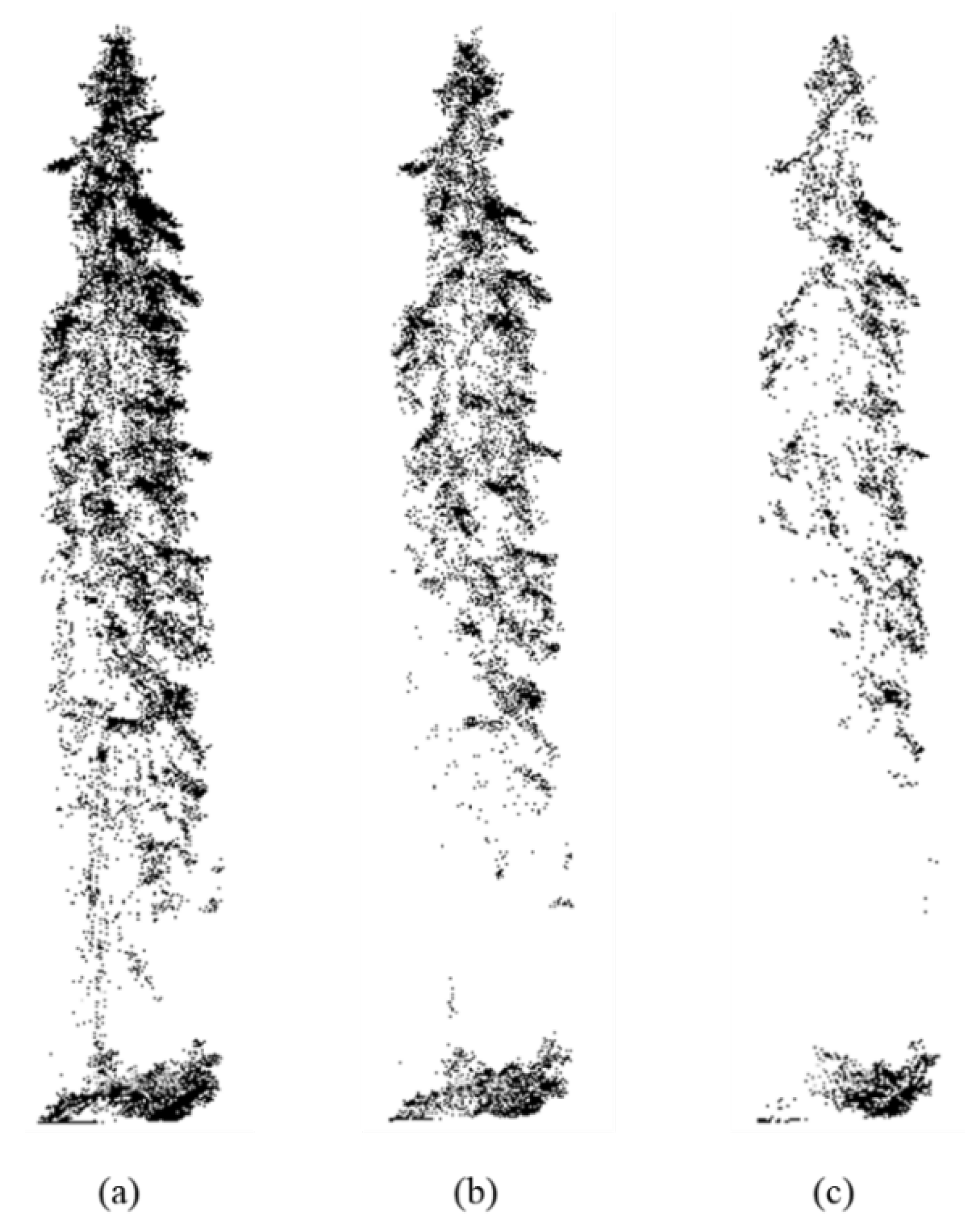
1. Introduction
Picea crassifolia (Kom., 1923) is a typical vegetation in Northwest China and is mainly distributed in the Qilian Mountains (approximately 1.3 × 10
5 ha), accounting for over 94.6% of the total area of
Picea crassifolia in this region [
1].
Picea crassifolia within the Qilian Mountains has been found mostly at altitudes of 2500–3200 m and in shady and semi-shady alpine areas with slopes of 12–24° [
2]. As one of the main species of water conservation forest in the Qilian Mountains, correctly monitoring the forest structural parameters of
Picea crassifolia is vital for the ecological protection and sustainable development of the Qilian Mountains.
Forest inventory in the mountainous regions of Northwest China is extremely challenging due to the presence of external constraints such as high altitude and inaccessibility. LiDAR is an active remote sensing method that has been utilized frequently in forests. LiDAR uses laser energy to determine the distance between the sensor and the target, providing accurate information regarding the forest structure, especially vertical structure information such as tree height. LiDAR can acquire forest structural parameters more conveniently and quickly than traditional field inventory methods [
3,
4,
5,
6,
7]. As one of the major parameters of forest structure, tree height can help provide forest aboveground biomass [
8,
9,
10,
11,
12]. Increasing the precision of tree height extraction in point cloud data can significantly improve the estimation of aboveground biomass [
8,
9,
10,
11].
Terrestrial laser scanning (TLS) provides dense point cloud data with millimeter-level resolution from which forest structure indicators can be accurately extracted [
13]. Although TLS is a good alternative to traditional forestry survey methods, as it is a nondestructive method, it is more difficult to use TLS in inaccessible mountainous areas, and the spatial extent of one performance acquisition by TLS is relatively small [
14]. TLS is often suitable for accurately acquiring forest structure features in small areas and the reconstruction of forest 3D structures. In terms of estimating tree height, TLS tends to be underestimated [
15,
16]. TLS is better for the bottom of the canopy measurement and is suitable for extracting parameters such as DBH (diameter at breast height) [
17,
18], but detecting the canopy top is problematic, as the accuracy of tree height extraction is lower compared to ALS or UAV-LiDAR [
7].
Compared to TLS, airborne laser scanning (ALS) has a clear advantage in estimating tree height with a much larger measurement range. However, ALS seems to be more suitable for areas with flat terrain. The point cloud density obtained with ALS is usually between 1 and 25 pts/m
2, and it is more expensive to use [
19,
20]. In recent years, with the increase in UAV loads and the availability of lightweight LiDAR sensors, some studies have integrated LiDAR sensors with UAV platforms [
21,
22]. Therefore, it is a wise choice to use UAV-LiDAR in mountainous areas, as it is cheaper and more flexible in flight. UAV-LiDAR can change flight parameters to better capture forest structure information for sample plots with different characteristics [
23,
24].
Nevertheless, there are still difficulties in using UAV-LiDAR for forest inventory in alpine mountains with relatively large terrain changes and dense forests. Using UAV-LiDAR to acquire forest parameters in mountain forests is a simple and quick approach; however, there are still limitations such as easy loss of signal and the possibility of hitting obstacles when flying the UAV. With the increase in flight height, the safety of the UAV is improved, while the point cloud density is lowered. Finally, this will cause the accuracy of the tree height estimation to decrease. Therefore, proper flight height and point cloud density are necessary to strike a balance between flight safety and data quality. In addition, flying UAVs in mountainous areas is also affected by the terrain, tree species, tree height, and performance of the LIDAR itself. Some studies have been conducted to investigate the relationship between the accuracy of tree height estimation and different point cloud densities [
25] or sensors [
26]. Furthermore, the majority of current studies on tree height extraction have been undertaken in areas with lower altitude and plain terrain [
27,
28,
29], with fewer studies on tree height in alpine mountainous regions. As a result, for operational mountainous forest management, dependable, cost-effective methods are required for practical tree attributes [
30].
In order to determine the UAV-LiDAR flight parameters suitable for collecting the tree heights of Picea crassifolia forests in the alpine mountains of Northwest China, we collected data at different flight heights and point cloud densities. These data were used to investigate (1) the optimal flight height for tree height estimation and (2) the appropriate point cloud density for tree height estimation in alpine mountainous areas in Northwest China.
4. Discussion
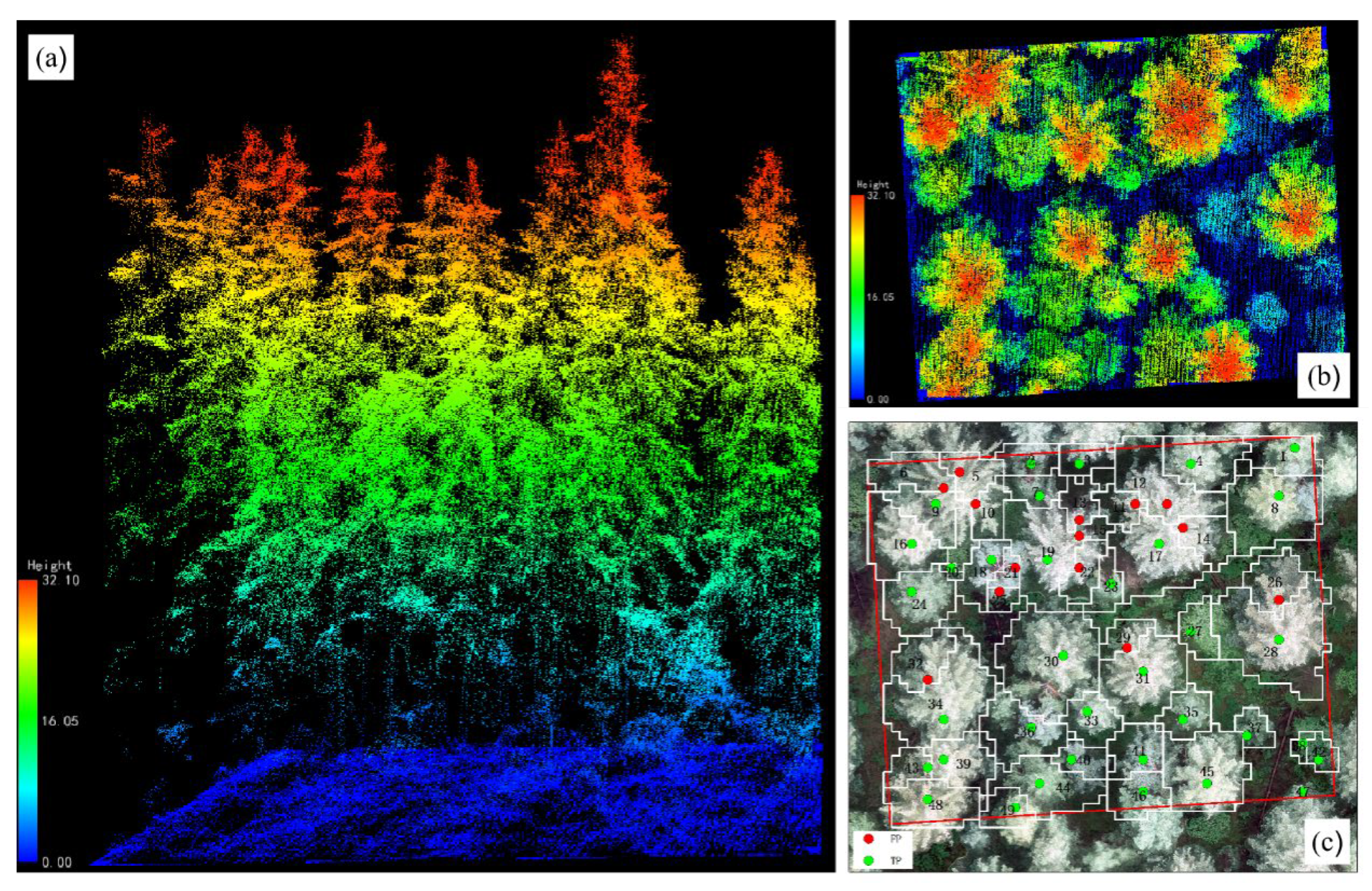
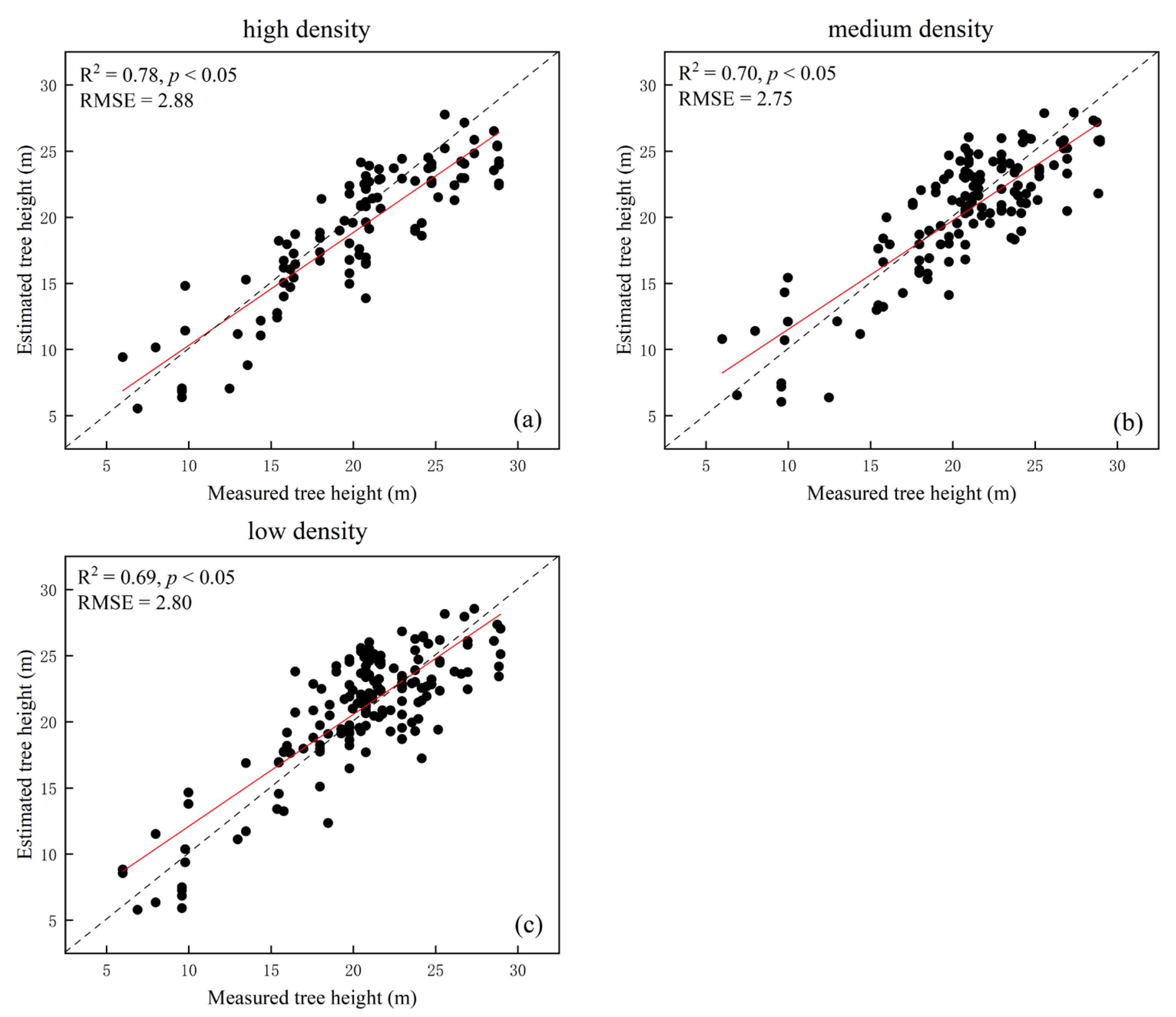
The flight height correlated with the accuracy of individual tree segmentation. As the flight height increased, the number of correct individual tree segmentations decreased, and the recall rates (r) decreased from 90.7% to 76.7%, indicating that the undersegmentation phenomenon increased. This was due to the decrease in point cloud density caused by the increase in flight height. However, too dense point clouds can also cause the oversegmentation phenomenon. Among them, the 145 m flight height had the best performance in oversegmentation.
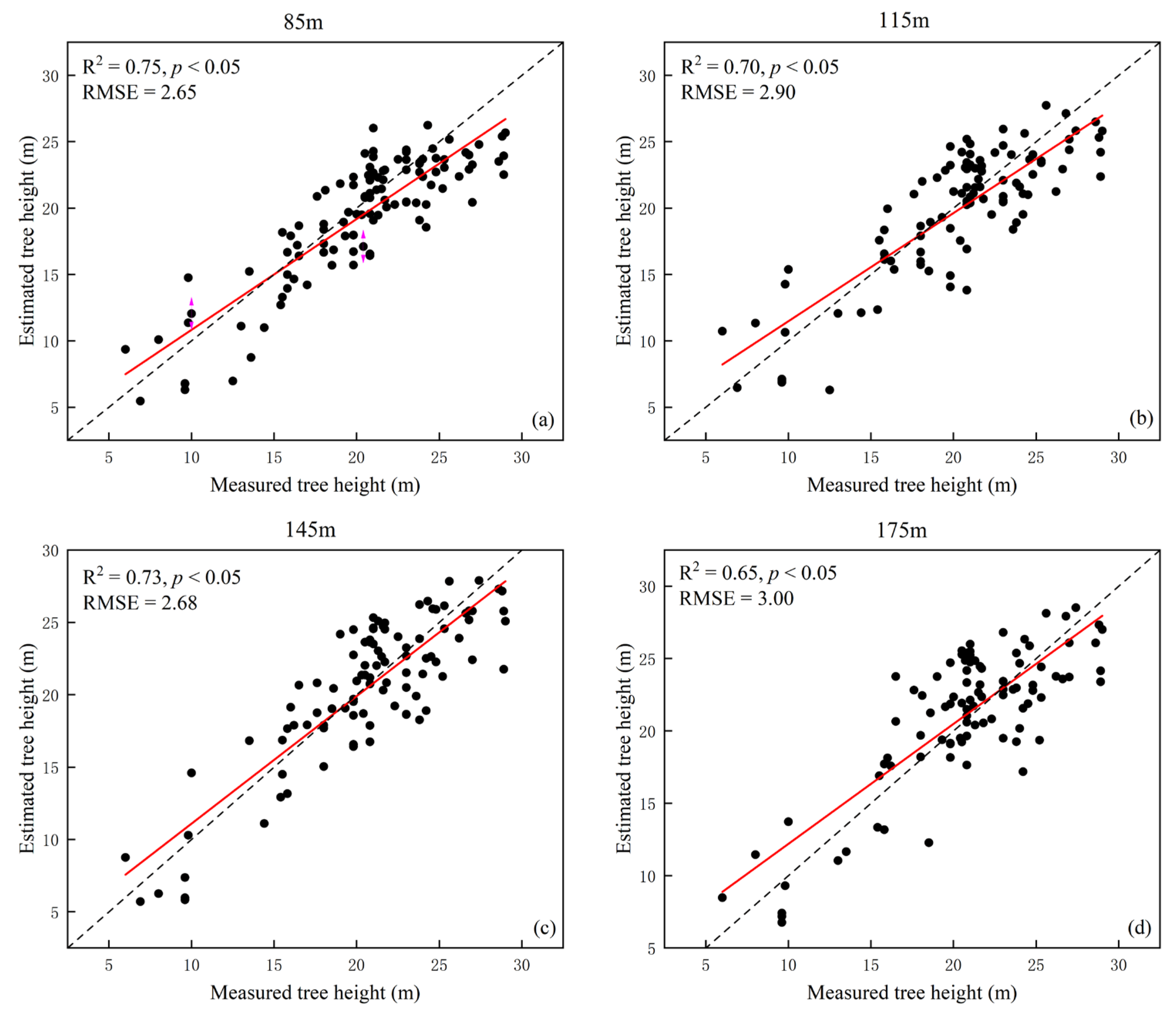
With the flight height increase, the oversegmentation phenomenon can be reduced to a certain extent. The precision rates (p) of the 145 m flight height was significantly smaller than that of 115 m, because the CHM identified neighboring and similar raster values as individual tree vertices at higher point cloud densities. The estimation accuracy of tree height was related to individual tree segmentation. Wu et al. [
8] found that improving the accuracy of individual tree segmentation can advance the calculation of forest biomass. In general, the lower the flight height, the more accurate the individual tree height [
26]. From a flight height of 145 m, the estimated tree height fit better than the 115 m height, probably because the number of individual trees extracted at the two flight heights was approximately the same. At a flight height of 115 m, individual trees were extracted with more severe oversegmentation (
Table 4), and the estimated tree height was more different than the measured tree height (
Figure 6). In terms of bias and mean difference (
Table 5), 145 m was the most suitable flight height and had a certain accuracy in fitting with the measured data (R
2 = 0.73).
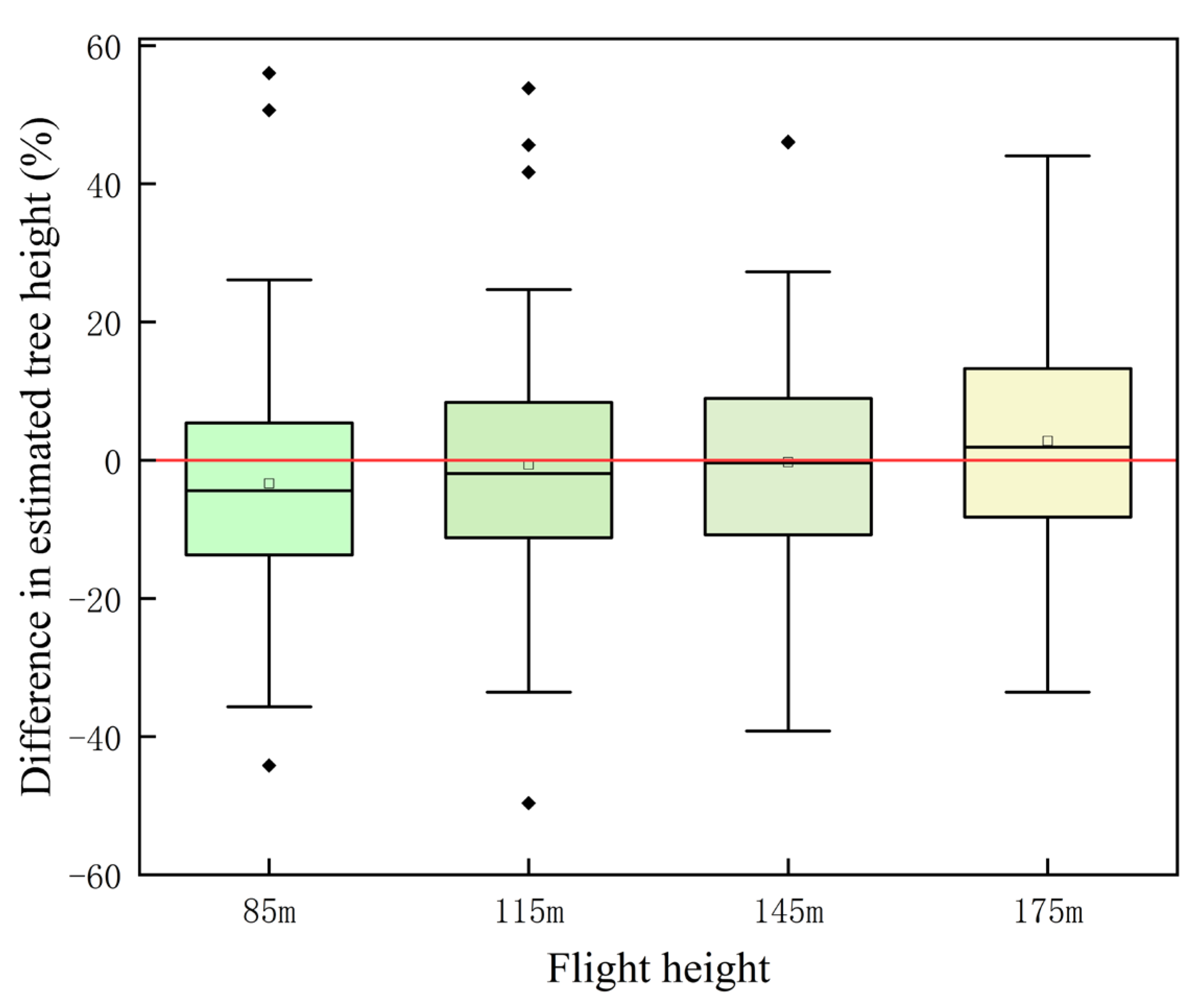
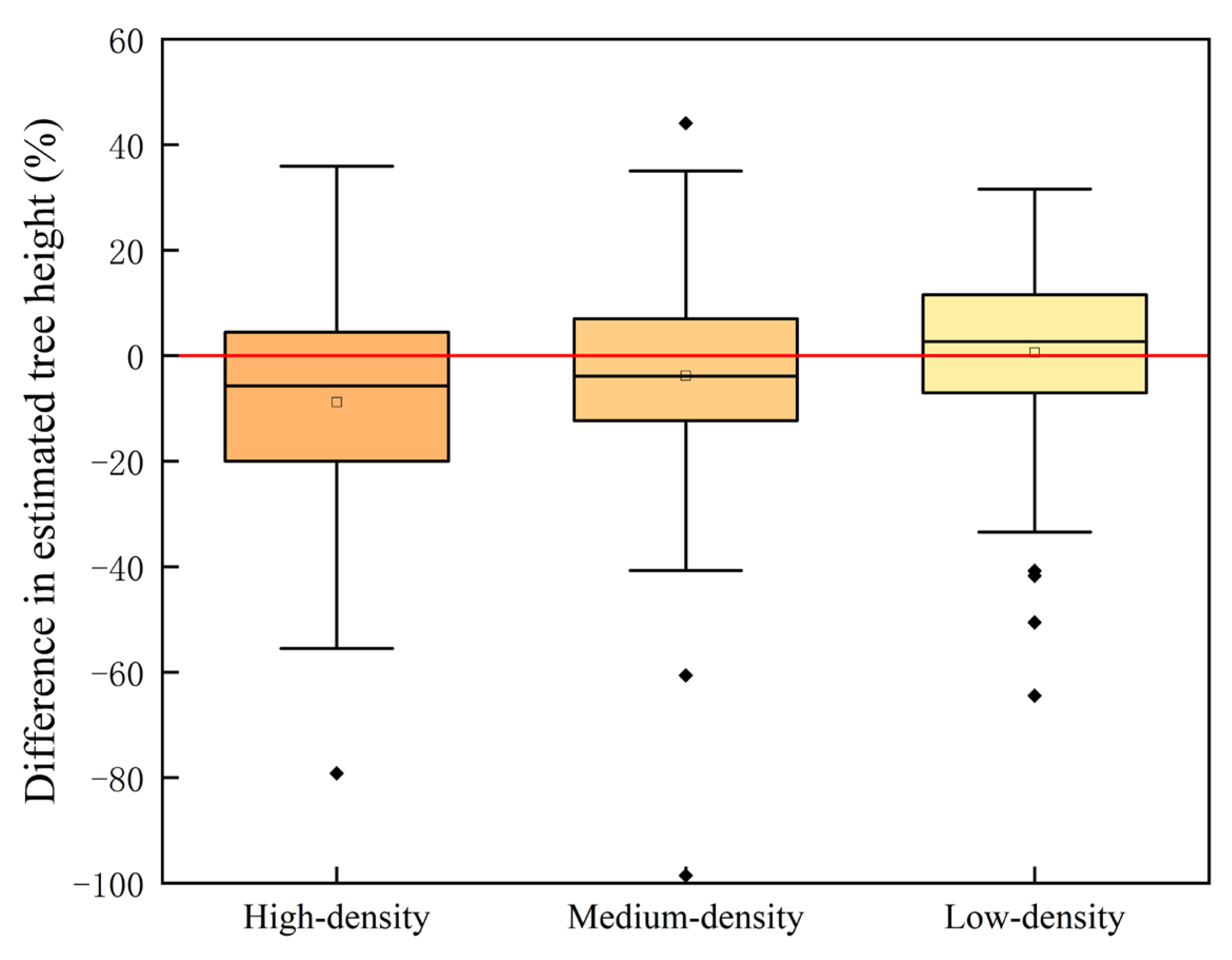
Tree height estimation at different flight heights (i.e., 85, 115, and 145 m) were lower than the measured tree height (
Figure 6,
Table 5). Various studies have found that individual tree segmentation-derived tree heights based on the CHM were significantly lower than those obtained from point cloud segmentation, because the interpolating process of point clouds into the CHM led to an overall reduction in tree height [
36,
37]. The Gaussian smoothing window was used in CHM-based individual tree segmentation, which took the average of the surrounding raster values to determine the maximum value of the top point of an individual tree, and this method caused the estimated tree height to be lower than the actual value [
38,
39,
40]. In other experiments that used UAV-LiDAR to obtain tree height, the phenomenon of tree height underestimation existed. Krause et al. [
41] obtained tree heights semi-automatically based on drones and found that measured tree heights were often overestimated, while drone measurements of tree heights were often underestimated. With the increase in flight height, the underestimation phenomenon was gradual and not obvious, which was probably because the acquired ground point cloud was becoming sparser, and the inaccuracy of the ground point separation caused the acquired tree height to become larger and the bias from the actual value to become smaller. The errors caused by ground points may be more obvious, especially in areas with large mountain slopes.
Various studies have found that different point cloud densities affect the estimation accuracy of tree height; the higher the point cloud density, the better the estimated tree height fits the measured tree height [
25,
26]. The medium-density point cloud extracted tree height with higher accuracy than the low-density point cloud, but the accuracy did not increase much, which indicates that the point cloud density at 125–185 pts/m
2 extracted tree height accurately. Therefore, it was not necessary to raise the point cloud density to 254–351 pts/m
2. Peng et al. [
25] found that when the point cloud density increased to 17 pts/m
2, the estimation accuracy of the tree height did not improve significantly. As the point cloud density decreased, the bias tree height relative to the measured value as well as the mean difference also decreased, and some studies have similar results [
42].
We found that the extracted accuracy of tree heights of
Picea crassifolia in alpine mountainous forests was inferior to that in several existing studies [
7,
28,
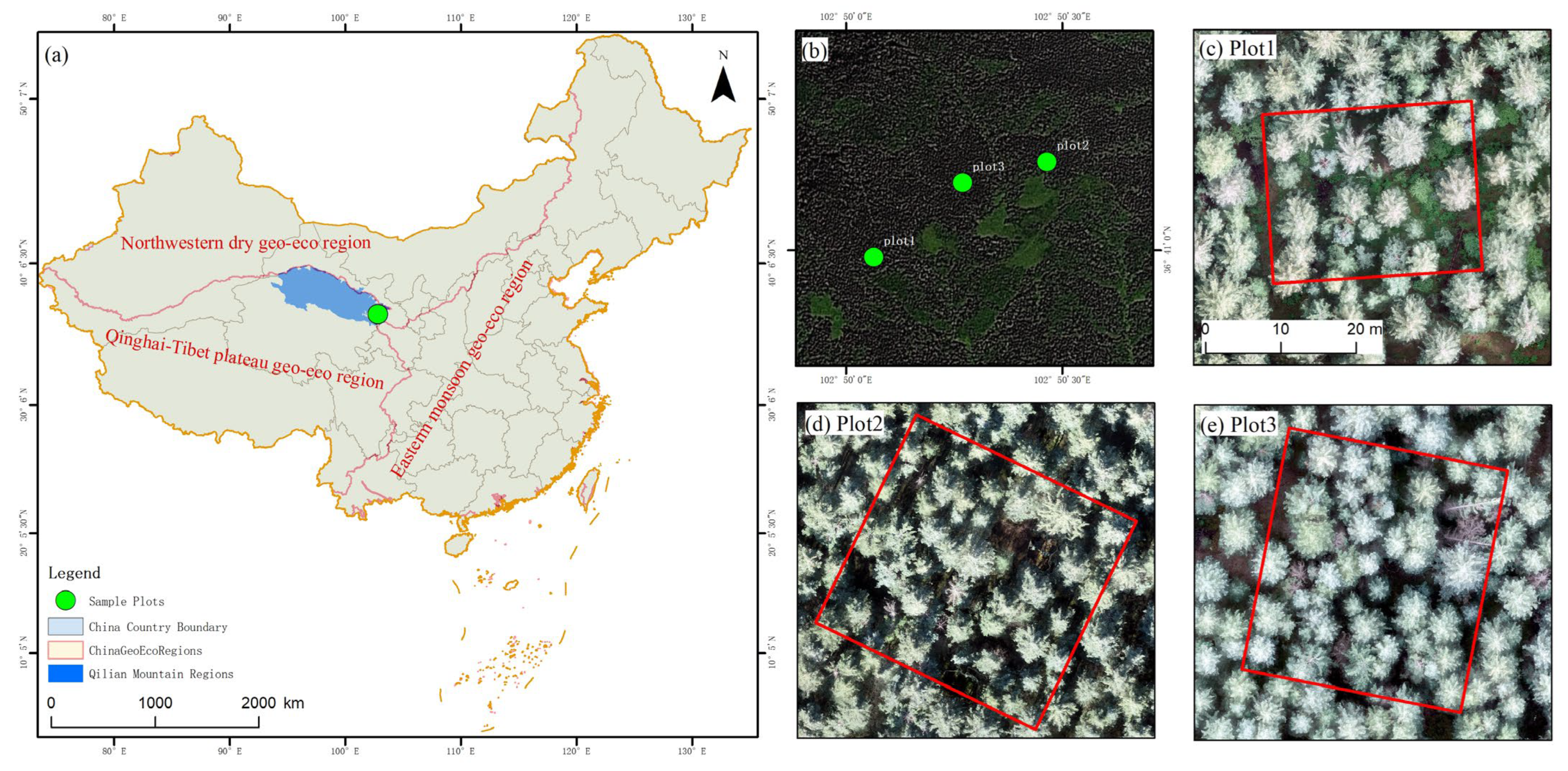
43]. Perhaps the slope of the mountainous terrain caused some uncertainty in distinguishing ground points and nonground points and then resulted in the lower extracted accuracy of tree height. In addition, the actual measurement data were obtained from rolling ground, and the measurement of tree height was affected by the slope of the mountain. Altitude and slope are the most important influencing factors when setting the flight heights of UAVs. Although we only set-up three
Picea crassifolia sample plots in the eastern Qilian Mountains, these plots were typically representative. Our results may be applicable to the survey of
Picea crassifolia mature forests with a certain slope. In future studies, we will add the effect of slope on the extraction of forest structure parameters in mountainous areas in Northwest China. In addition to the use of UAV-LiDAR, there is currently research on the use of the more cost-effective unpiloted digital aerial photogrammetry (UAS DAP) to predict forest attributes [
44], and we can also try to use UAS DAP for forest monitoring in alpine mountain areas in the future.
5. Conclusions
UAV-LIDAR was utilized to acquire point cloud data in alpine mountainous areas. We found that a flight height of 145 m was more applicable to maintain tree height estimation accuracy and assuring the safety of UAVs flying in alpine mountain regions. A point cloud density of 125–185 pts/m2 can guarantee tree height estimation accuracy. These results can serve as a good reference for alpine forest management. In retrospect, our study employing UAV-LiDAR might contribute to a more automated forest inventory, which could help to reduce the amount of field activities performed in the future.
In general, the estimation of tree height only requires LiDAR to detect the point cloud at the top of the tree, and the accuracy difference between different flight heights was not significant. If the ground point could be separated more accurately, the tree height would be extracted closer to the measured value. This study may improve the flight efficiency of UAV-LiDAR in alpine mountain forests in Northwest China. By selecting the appropriate flying height and point cloud density, considerable cost efficiency and time savings may be realized, benefiting forestry employees.