu | ||
| Liuliumei No. 1 | SFB2/SFB46 | SFB46 | MW186462 | Jiangsu |
| Liuliumei No. 2 | SFB24/SFB56 | SFB56 | MW186472 | Jiangsu |
| Nannongfengyan | SFB14/SFB31 | Jiangsu | ||
| Nannongfengmao | SFB12/SFB43 | Jiangsu | ||
| Nannonglongfeng | SFB31/SFB43 | Jiangsu | ||
| Wanhong | SFB2/SFB57 | SFB57 | MW786959 | Jiangsu |
| Yunnankumei | SFB50/SFB59 | SFB59 | MW786961 | Yunnan |
| Hangzhoubaimei | SFB42/SFB58 | SFB58 | MW786960 | Zhejiang |
| Zhizhimei | SFB12/SFB24 | Jiangsu | ||
| Fenbanguomei | SFB2/SFB7 | Hubei | ||
| Dongshanlimei | SFB14/SFB24 | Jiangsu | ||
| Dongqing | SFB7/SFB42 | Zhejiang | ||
| Taihu No. 1 | SFB24/SFB46 | Jiangsu | ||
| Taihu No. 3 | SFB2/SFB24 | Jiangsu | ||
| Huangxiaoda | SFB1/SFB43 | Zhejiang | ||
| Lvmei | SFB14/SFB31 | Jiangsu |
| SFB | PmSFB | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pm | SFB1 | SFB2 | SFB12 | SFB14 | SFB18 | SFB24 | SFB31 | SFB41 | SFB42 | SFB43 | SFB44 | SFB45 | SFB46 | SFB47 | SFB48 | SFB49 | SFB50 | SFB51 | SFB52 | SFB53 | SFB54 | SFB55 | SFB56 | SFB57 | SFB58 | SFB59 |
| SFB1 | / | 82.41 | 80.19 | 80.00 | 82.10 | 81.73 | 78.95 | 83.25 | 79.94 | 83.28 | 60.06 | 96.93 | 79.26 | 81.73 | 82.35 | 80.80 | 81.11 | 82.10 | 80.50 | 80.31 | 79.88 | 81.42 | 79.94 | 82.41 | 80.25 | 81.73 |
| SFB2 | 82.41 | / | 79.32 | 79.08 | 81.17 | 81.17 | 77.78 | 79.32 | 78.09 | 79.63 | 62.96 | 82.72 | 80.25 | 79.63 | 80.25 | 76.23 | 79.01 | 80.31 | 77.78 | 76.99 | 79.01 | 78.09 | 79.38 | 95.06 | 76.54 | 76.54 |
| SFB12 | 80.19 | 79.32 | / | 79.69 | 77.78 | 80.80 | 78.95 | 84.83 | 77.78 | 82.04 | 64.71 | 80.50 | 78.02 | 78.64 | 81.12 | 81.73 | 76.47 | 79.94 | 89.69 | 76.37 | 74.54 | 76.47 | 79.63 | 79.01 | 76.54 | 82.04 |
| SFB14 | 80.00 | 79.08 | 79.69 | / | 78.46 | 82.46 | 79.69 | 84.31 | 76.62 | 79.38 | 63.69 | 79.69 | 81.54 | 82.46 | 81.58 | 77.85 | 81.54 | 80.98 | 79.08 | 77.68 | 79.38 | 78.46 | 77.30 | 79.69 | 76.31 | 77.85 |
| SFB18 | 82.10 | 81.17 | 77.78 | 78.46 | / | 79.63 | 78.40 | 78.09 | 81.48 | 78.09 | 63.27 | 81.79 | 77.16 | 80.86 | 79.01 | 78.09 | 78.70 | 78.77 | 77.47 | 80.06 | 80.86 | 77.47 | 78.15 | 80.86 | 80.56 | 78.40 |
| SFB24 | 81.73 | 81.17 | 80.80 | 82.46 | 79.63 | / | 83.28 | 81.11 | 79.94 | 81.73 | 63.04 | 82.04 | 81.42 | 81.73 | 82.61 | 79.57 | 80.80 | 86.38 | 79.88 | 80.86 | 81.42 | 81.11 | 80.19 | 81.17 | 78.70 | 78.95 |
| SFB31 | 78.95 | 77.78 | 78.95 | 79.69 | 78.40 | 83.28 | / | 78.95 | 76.85 | 79.57 | 64.09 | 78.64 | 78.02 | 83.59 | 82.66 | 77.71 | 78.95 | 82.10 | 76.78 | 77.54 | 78.02 | 79.26 | 76.85 | 78.09 | 75.93 | 77.71 |
| SFB41 | 83.25 | 79.32 | 84.83 | 84.31 | 78.09 | 81.11 | 78.95 | / | 79.63 | 82.35 | 65.33 | 82.04 | 80.05 | 80.80 | 82.04 | 83.59 | 82.35 | 80.25 | 83.90 | 76.92 | 79.26 | 77.40 | 80.86 | 79.94 | 79.01 | 83.59 |
| SFB42 | 79.94 | 78.09 | 77.78 | 76.62 | 81.48 | 79.94 | 76.85 | 79.63 | / | 78.09 | 62.96 | 80.25 | 76.85 | 78.40 | 77.47 | 77.16 | 76.85 | 77.23 | 75.00 | 75.46 | 77.16 | 75.31 | 77.54 | 78.09 | 96.91 | 78.40 |
| SFB43 | 83.28 | 79.63 | 82.04 | 79.38 | 78.09 | 81.73 | 79.57 | 82.35 | 78.09 | / | 64.71 | 83.59 | 78.33 | 82.04 | 80.50 | 83.59 | 80.19 | 82.10 | 79.88 | 78.15 | 76.47 | 79.88 | 80.25 | 79.01 | 76.85 | 84.21 |
| SFB44 | 60.06 | 62.96 | 64.71 | 63.69 | 63.27 | 63.04 | 64.09 | 65.33 | 62.96 | 64.71 | / | 62.85 | 65.63 | 65.02 | 66.46 | 63.78 | 61.92 | 62.85 | 62.45 | 61.11 | 60.68 | 62.85 | 65.63 | 63.27 | 62.35 | 63.78 |
| SFB45 | 96.93 | 82.72 | 80.50 | 79.69 | 81.79 | 82.04 | 78.64 | 82.04 | 80.25 | 83.59 | 62.85 | / | 79.57 | 81.42 | 82.04 | 80.50 | 80.80 | 81.79 | 80.19 | 80.00 | 80.19 | 81.11 | 80.25 | 82.10 | 79.94 | 81.42 |
| SFB46 | 79.26 | 80.25 | 78.02 | 81.54 | 77.16 | 81.42 | 78.02 | 80.05 | 76.85 | 78.33 | 65.63 | 79.57 | / | 79.88 | 79.88 | 76.16 | 81.73 | 78.70 | 76.47 | 75.83 | 81.73 | 77.09 | 78.40 | 79.32 | 75.62 | 76.47 |
| SFB47 | 81.73 | 79.63 | 78.64 | 82.46 | 80.86 | 81.73 | 83.59 | 80.80 | 78.40 | 82.04 | 65.02 | 81.42 | 79.88 | / | 82.97 | 80.50 | 81.73 | 81.79 | 78.95 | 78.46 | 79.88 | 78.33 | 77.16 | 80.56 | 78.09 | 79.88 |
| SFB48 | 82.35 | 80.25 | 81.12 | 81.58 | 79.01 | 82.61 | 82.66 | 82.04 | 77.47 | 80.50 | 66.46 | 82.04 | 79.88 | 82.97 | / | 79.26 | 80.19 | 82.35 | 82.04 | 78.07 | 79.26 | 81.73 | 80.08 | 80.25 | 77.16 | 79.26 |
| SFB49 | 80.80 | 76.23 | 81.73 | 77.85 | 78.09 | 79.57 | 77.71 | 83.59 | 77.16 | 83.59 | 63.78 | 80.50 | 76.16 | 80.50 | 79.26 | / | 77.71 | 79.63 | 82.35 | 79.08 | 76.47 | 77.71 | 78.09 | 76.85 | 77.16 | 96.56 |
| SFB50 | 81.11 | 79.01 | 76.47 | 81.54 | 78.70 | 80.80 | 78.95 | 82.35 | 76.85 | 80.19 | 61.92 | 80.80 | 81.73 | 81.73 | 80.19 | 77.71 | / | 82.10 | 77.71 | 78.77 | 81.11 | 78.33 | 75.93 | 79.63 | 76.85 | 78.02 |
| SFB51 | 82.10 | 80.31 | 79.94 | 80.98 | 78.77 | 86.38 | 82.10 | 80.25 | 77.23 | 82.10 | 62.85 | 81.79 | 78.70 | 81.79 | 82.35 | 79.63 | 82.10 | / | 79.94 | 83.02 | 77.47 | 82.10 | 78.38 | 81.23 | 77.23 | 79.94 |
| SFB52 | 80.50 | 77.78 | 89.69 | 79.08 | 77.47 | 79.88 | 76.78 | 83.90 | 75.00 | 79.88 | 62.45 | 80.19 | 76.47 | 78.95 | 82.04 | 82.35 | 77.71 | 79.94 | / | 76.00 | 75.54 | 76.16 | 77.47 | 79.01 | 75.00 | 82.35 |
| SFB53 | 80.31 | 76.99 | 76.37 | 77.68 | 80.06 | 80.86 | 77.54 | 76.92 | 75.46 | 78.15 | 61.11 | 80.00 | 75.83 | 78.46 | 78.07 | 79.08 | 78.77 | 83.02 | 76.00 | / | 79.08 | 77.85 | 77.47 | 78.22 | 75.46 | 80.00 |
| SFB54 | 79.88 | 79.01 | 74.54 | 79.38 | 80.86 | 81.42 | 78.02 | 79.26 | 77.16 | 76.47 | 60.68 | 80.19 | 81.73 | 79.88 | 79.26 | 76.47 | 81.11 | 77.47 | 75.54 | 79.08 | / | 74.92 | 75.62 | 78.40 | 75.93 | 75.85 |
| SFB55 | 81.42 | 78.09 | 76.47 | 78.46 | 77.47 | 81.11 | 79.26 | 77.40 | 75.31 | 79.88 | 62.85 | 81.11 | 77.09 | 78.33 | 81.73 | 77.71 | 78.33 | 82.10 | 76.16 | 77.85 | 74.92 | / | 75.93 | 78.09 | 75.31 | 78.02 |
| SFB56 | 79.94 | 79.38 | 79.63 | 77.30 | 78.15 | 80.19 | 76.85 | 80.86 | 77.54 | 80.25 | 65.63 | 80.25 | 78.40 | 77.16 | 80.08 | 78.09 | 75.93 | 78.38 | 77.47 | 77.47 | 75.62 | 75.93 | / | 78.77 | 75.69 | 79.32 |
| SFB57 | 82.41 | 95.06 | 79.01 | 79.69 | 80.86 | 81.17 | 78.09 | 79.94 | 78.09 | 79.01 | 63.27 | 82.10 | 79.32 | 80.56 | 80.25 | 76.85 | 79.63 | 81.23 | 79.01 | 78.22 | 78.40 | 78.09 | 78.77 | / | 77.78 | 76.85 |
| SFB58 | 80.25 | 76.54 | 76.54 | 76.31 | 80.56 | 78.70 | 75.93 | 79.01 | 96.91 | 76.85 | 62.35 | 79.94 | 75.62 | 78.09 | 77.16 | 77.16 | 76.85 | 77.23 | 75.00 | 75.46 | 75.93 | 75.31 | 75.69 | 77.78 | / | 78.40 |
| SFB59 | 81.73 | 76.54 | 82.04 | 77.85 | 78.40 | 78.95 | 77.71 | 83.59 | 78.40 | 84.21 | 63.78 | 81.42 | 76.47 | 79.88 | 79.26 | 96.56 | 78.02 | 79.94 | 82.35 | 80.00 | 75.85 | 78.02 | 79.32 | 76.85 | 78.40 | / |
| SFB | Par | Pa | Pd | Ps | Pspe | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pm | SFB2 | SFB24 | SFB60 | SFB3 | SFB6 | SFB13 | SFBc | SFBd | SFBe | SFB7 | SFB10 | SFBh | SFB22 | SFB31 | SFB51 |
| F1-box | 63.94 | 65.15 | 65.76 | 61.33 | 62.12 | 61.03 | 61.93 | 64.24 | 61.33 | 62.54 | 63.25 | 61.03 | 61.52 | 64.55 | 63.55 |
| F2-box | 64.55 | 65.15 | 65.76 | 61.93 | 62.12 | 61.33 | 61.33 | 63.94 | 61.14 | 62.65 | 63.86 | 61.33 | 61.21 | 65.15 | 64.15 |
| SFB1 | 81.73 | 84.52 | 83.90 | 81.73 | 80.50 | 80.19 | 76.78 | 82.35 | 78.95 | 77.23 | 85.49 | 79.23 | 65.53 | 80.80 | 70.80 |
| SFB2 | 78.70 | 79.94 | 79.94 | 76.85 | 79.01 | 79.01 | 74.38 | 80.25 | 75.62 | 75.00 | 81.23 | 75.00 | 69.14 | 79.63 | 69.63 |
| SFB12 | 81.42 | 80.19 | 79.88 | 82.04 | 78.64 | 75.54 | 75.23 | 78.64 | 84.21 | 79.26 | 78.95 | 79.32 | 73.46 | 79.88 | 78.88 |
| SFB14 | 80.00 | 79.38 | 79.08 | 79.08 | 81.54 | 80.92 | 77.54 | 82.15 | 79.08 | 76.31 | 78.46 | 78.77 | 69.33 | 77.54 | 73.54 |
| SFB18 | 77.16 | 79.94 | 79.63 | 76.23 | 77.78 | 82.72 | 73.15 | 78.70 | 76.23 | 75.31 | 79.94 | 75.00 | 69.75 | 77.16 | 72.16 |
| SFB24 | 80.12 | 83.59 | 83.28 | 80.50 | 97.20 | 79.57 | 76.47 | 82.04 | 79.26 | 77.09 | 82.35 | 75.31 | 71.83 | 80.43 | 77.43 |
| SFB31 | 76.16 | 83.59 | 83.28 | 77.40 | 81.73 | 77.09 | 74.92 | 78.95 | 79.57 | 75.23 | 78.95 | 73.46 | 72.22 | 76.78 | 74.78 |
| SFB41 | 82.97 | 80.50 | 80.19 | 83.59 | 79.26 | 81.42 | 79.88 | 81.73 | 83.28 | 82.04 | 79.26 | 82.10 | 69.66 | 81.11 | 71.11 |
| SFB42 | 75.62 | 80.25 | 79.63 | 75.93 | 77.47 | 77.47 | 76.54 | 76.85 | 76.23 | 75.31 | 78.09 | 73.77 | 67.59 | 77.78 | 70.78 |
| SFB43 | 79.26 | 82.35 | 81.73 | 81.73 | 79.88 | 78.02 | 75.85 | 80.19 | 79.88 | 81.42 | 81.11 | 76.54 | 72.53 | 80.50 | 78.64 |
| SFB44 | 63.94 | 64.85 | 65.45 | 61.03 | 62.12 | 60.73 | 61.93 | 64.24 | 60.54 | 62.24 | 63.14 | 60.73 | 61.28 | 64.24 | 61.03 |
| SFB45 | 81.42 | 84.21 | 83.59 | 82.04 | 80.19 | 79.88 | 76.47 | 82.66 | 78.64 | 77.40 | 86.07 | 79.01 | 72.22 | 81.11 | 82.04 |
| SFB46 | 77.40 | 80.19 | 79.57 | 77.71 | 78.95 | 79.57 | 77.40 | 82.04 | 74.92 | 74.30 | 79.26 | 77.47 | 68.50 | 78.64 | 81.11 |
| SFB47 | 79.57 | 84.21 | 83.59 | 77.40 | 81.42 | 80.50 | 74.92 | 80.19 | 82.04 | 77.71 | 78.02 | 75.62 | 73.15 | 78.02 | 80.19 |
| SFB48 | 81.37 | 83.90 | 83.28 | 78.95 | 81.68 | 78.02 | 74.61 | 81.73 | 81.42 | 77.40 | 81.42 | 75.62 | 73.99 | 81.73 | 80.50 |
| SFB49 | 79.57 | 81.11 | 80.50 | 83.28 | 78.95 | 76.47 | 77.71 | 78.02 | 82.35 | 86.69 | 78.95 | 80.25 | 69.66 | 78.95 | 78.33 |
| SFB50 | 78.64 | 79.88 | 79.26 | 76.47 | 80.50 | 90.40 | 74.92 | 81.11 | 75.23 | 76.23 | 79.57 | 76.85 | 70.06 | 77.40 | 78.95 |
| SFB51 | 80.19 | 83.90 | 83.99 | 80.25 | 86.07 | 80.25 | 75.93 | 81.79 | 79.01 | 77.78 | 83.02 | 74.85 | 72.53 | 79.26 | 78.40 |
| SFB52 | 81.42 | 79.02 | 78.64 | 82.97 | 79.88 | 75.85 | 75.23 | 78.64 | 87.00 | 79.88 | 77.71 | 77.47 | 71.91 | 78.33 | 77.71 |
| SFB53 | 76.85 | 80.31 | 80.00 | 77.23 | 79.94 | 81.23 | 76.31 | 79.69 | 76.62 | 75.69 | 79.69 | 75.15 | 69.23 | 78.09 | 79.08 |
| SFB54 | 76.47 | 79.88 | 79.57 | 74.92 | 78.95 | 81.73 | 75.23 | 81.42 | 70.23 | 74.30 | 78.33 | 76.23 | 69.44 | 75.54 | 82.66 |
| SFB55 | 75.85 | 82.04 | 81.73 | 79.57 | 81.68 | 77.09 | 72.76 | 79.26 | 77.71 | 75.54 | 84.21 | 74.38 | 69.44 | 76.78 | 78.33 |
| SFB56 | 78.33 | 79.63 | 79.32 | 77.47 | 77.71 | 76.85 | 76.23 | 79.63 | 76.85 | 77.16 | 79.01 | 75.38 | 71.30 | 98.14 | 78.40 |
| SFB57 | 74.32 | 79.56 | 84.33 | 79.45 | 80.27 | 81.36 | 74.36 | 78.54 | 77.86 | 78.36 | 78.25 | 79.36 | 80.22 | 85.31 | 78.12 |
| SFB58 | 84.32 | 75.23 | 77.56 | 85.32 | 74.32 | 76.23 | 77.23 | 75.32 | 78.86 | 78.64 | 79.62 | 83.25 | 84.32 | 86.32 | 78.23 |
| SFB59 | 83.23 | 75.23 | 78.63 | 76.51 | 77.53 | 78.54 | 78.24 | 79.53 | 86.23 | 75.23 | 77.12 | 76.24 | 75.23 | 80.23 | 81.32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coulibaly, D.; Hu, G.; Ni, Z.; Ouma, K.O.; Huang, X.; Iqbal, S.; Ma, C.; Shi, T.; Hayat, F.; Karikari, B.; et al. A Key Study on Pollen-Specific SFB Genotype and Identification of Novel SFB Alleles from 48 Accessions in Japanese Apricot (Prunus mume Sieb. et Zucc.). Forests 2022, 13, 1388. https://doi.org/10.3390/f13091388
Coulibaly D, Hu G, Ni Z, Ouma KO, Huang X, Iqbal S, Ma C, Shi T, Hayat F, Karikari B, et al. A Key Study on Pollen-Specific SFB Genotype and Identification of Novel SFB Alleles from 48 Accessions in Japanese Apricot (Prunus mume Sieb. et Zucc.). Forests. 2022; 13(9):1388. https://doi.org/10.3390/f13091388
Chicago/Turabian StyleCoulibaly, Daouda, Guofeng Hu, Zhaojun Ni, Kenneth Omondi Ouma, Xiao Huang, Shahid Iqbal, Chengdong Ma, Ting Shi, Faisal Hayat, Benjamin Karikari, and et al. 2022. "A Key Study on Pollen-Specific SFB Genotype and Identification of Novel SFB Alleles from 48 Accessions in Japanese Apricot (Prunus mume Sieb. et Zucc.)" Forests 13, no. 9: 1388. https://doi.org/10.3390/f13091388
APA StyleCoulibaly, D., Hu, G., Ni, Z., Ouma, K. O., Huang, X., Iqbal, S., Ma, C., Shi, T., Hayat, F., Karikari, B., & Gao, Z. (2022). A Key Study on Pollen-Specific SFB Genotype and Identification of Novel SFB Alleles from 48 Accessions in Japanese Apricot (Prunus mume Sieb. et Zucc.). Forests, 13(9), 1388. https://doi.org/10.3390/f13091388







